Synthesis, Characterization and Biological Activity of Novel Cu(II) Complexes of 6-Methyl-2-Oxo-1,2-Dihydroquinoline-3-Carbaldehyde-4n-Substituted Thiosemicarbazones
Abstract
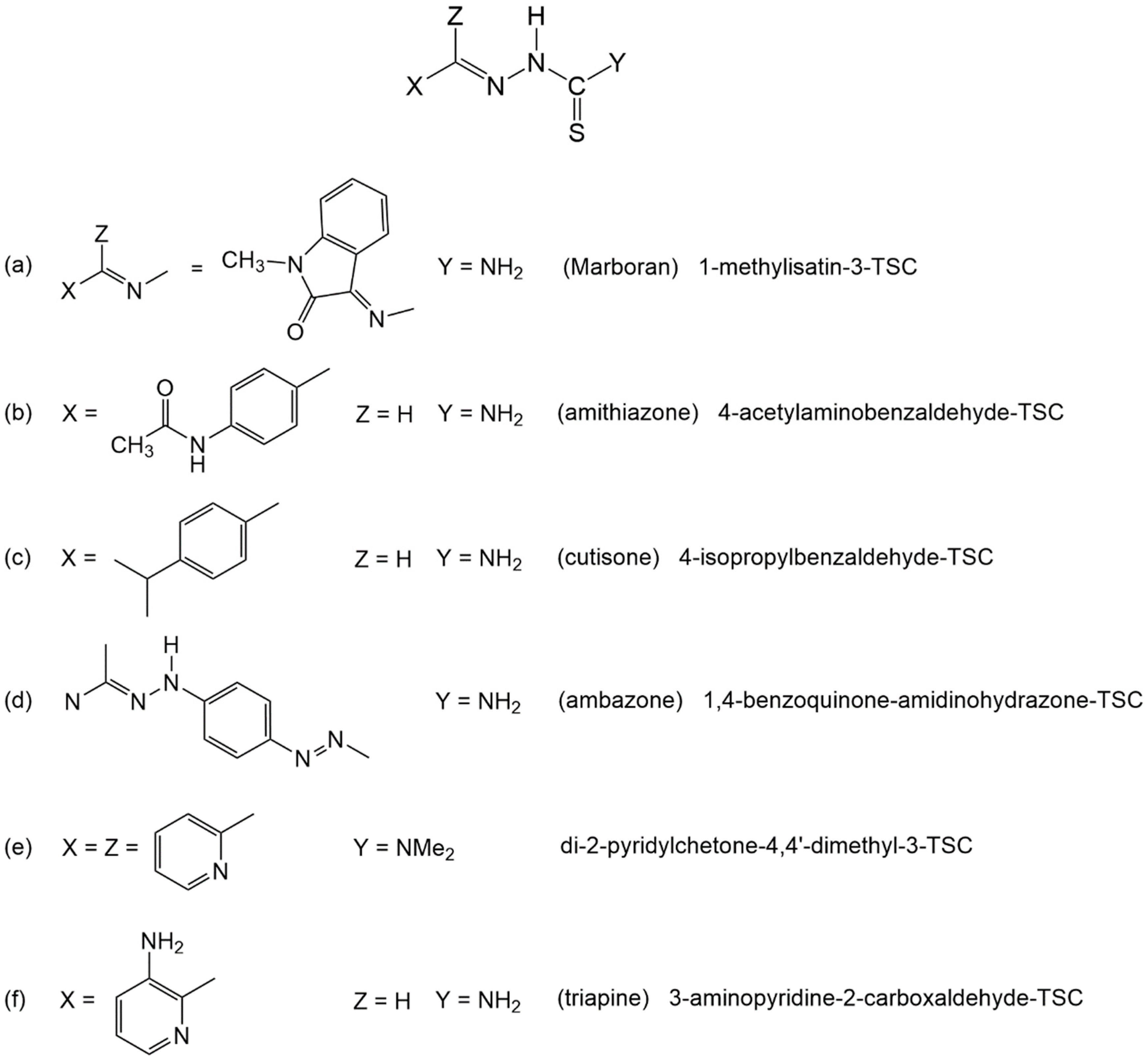
1. Introduction
2. Results and Discussion
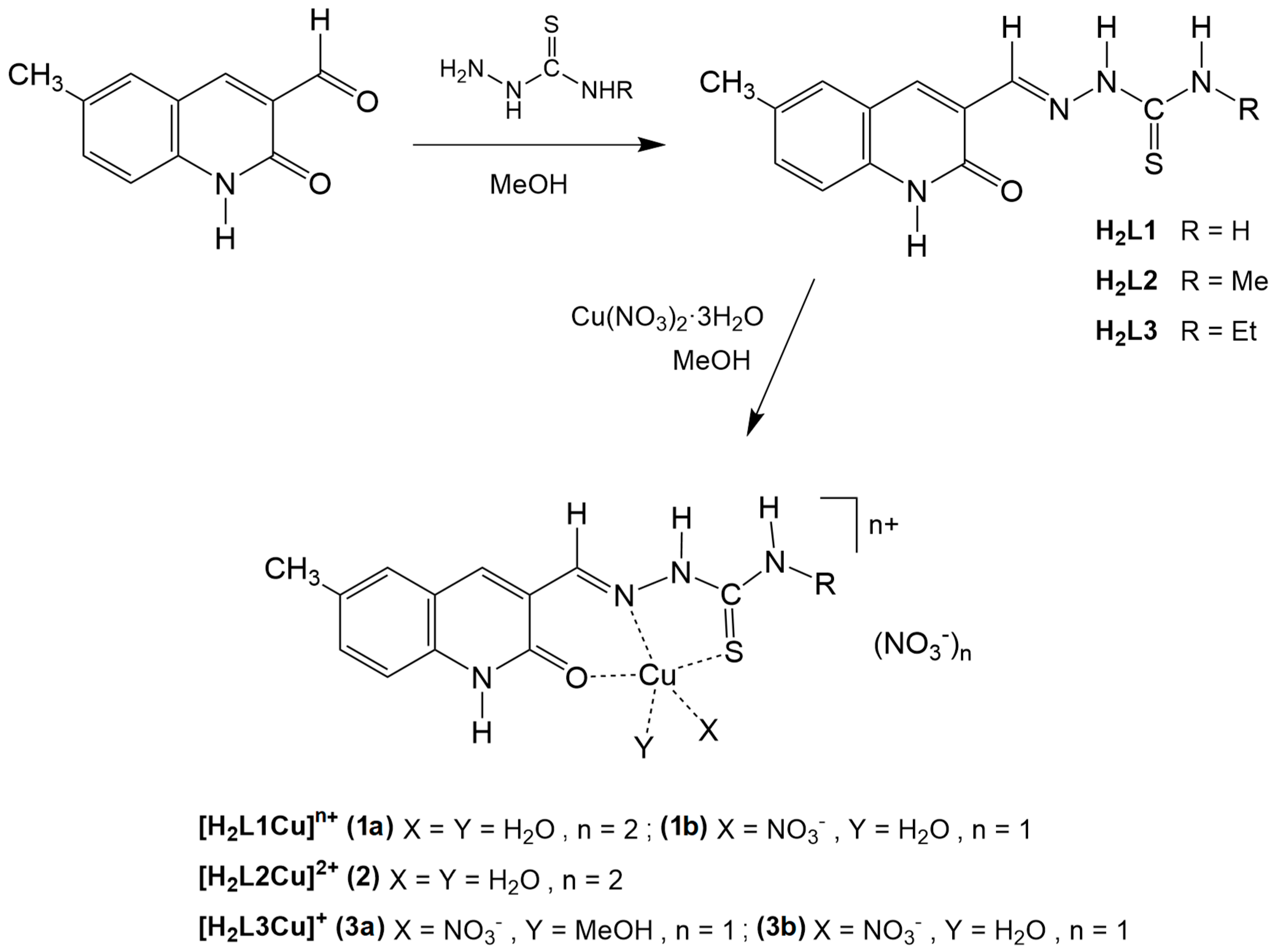
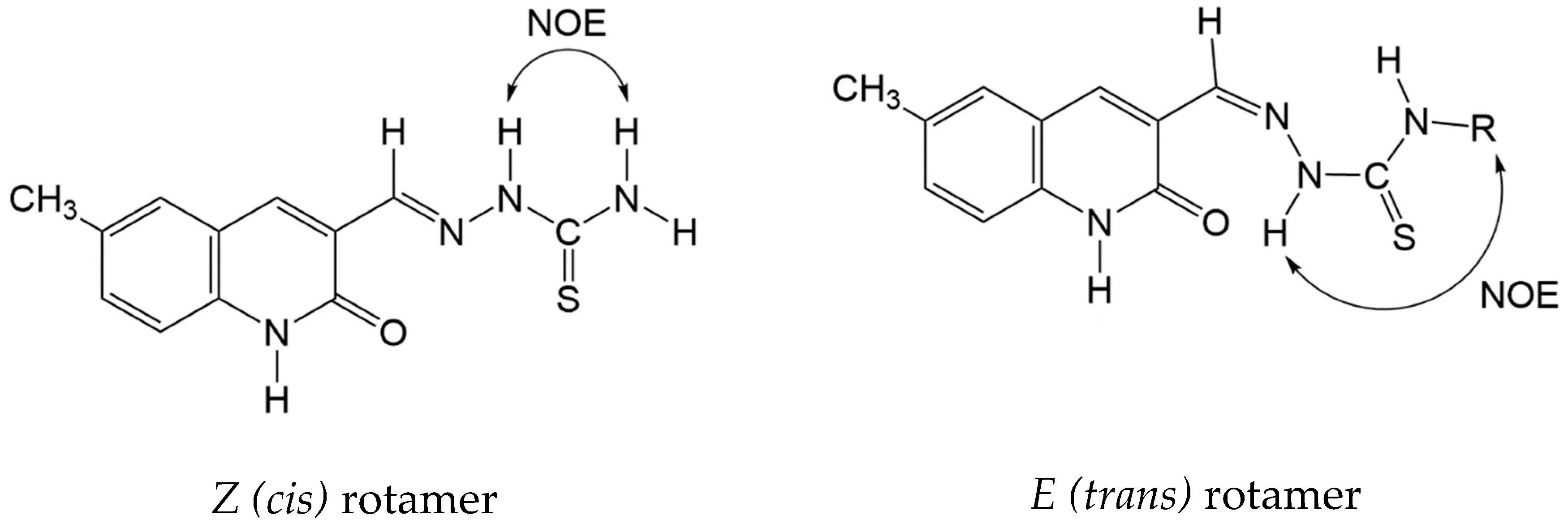
2.1. Synthesis and Characterization of the Pro-Ligands
2.2. Synthesis and Characterization of the Complexes
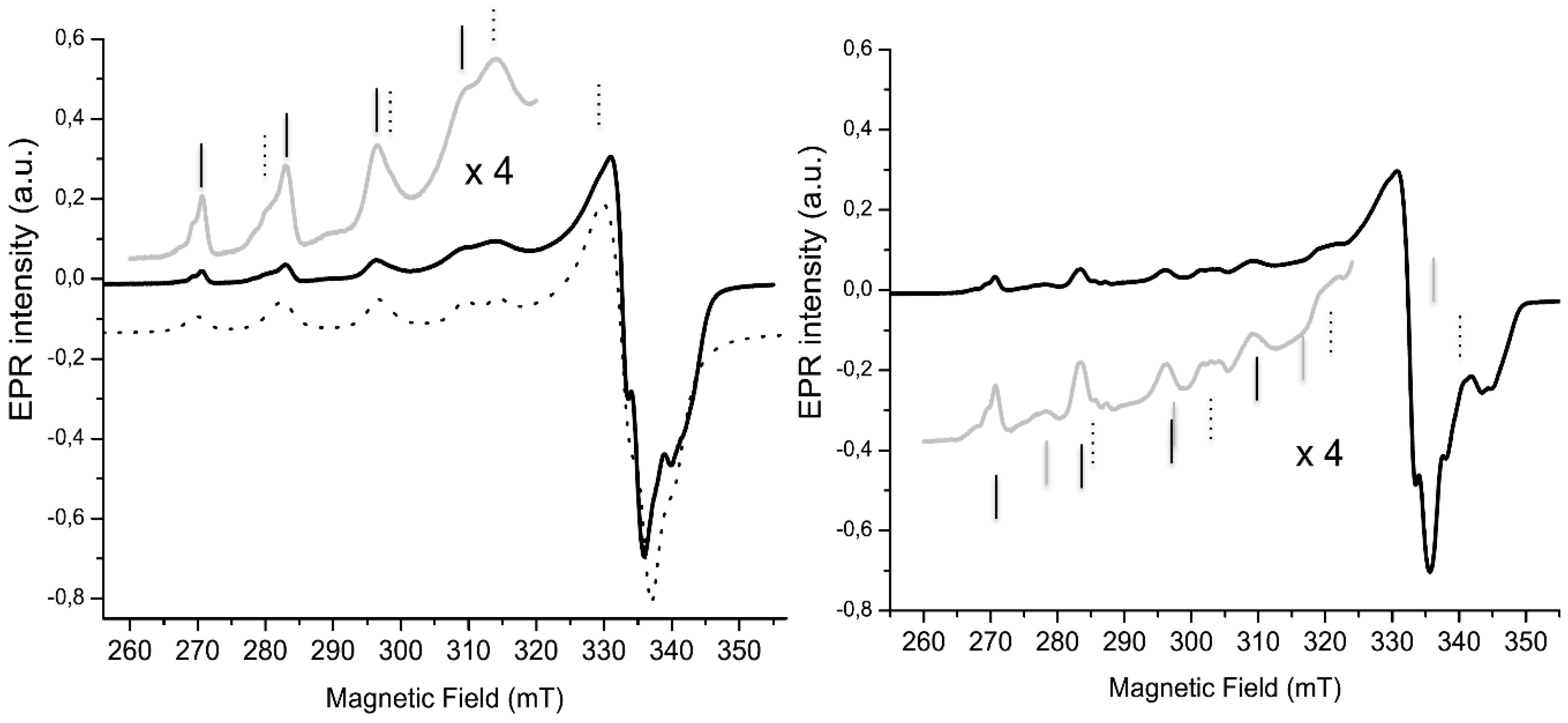
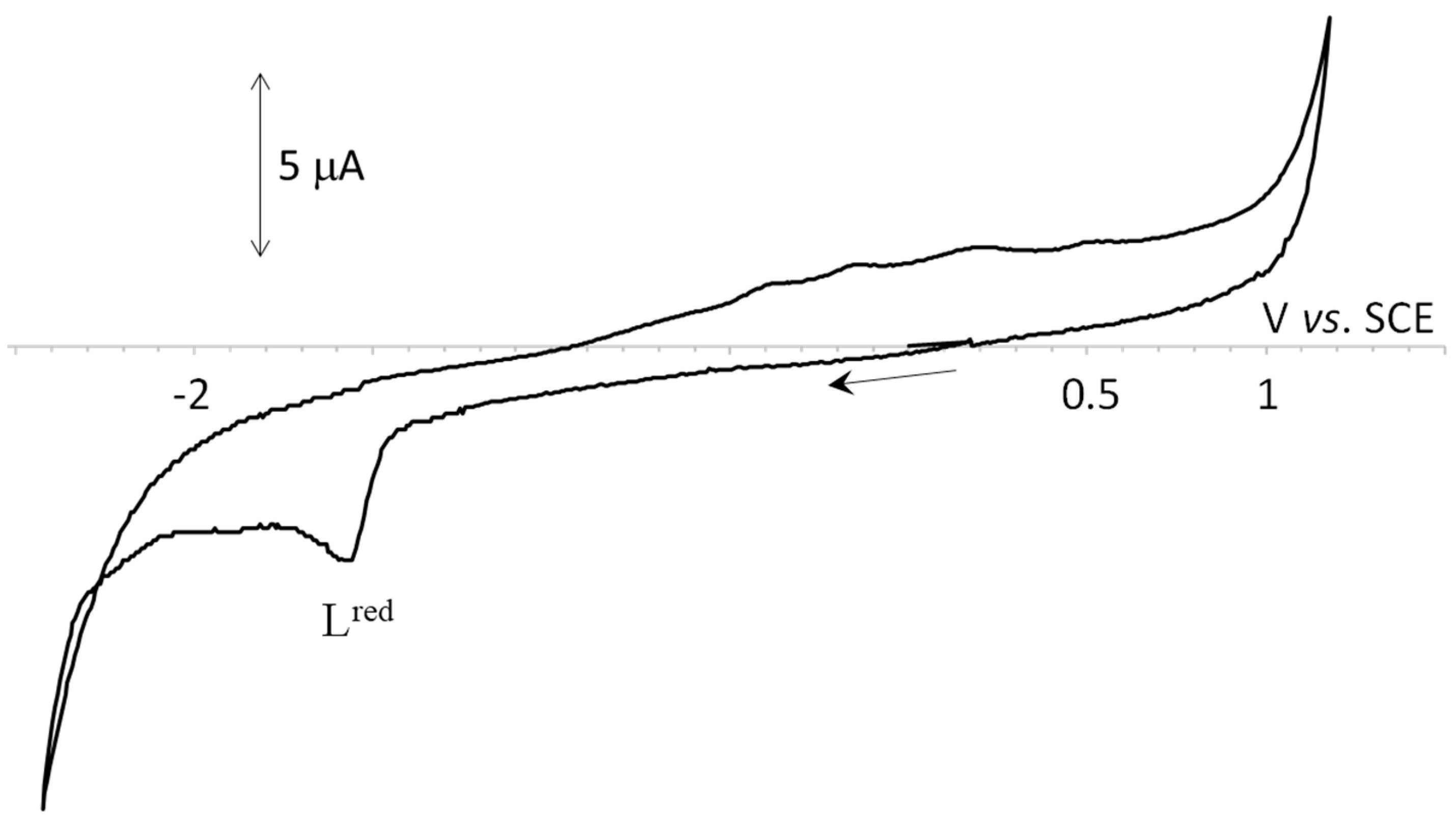
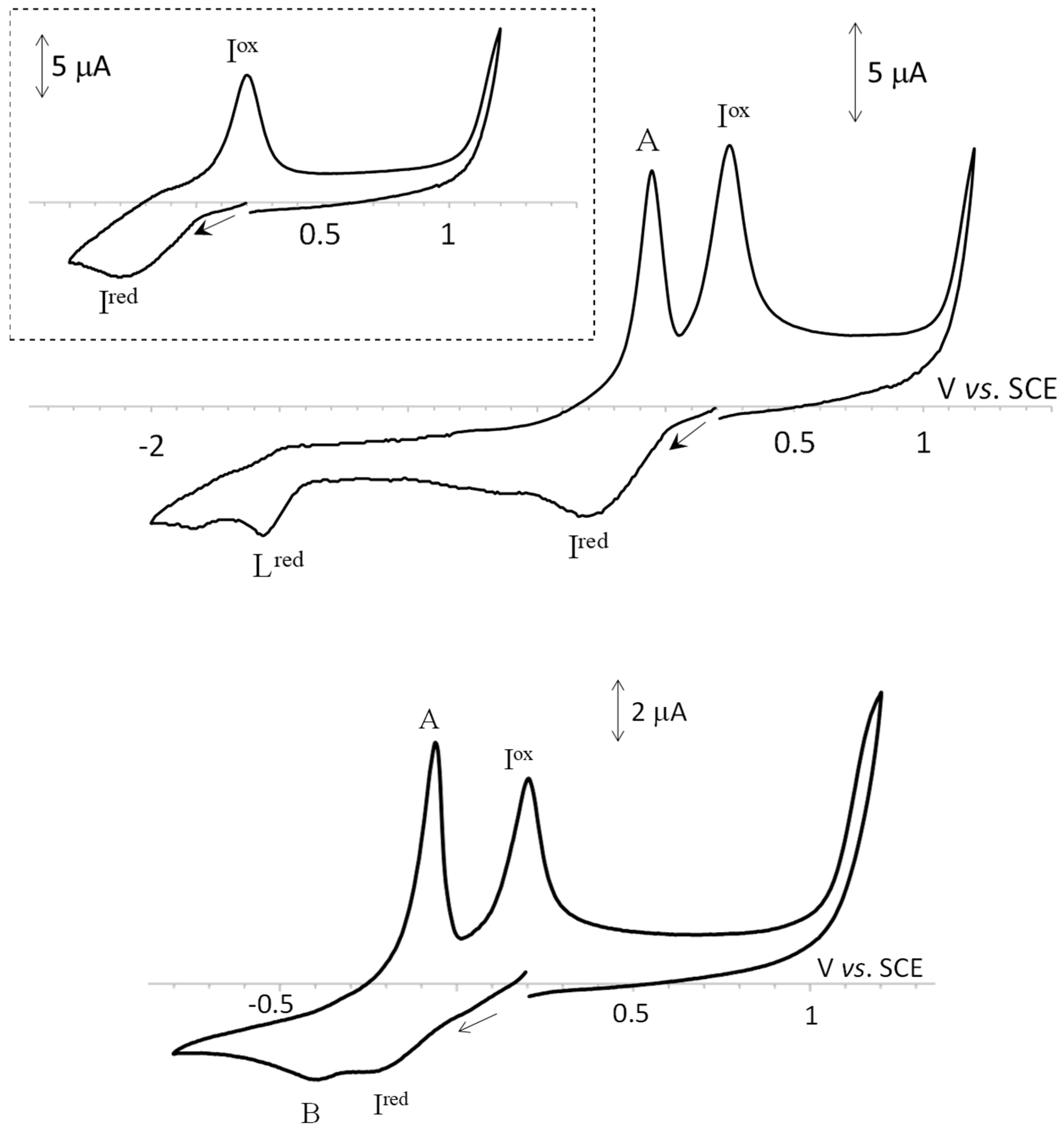
2.3. Electrochemical Properties
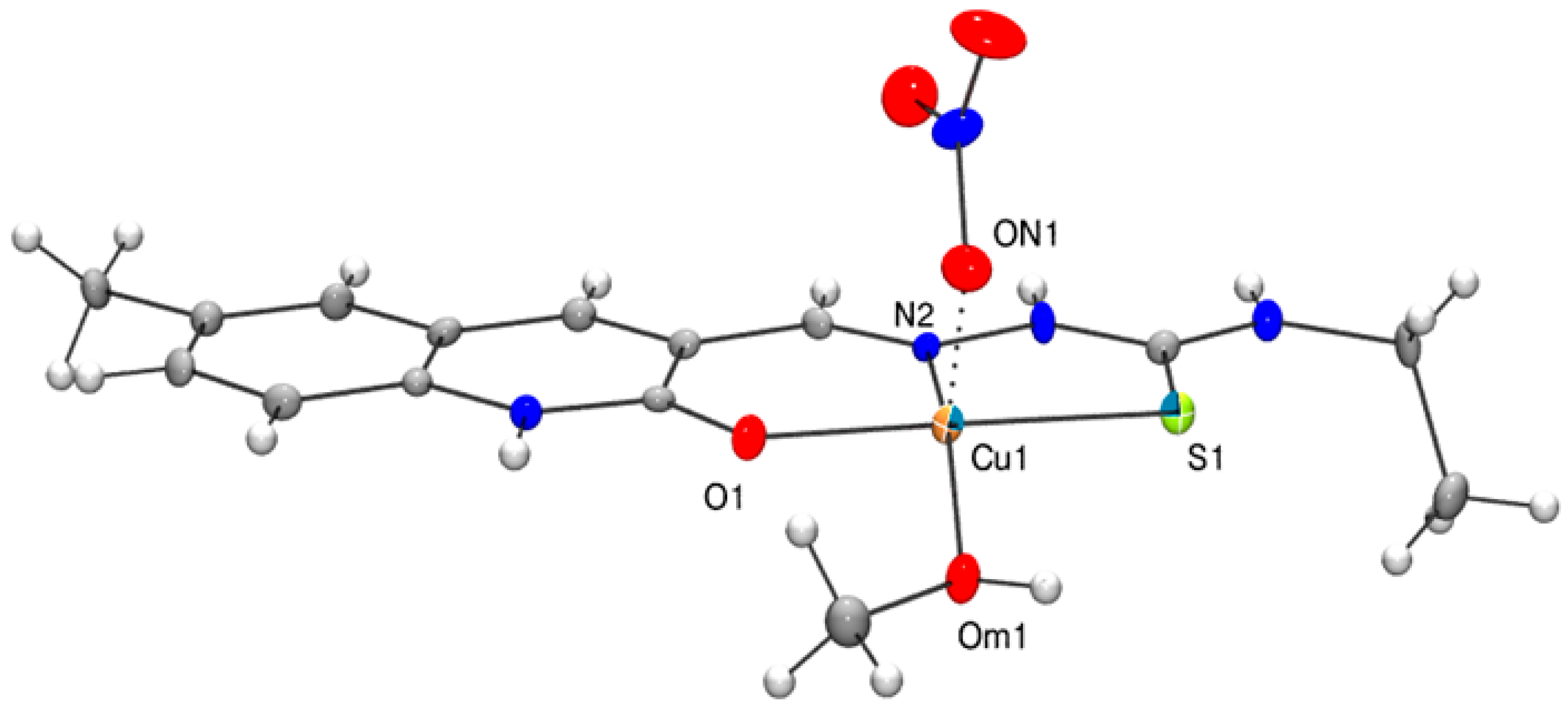
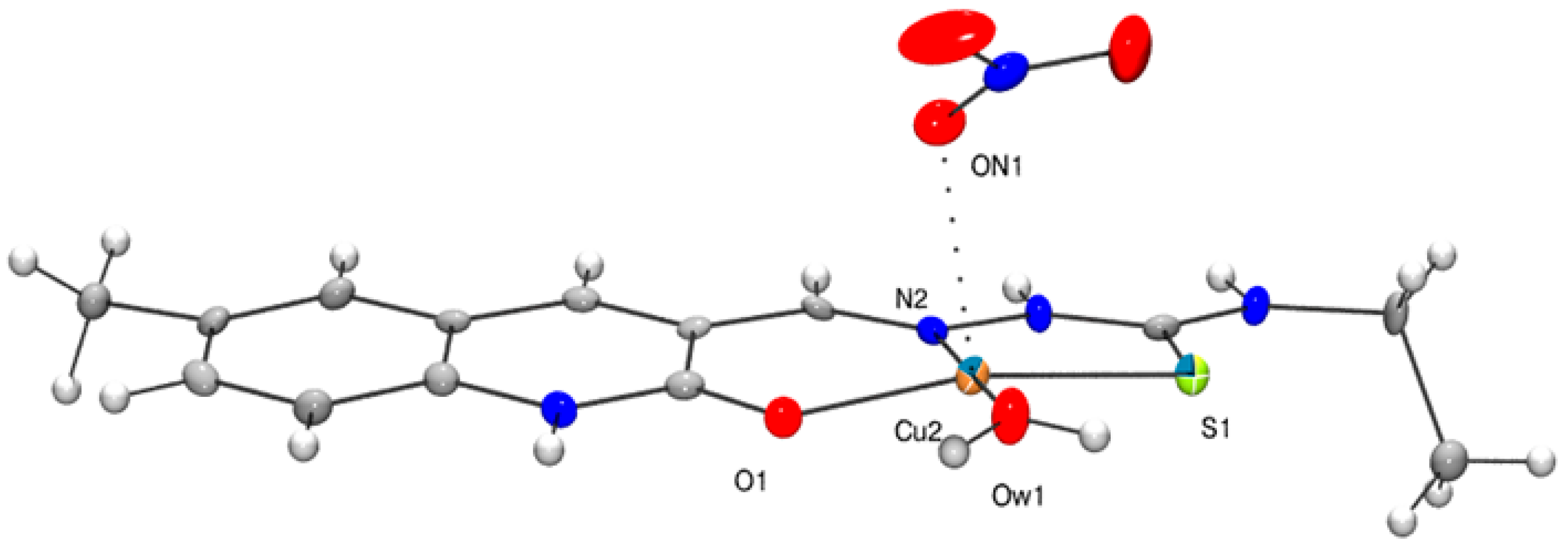
3. Crystal Structures of the Complexes 1, 2 and 3
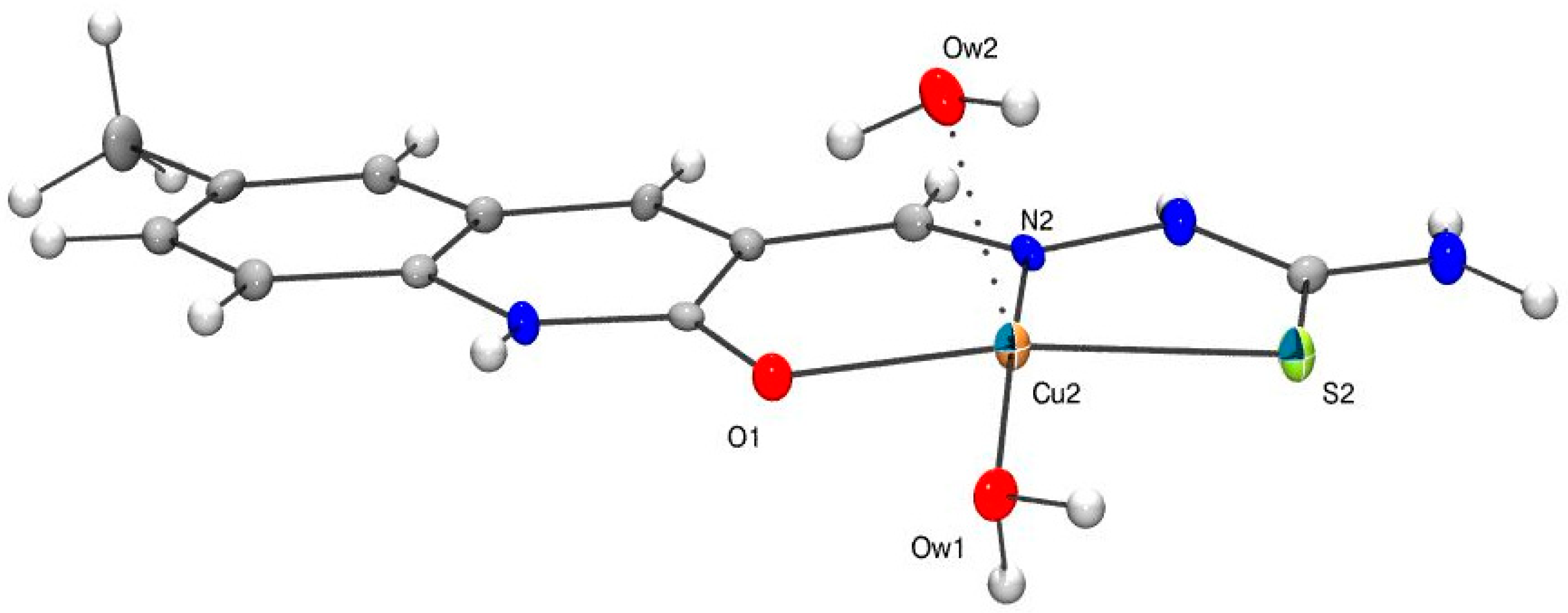
3.1. Solid State Structure of 2
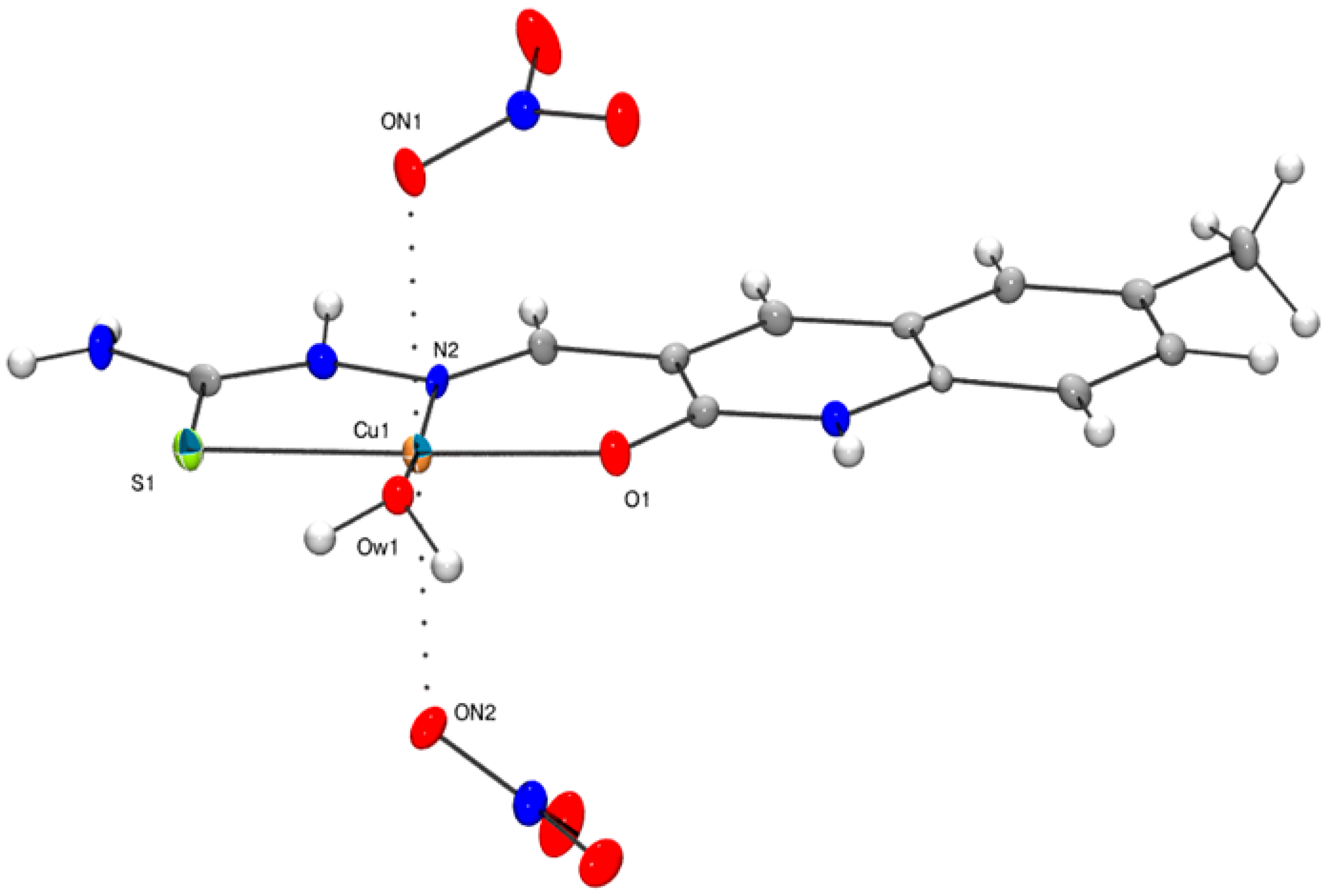
3.2. Solid State Structure of 1
3.3. Solid State Structure of 3
4. Biological Activity
5. Materials and Methods
5.1. General
5.2. Preparation of the Compounds
5.2.1. Synthesis of 6-Methyl-2-oxo-1,2-dihydro-quinoline-3-carbaldehyde thiosemicarbazone (H2L1)
5.2.2. Synthesis of 6-Methyl-2-oxo-1,2-dihydroquinoline-3-carbaldehyde-4N-methylthiosemicarbazone (H2L2)
5.2.3. Synthesis of 6-Methyl-2-oxo-1,2-dihydro-quinoline-3-carbaldehyde-4N-ethylthiosemicarbazone (H2L3)
5.2.4. Synthesis of the Complex 1 (H2L1Cu)
5.2.5. Synthesis of the Complex 2 (H2L2Cu)
5.2.6. Synthesis of the Complex 3 (H2L3Cu)
5.3. X-ray Crystallography
5.4. Experiments with Human Cells
5.4.1. Cell Cultures
5.4.2. Cytotoxic Activity
5.4.3. Spheroid Cultures and Acid Phosphatase (APH) Assay
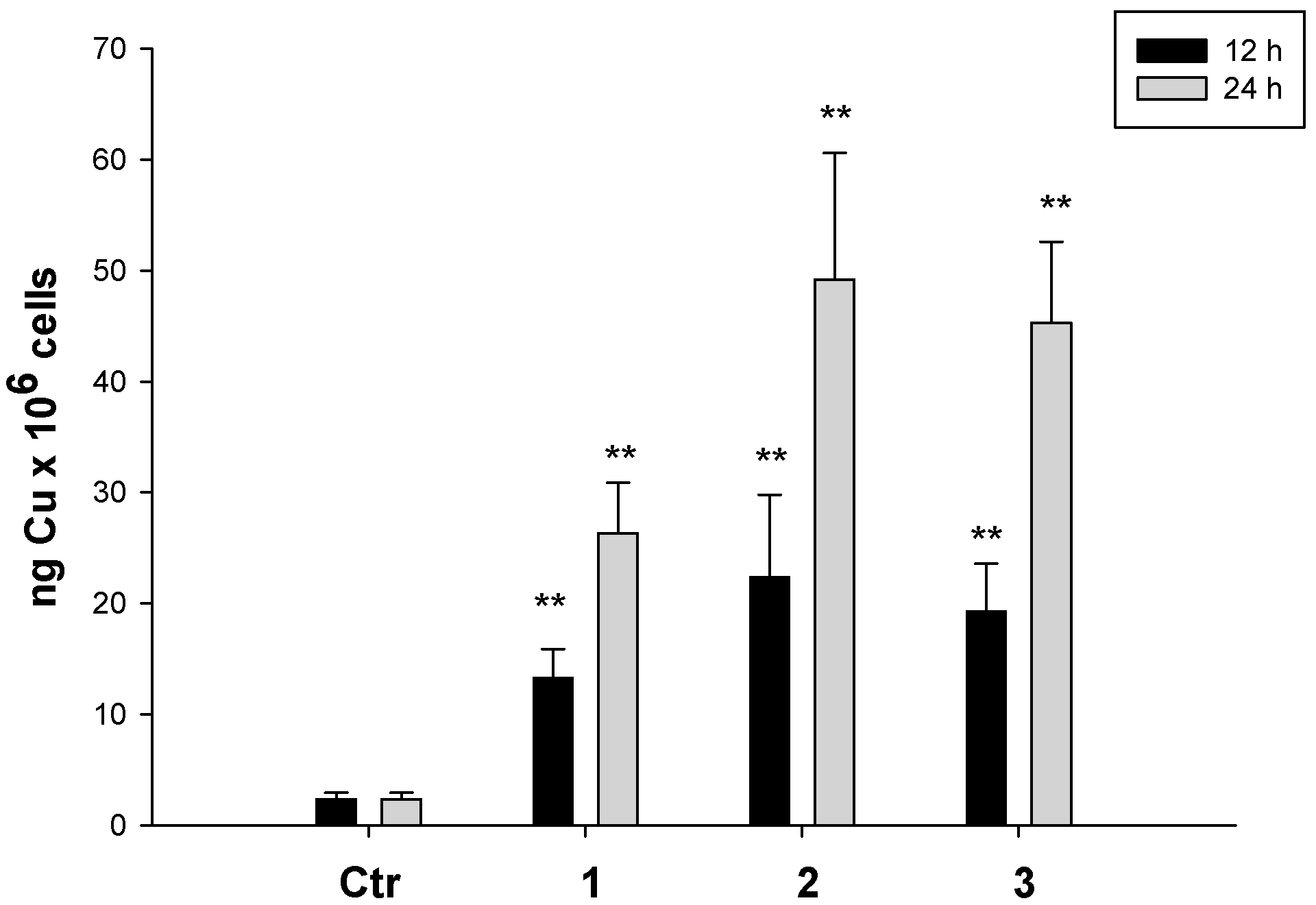
5.4.4. Cellular Accumulation of Cu
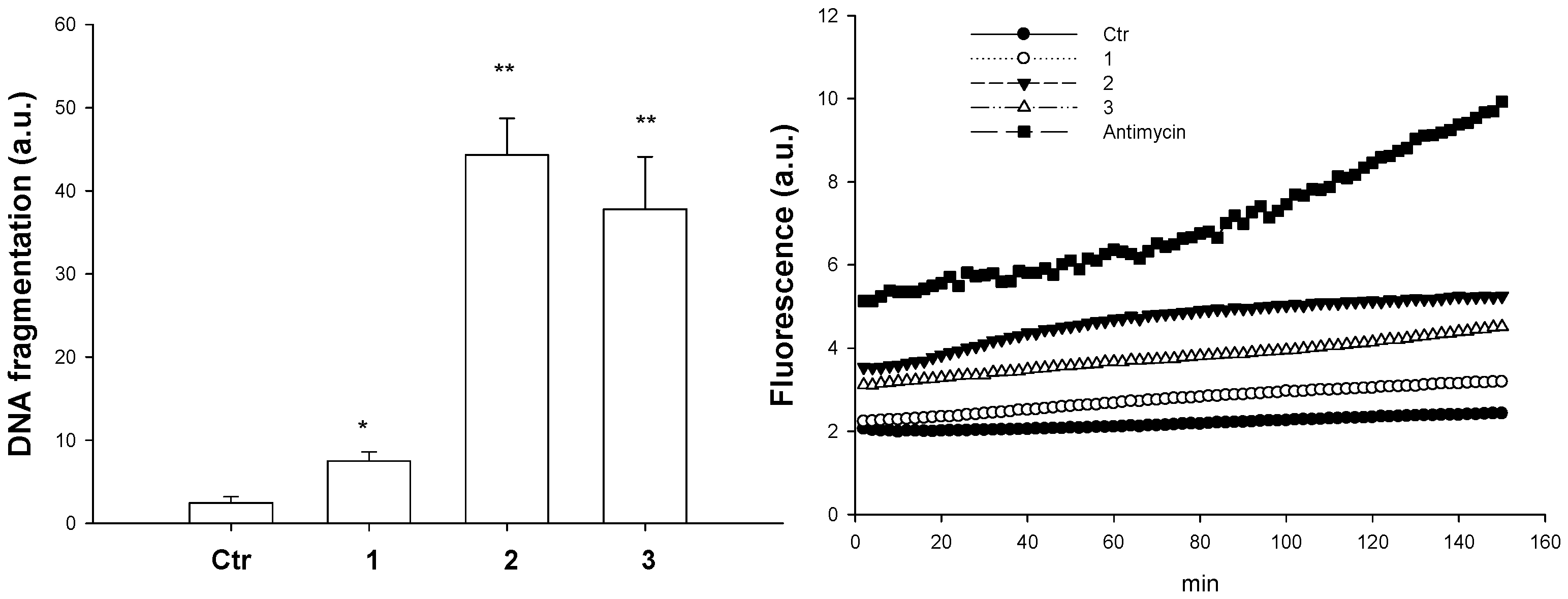
5.4.5. DNA Fragmentation
5.4.6. Reactive Oxygen Species (ROS) Production
5.4.7. Transmission Electron Microscopy (TEM) Analyses
5.4.8. Statistical Analysis
6. Conclusions
- the IC50 values against some human tumor cell lines derived from solid tumors are significantly lower (at nanomolar level) than those elicited by cisplatin; it is worth noting that a higher cytotoxicity, with respect to the reference compound, was retained in the 3D spheroids of colon cancer and melanoma cells, which is undoubtedly more predictive of in vivo results than standard 2D monolayer cultures;
- the remarkable cancer cell-killing effect is consistent with their ability to accumulate in human melanoma cells, which may be favored by a suitable balance between the cationic nature of the overall complexes [126] and the hydrophobicity of the pro-ligand skeleton;
- complexes 1–3 are markedly effective against human cancer cells with a developed resistance to cisplatin, with an RF factor of 1,8, 1.1 and 1.6 for 1, 2 and 3, respectively;
- complexes 2 and 3 exhibit a significant preferential cytotoxicity against human melanoma cancer cells with respect to non-cancerous HEK293 cells;
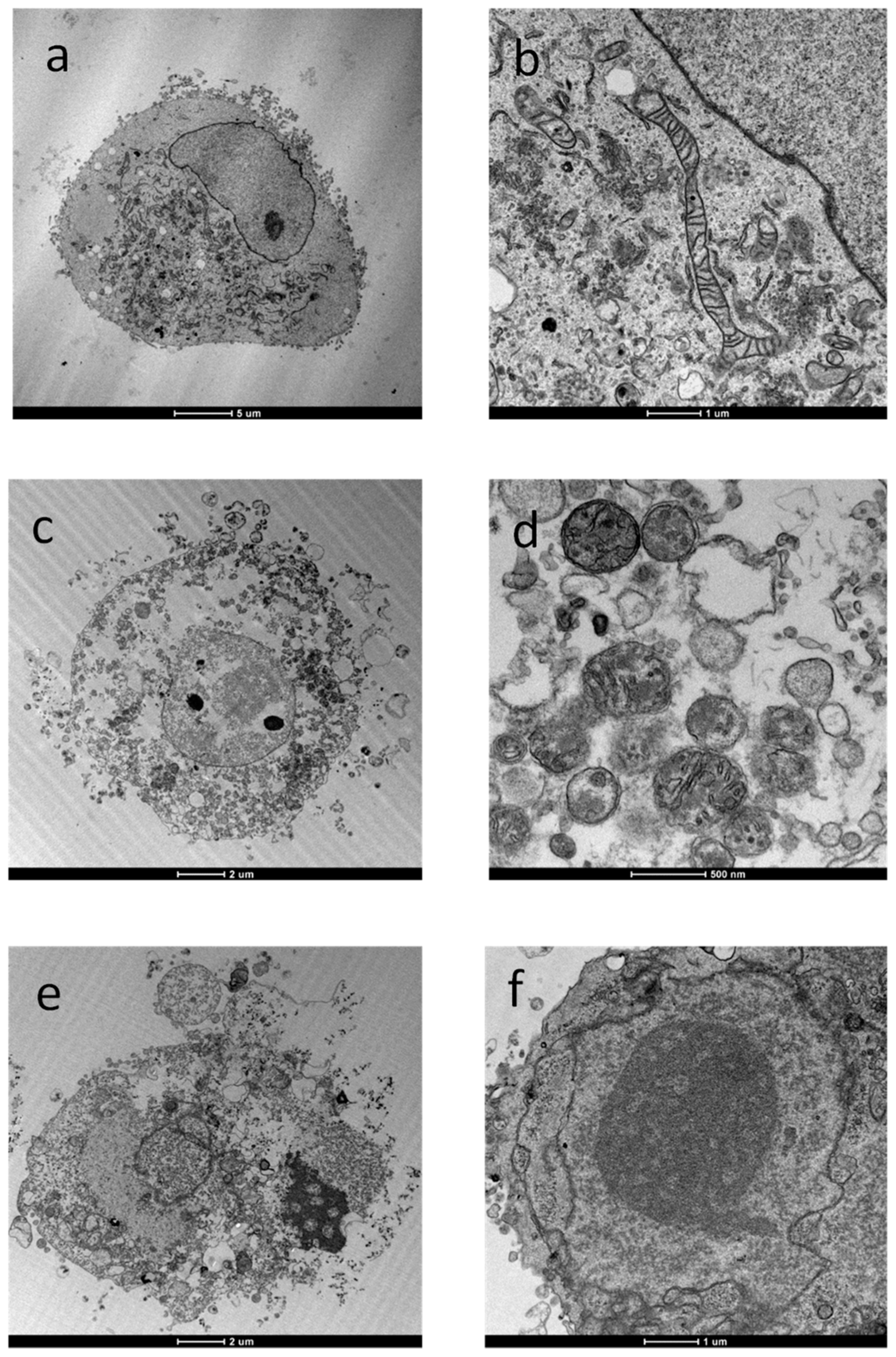
- Despite other copper thiosemicarbazone complexes having been shown to be primarily localized in mitochondria, thus compromising mitochondrial functions [127], TEM images of treated A375 cells indicated that mitochondria were quite conserved and that no alterations in internal structures were detected. Hence, the modest ROS formation induced by the tested complexes do not contribute in a significant manner to the DNA damage induced by the tested complexes. In addition, the morphological changes such as cell shrinkage and the formation of apoptotic bodies suggest the occurrence of the apoptotic process, likely due to the induction of DNA damage as revealed by DNA fragmentation. Further studies will be performed in order to better characterize the mechanism of cell death induced by the tested complexes.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pape, V.P.S.; Toth, S.; Furedi, A.; Szebenyi, K.; Lovrics, A.; Szabo, P.; Wiese, M.; Szakacs, G. Design, synthesis and biological evaluation of thiosemicarbazones, hydrazinobenzothiazoles and arylhydrazones as anticancer agents with a potential overcome multidrug resistance. Eur. J. Med. Chem. 2016, 117, 335–354. [Google Scholar] [CrossRef] [PubMed]
- Linciano, P.; Moraes, C.B.; Alcantara, L.M.; Franco, C.H.; Pascoalino, B.; Freitas-Junior, L.H.; Macedo, S.; Santarem, N.; Cordeiro-da-Silva, A.; Gul, S.; et al. Aryl thiosemicarbazones for the treatment of trypanosomatidic infections. Eur. J. Med. Chem. 2018, 146, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Pelivan, K.; Frensemeier, L.M.; Karst, U.; Koellensperger, G.; Heffeter, P.; Keppler, B.K.; Kowol, C.R. Comparison of metabolic pathways of different α-N-heterocyclic thiosemicarbazones. Anal. Bioanal. Chem. 2018, 410, 2343–2361. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, G. Thiosemicarbazone Metal Complexes: From structure to activity. Open Crystallogr. J. 2010, 3, 16–28. [Google Scholar] [CrossRef]
- Rahman, L.N.A.; Haribabu, J.; Balachandran, C.; Bhuvanesh, N.S.P.; Karvembu, R.; Sreekanth, A. Copper, nickel and zinc complexes of 3-acetyl coumarin thiosemicarbazone: Synthesis, characterization and in vitro evaluation of cytotoxicity and DNA/protein binding properties. Polyhedron 2017, 135, 26–35. [Google Scholar] [CrossRef]
- Palanimuthu, D.; Poon, R.; Sahni, S.; Anjum, R.; Hibbs, D.; Lin, H.Y.; Bernhardt, P.V.; Kalinowski, D.S.; Richardson, D.R. A novel class of thiosemicarbazones show multi-functional activity for the treatment of Alzheimer’s disease. Eur. J. Med. Chem. 2017, 139, 612–632. [Google Scholar] [CrossRef]
- Campbell, M.J.M. Transition metal complexes of thiosemicarbazide and thiosemicarbazones. Coord. Chem. Rev. 1975, 15, 279–319. [Google Scholar] [CrossRef]
- Patel, S.R.; Gangwal, R.; Sangamwar, A.T.; Jain, R. Synthesis, biological evaluation and 3D-QSAR study of hydrazide, semicarbazide and thiosemicarbazide derivatives of 4-(adamant-1-yl)quinoline as antituberculosis agents. Eur. J. Med. Chem. 2014, 85, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Manzano, J.I.; Cochet, F.; Boucherle, B.; Gomez-Perez, V.; Boumendjel, A.; Gamarro, F.; Peuchmaur, M. Arylthiosemicarbazones as antileishmanial agents. Eur. J. Med. Chem. 2016, 123, 161–170. [Google Scholar] [CrossRef]
- Thanh, N.D.; Giang, N.T.K.; Quyen, T.H.; Huong, D.T.; Toan, V.N. Synthesis and evaluation of in vivo antioxidant, in vitro antibacterial, MRSA and antifungal activity of novel substituted isatin N-(2,3,4,6-tetra-O-acetyl-β-D-glucopyranosyl)thiosemicarbazones. Eur. J. Med. Chem. 2016, 123, 532–543. [Google Scholar] [CrossRef]
- Santos, M.A.; Chand, K.; Chaves, S. Recent progress in multifunctional metal chelators as potential drugs for Alzheimer’s disease. Coord. Chem. Rev. 2016, 327–328, 287–303. [Google Scholar] [CrossRef]
- Bhalerao, M.B.; Dhumal, S.T.; Deshmukh, A.R.; Nawale, L.U.; Khedkar, V.; Sarkar, D.; Mane, R.A. New bithiazolyl hydazones: Novel synthesis, characterization and antitubercular evaluation. Bioorg. Med. Chem. Lett. 2017, 27, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Quenelle, D.C.; Keith, K.A.; Kern, E.R. In vitro and in vivo evaluation of isatin-β-thiosemicarbazone and marboran against vaccinia and cowpox virus infections. Antivir. Res. 2006, 71, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Tirunarayanam, M.O.; Vischer, W.A.; Bruhin, H. Amithiozone: Mechanism of action and resistance development by mycobacteria. Am. Rev. Respir. Dis. 1959, 80, 559–568. [Google Scholar]
- Alahari, A.; Trivelli, X.; Guerardel, Y.; Dover, L.G.; Besra, G.S.; Sacchettini, J.C.; Reynolds, R.C.; Coxon, G.D.; Kremer, L. Thiacetazone, an antitubercular drug that inhibits cyclopropanation of cell wall mycolic acids in Mycobacteria. PLoS ONE 2007, 2, e1343. [Google Scholar] [CrossRef]
- Caires, A.C. Recent advances involving palladium(II) complexes for the cancer therapy. Anticancer Agents Med. Chem. 2007, 7, 484–491. [Google Scholar] [CrossRef]
- Lober, G.; Hoffmann, H. Ambazone as a membrane active antitumor drug. Biophys. Chem. 1990, 35, 287–300. [Google Scholar] [CrossRef]
- Yuan, J.; Lovejoy, D.B.; Richardson, D.R. Novel di-2-pyridyl-derived iron chelators with marked and selective antitumor activity: In vitro and in vivo assessment. Blood 2004, 104, 1450–1458. [Google Scholar] [CrossRef]
- Kowol, C.R.; Trondl, R.; Heffeter, P.; Arion, V.B.; Jakupec, M.A.; Roller, A.; Galansli, M.; Berger, W.; Keppler, B.K. Impact of metal coordination on cytotoxicity of 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (Triapine) and novel insights into terminal dimethylation. J. Med. Chem. 2009, 52, 5032–5043. [Google Scholar] [CrossRef]
- Nutting, C.M.; van Herpen, C.M.L.; Miah, A.B.; Bhide, S.A.; Machiels, J.P.; Buter, J.; Kelly, C.; de Raucourt, D.; Harrington, K.J. Phase II study of 3-AP Triapine in patients with recurrent or metastatic head and neck squamous cell carcinoma. Ann. Oncol. 2009, 20, 1275–1279. [Google Scholar] [CrossRef]
- Hancock, C.N.; Stockwin, L.H.; Han, B.; Divelbiss, R.D.; Jun, J.H.; Malhotra, S.V.; Hollingshead, M.G.; Newton, D.L. A copper chelate of thiosemicarbazone NSC 689534 induces oxidative/ER stress and inhibits tumor growth in vitro and in vivo. Free Radic. Biol. Med. 2011, 50, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Raman, N.; Jeyamurugan, R.; Rajkapoor, B.; Magesh, V. Metal-based antitumor, cytotoxic and antimicrobial activity: Pharmacological evaluation of Knoevenagel condensate β-diketone Schiff base thiosemicarbazone Cu(II) and Zn(II) complexes. Appl. Organomet. Chem. 2009, 23, 283–290. [Google Scholar] [CrossRef]
- Sathisha, M.P.; Budagumpi, S.; Kulkarni, N.V.; Kurdekar, G.S.; Revankar, V.K.; Pai, K.S.R. Synthesis, structure, electrochemistry and spectral characterization of (D-glucopyranose)-4-phenylthiosemicarbazide metal complexes and their antitumor activity against Ehrlich Ascites carcinoma in Swiss albino mice. Eur. J. Med. Chem. 2010, 45, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Sathisha, M.P.; Shetti, U.N.; Revankar, V.K.; Pai, K.S.R. Synthesis and antitumor studies on novel Co(II), Ni(II) and Cu(II) metal complexes of bis(3-acetylcoumarin)thiocarbohydrazone. Eur. J. Med. Chem. 2008, 43, 2338–2346. [Google Scholar] [CrossRef]
- Akam, E.A.; Utterback, R.D.; Marcero, J.R.; Dailey, H.A.; Tomat, E. Disulfide-masked iron prochelators: Effects on cell death, proliferation and hemoglobin production. J. Inorg. Biochem. 2018, 180, 186–193. [Google Scholar] [CrossRef]
- Whitnall, M.; Howard, J.; Ponka, P.; Richardson, D.R. A class ofiron chelators with a wide spectrum of potent antitumor activity that overcomes resistance to chemotherapeutics. Proc. Natl. Acad. Sci. USA 2006, 103, 14901–14906. [Google Scholar] [CrossRef]
- Sirbu, A.; Palamarciuc, O.; Babak, M.V.; Lim, J.M.; Ohui, K.; Enyedy, E.A.; Shova, S.; Darvasiova, D.; Rapta, P.; Ang, W.H.; et al. Copper(II) thiosemicarbazone complexes induce marked ROS accumulation and promote nrf2-mediated antioxidant response in highly resistant breast cancer cells. Dalton Trans. 2017, 46, 3833–3847. [Google Scholar] [CrossRef]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in copper complexes as anticancer agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef]
- Song, S.; You, A.; Chen, Z.; Zhu, G.; Wen, H.; Song, H.; Yi, W. Study on the design, synthesis and structure-activity relationships of new thiosemicarbazone compounds as tyrosinase inhibitors. Eur. J. Med. Chem. 2017, 139, 815–825. [Google Scholar] [CrossRef]
- Cai, P.; Xiong, Y.; Yao, Y.; Chen, W.; Dong, X. Synthesis, screening and biological activity of potent thiosemicarbazone compounds as a tyrosinase inhibitor. New J. Chem. 2019, 43, 14102–14111. [Google Scholar] [CrossRef]
- Dong, H.; Liu, J.; Liu, X.; Yu, Y.; Cao, S. Combining molecular docking and QSAR studies for modeling the anti-tyrosinase activity of aromatic heterocycle thiosemicarbazone analogues. J. Mol. Struct. 2018, 1151, 353–365. [Google Scholar] [CrossRef]
- Ribeiro, A.G.; de Almeida, S.N.V.; de Oliveira, J.F.; de Lima Souza, T.R.C.; dos Santos, K.L.; de Barros Albuquerque, A.P.; de Britto Lira Nogueira, M.C.; de Carvalho Junior, L.B.; de Moura, R.O.; da Silva, A.C.; et al. Novel 4-quinoline-thiosemicarbazone derivatives: Synthesis, antiproliferative activity, in vitro and in silico biomacromolecule interaction studies and topoisomerase inhibition. Eur. J. Med. Chem. 2019, 182, 111592–111607. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Bao, G.; Wang, L.; Cheng, M.; Zhao, M.; Zhao, S.; Zhou, H.; Gong, P. Design, synthesis and biological evaluation of novel 4-phenoxy-6,7-disubstuìitted quinoline possessing (thio)semicarbazone as c-Met kinase inhibitors. Bioorg. Med. Chem. 2016, 14, 1331–1345. [Google Scholar] [CrossRef] [PubMed]
- Cushing, T.D.; Baichwal, V.; Berry, K.; Billedeau, R.; Bordunov, V.; Broka, C.; Cardozo, M.; Cheng, P.; Clark, D.; Dalrymple, S.; et al. A novel series of IKKβ inhibitors part I: Initial SAR studies of a HTS hit. Bioorg. Med. Chem. Lett. 2011, 21, 417–422. [Google Scholar] [CrossRef]
- Yu, Y.; Kalinowski, D.S.; Kovacevic, Z.; Siafakas, A.R.; Jansson, P.J.; Stefani, C.; Lovejoy, D.B.; Sharpe, P.C.; Bernhardt, P.V.; Richardson, D.R. Thiosemicarbazones from the old to new: Iron chelators that are more than just Ribonucleotide Reductase inhibitors. J. Med. Chem. 2009, 52, 5271–5294. [Google Scholar] [CrossRef]
- Ferreira, F.B.; Pereira, T.M.; Souza, D.L.N.; Lopes, D.S.; Freitas, V.; Avila, V.M.R.; Kummerle, A.E.; Sant’Anna, C.M.R. Structure-based discovery of thiosemicarbazone metalloproteinase inhibitors for hemorrhage treatment in snakebites. ACS Med. Chem. Lett. 2017, 8, 1136–1141. [Google Scholar] [CrossRef]
- Haribabu, J.; Tamizh, M.M.; Balachandran, C.; Arun, Y.; Bhuvanesh, N.S.P.; Endo, A.; Karvembu, R. Synthesis, structures and mechanistic pathways of anticancer activity of palladium(II) complexes with indole-3-carbaldehyde thiosemicarbazones. New J. Chem. 2018, 42, 10818–10832. [Google Scholar] [CrossRef]
- Kowol, C.R.; Berger, R.; Eichinger, R.; Roller, A.; Jakupec, M.A.; Schmidt, P.P.; Arion, V.B.; Keppler, B.K. Gallium(III) and Iron (III) complexes of α-N-heterocyclic thiosemicarbazones: Synthesis, characterization, cytotoxicity and interaction with Ribonucleotide Reductase. J. Med. Chem. 2007, 50, 1254–1265. [Google Scholar] [CrossRef]
- Salam, M.A.; Alam, M.; Sarker, S.; Rahman, M.M. Synthesis, spectroscopic characterization, crystal structure and anti-bacterial activity of diorganotin(IV) complexes with 5-bromo-2-hydroxybenzaldehyde-N(4)-ethylthiosemicarbazone. J. Coord. Chem. 2018, 71, 1593–1605. [Google Scholar] [CrossRef]
- Yousef, T.A.; El-Reash, G.M.A. Synthesis and biological evaluation of complexes based on thiosemicarbazone ligand. J. Mol. Struct. 2020, 1201, 127180–127190. [Google Scholar] [CrossRef]
- Rosu, T.; Pahontu, E.; Pasculescu, S.; Georgescu, R.; Stanica, N.; Curaj, A.; Popescu, A.; Leabu, M. Synthesis, characterization antibacterial and antiproliferative activity of novel Cu(II) and Pd(II) complexes with 2-hydroxy-8-R-tricyclo [7.3.1.0.2,7]tridecane-13-one thiosemicarbazone. Eur. J. Med. Chem. 2010, 45, 1627–1634. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.C.; Wang, B.D.; Yang, Z.Y.; Li, Y.; Qin, D.D.; Li, T.R. Synthesis, crystal structure, DNA interaction and antioxidant activities of two novel water-soluble Cu(2+) complexes derived from 2-oxo-quinoline-3-carbaldehyde Schiff-bases. Eur. J. Med. Chem. 2009, 44, 4477–4484. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, R.; Garcia, B.; Tojal, J.G.; Busto, N.; Ibeas, S.; Leal, J.M.; Martins, C.; Gaspar, J.; Borras, J.; Gil-Garcia, R.; et al. Biological assays and non covalent interactions of pyridine-2-carbaldehyde thiosemicarbazone copper(II) drugs with [poly(dA-dT)]2, [poly(dG-dC)]2 and calf thymus DNA. J. Biol. Inorg. Chem. 2010, 15, 515–532. [Google Scholar] [CrossRef] [PubMed]
- Rogolino, D.; Cavazzoni, A.; Gatti, A.; Tegoni, M.; Pelosi, G.; Verdolino, V.; Fumarola, C.; Cretella, D.; Petronini, P.G.; Carcelli, M. Anti-proliferative effects of copper(II) complexes with hydroxyquinoline-thiosemicarbazone ligands. Eur. J. Med. Chem. 2017, 128, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Belicchi-Ferrari, M.; Bisceglie, F.; Pelosi, G.; Tarasconi, P. Heterocyclic substituted thiosemicarbazones and their Cu(II) complexes. Synthesis, characterization and studies of substituent effects on coordination and DNA binding. Polyhedron 2008, 27, 1361–1367. [Google Scholar] [CrossRef]
- Quiroga, A.G.; Ranninger, C.N. Contribution to the SAR field of metallated and coordination complexes. Studies of the palladium and platinum derivatives with selected thiosemicarbazones as antitumoral drugs. Coord. Chem. Rev. 2004, 248, 119–133. [Google Scholar] [CrossRef]
- Prabhakaran, R.; Kalaivani, P.; Poornima, P.; Dallemer, F.; Huang, R.; Vijaya Padma, V.; Natarajan, K. Synthesis, DNA/protein binding and in vitro cytotoxic studies of new palladium metallothiosemicarbazones. Bioorg. Med. Chem. 2013, 21, 6742–6752. [Google Scholar] [CrossRef]
- Hosseinpour, S.; Hosseini-Yazdi, S.A.; White, J. Binuclear zinc(II) complexes of N(4)-substituted bis(thiosemicarbazone) ligands incorporating hydroxyl group and their non-hydroxyl analogues. Inorg. Chim. Acta 2017, 461, 150–160. [Google Scholar] [CrossRef]
- Rui, W.; Tian, X.; Zeng, P.; Liu, W.; Ying, P.; Chen, H.; Lu, J.; Yang, N.; Chen, H. The antit-tumor activity of novel oxovanadium complexes derived from thiosemicarbazones and fluoro-phenanthroline derivatives. Polyhedron 2016, 117, 803–816. [Google Scholar] [CrossRef]
- Bargujar, S.; Chandra, S.; Chauhan, R.; Rajor, H.K.; Bhardwaj, J. Synthesis, spectroscopic evaluation, molecular modelling, thermal study and biological evaluation of manganese(II) complexes derived from bidentate N,O and N,S donor Schiff base ligands. Appl. Organomet. Chem. 2018, 32, e4149. [Google Scholar] [CrossRef]
- Gou, Y.; Wang, J.; Chen, S.; Zhang, Z.; Zhang, Y.; Zhang, W.; Yang, F. α-N-heterocyclic thiosemicarbazone Fe(III) complex: Characterization of its antitumor activity and identification of anticancer mechanism. Eur. J. Med. Chem. 2016, 123, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, E.; Thomas, S.P.; Poornima, P.; Kalaivani, P.; Prabhakaran, R.; Vijaya Padma, V.; Natarajan, K. Evaluation of DNA binding, antioxidant and cytotoxic activity of mononuclear Co(III) complexes of 2-oxo-1,2-dihydrobenzo[h]quinolone-3-carbaldehyde thiosemicarbazones. Eur. J. Med. Chem. 2012, 50, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.G.; Su, G.; Chen, P.; Du, Y.; Gou, Y.; Liu, Y. Evaluation of DNA binding and DNA cleavage of nickel(II) complexes with tridentate α-N-heterocyclic thiosemicarbazone ligands. Inorg. Chim. Acta 2018, 471, 194–202. [Google Scholar] [CrossRef]
- Haribabu, J.; Jeyalakshmi, K.; Arun, Y.; Bhuvanesh, N.S.P.; Perumal, P.T.; Karvembu, R. Synthesis of Ni(II) complexes bearing indole-based thiosemicarbazone ligands for interaction with biomolecules and some biological applications. J. Biol. Inorg. Chem. 2017, 22, 461–480. [Google Scholar] [CrossRef] [PubMed]
- Kalaiarasi, G.; Umadevi, C.; Shanmugapriya, A.; Kalaivani, P.; Dallemer, F.; Prabhakaran, R. DNA(CT), protein (BSA) binding studies, anti-oxidant and cytotoxicity studies of new binuclear Ni(II) complexes containing 4(N)-substituted thiosemicarbazones. Inorg. Chim. Acta 2016, 453, 547–558. [Google Scholar] [CrossRef]
- Ramachandran, E.; Raja, D.S.; Bhuvanesh, N.S.P.; Natarajan, K. Synthesis, characterization and in vitro pharmacological evaluation of new water soluble Ni(II) complexes of 4N-substituted thiosemicarbazones of 2-oxo-1,2-dihydroquinoline-3-carbaldehyde. Eur. J. Med. Chem. 2013, 64, 179–189. [Google Scholar] [CrossRef]
- Kalaiarasi, G.; Rajkumar, S.R.J.; Dharani, S.; Lynch, V.M.; Prabhakaran, R. Synthesis, spectral characterization and biological evaluation of some copper(II) complexes containing 4-oxo-4H-chromene-3-carbaldehyde-4(N)-substituted thiosemicarbazones. Inorg. Chim. Acta 2018, 471, 759–776. [Google Scholar] [CrossRef]
- Lobana, T.S.; Indoria, S.; Sood, H.; Arora, D.S.; Randhawa, B.S.; Garcia-Santos, I.; Smolinski, V.A.; Jasinski, J.P. Synthesis of 5-nitro-salicylaldehyde-N-substituted thiosemicarbazones of copper(II): Molecular structures, spectroscopy, ESI-mass studies and antimicrobial activity. Inorg. Chim. Acta 2017, 461, 248–260. [Google Scholar] [CrossRef]
- Beckford, F.A.; Brock, A.; Gonzalez-Sarrias, A.; Seeram, N.P. Cytotoxic gallium complexes containing thiosemicarbazones derived from 9-anthraldehyde: Molecular docking with biomolecules. J. Mol. Struct. 2016, 1121, 156–166. [Google Scholar] [CrossRef]
- Qi, J.; Deng, J.; Qian, K.; Tian, L.; Li, J.; He, K.; Huang, X.; Cheng, Z.; Zheng, Y.; Wang, Y. Novel 2-pyridinecarboxaldehyde thiosemicarbazones Ga(III) complexes with a high antiproliferative activity by promoting apoptosis and inhibiting cell cycle. Eur. J. Med. Chem. 2017, 134, 34–42. [Google Scholar] [CrossRef]
- Mahalingam, V.; Chitrapriya, N.; Fronczek, F.R.; Natarajan, K. New Ru(II)-DMSO complexes of ON/SN chelates: Synthesis, behaviour of Schiff bases towards hydrolitic cleavage of C=N bond, electrochemistry and biological activities. Polyhedron 2010, 29, 3363–3371. [Google Scholar] [CrossRef]
- Ghosh, B.; Adak, P.; Naskar, S.; Pakhira, B.; Mitra, P.; Chattopadhyay, S.K. Ruthenium(II/III) complexes of redox non-innocent bis(thiosemicarbazone)ligands: Synthesis, X-ray crystal structures, electrochemical, DNA binding and DFT studies. Polyhedron 2017, 131, 74–85. [Google Scholar] [CrossRef]
- Selvamurugan, S.; Ramachandran, R.; Prakash, G.; Nirmala, M.; Viswanathamurthy, P.; Fujiwara, S.; Endo, A. Ruthenium(II) complexes encompassing 2-oxo-1,2-dihydroquinoline-3-carbaldehyde thiosemicarbazone hybrid ligand: A new versatile potential catalyst for dehydrogenative amide synthesis. Inorg. Chim. Acta 2017, 454, 46–53. [Google Scholar] [CrossRef]
- Prabhakaran, R.; Huang, R.; Karvembu, R.; Jayabalakrishnan, C.; Natarajan, K. Formation of unusual ruthenium(III) carbonyl complex through ONS tricoordination of salicyladehyde-N-phenylthiosemicarbazone. Inorg. Chim Acta 2007, 360, 691–694. [Google Scholar] [CrossRef]
- Mahalingam, V.; Chitrapriya, N.; Fronczek, F.R.; Natarajan, K. Dimethyl sulfoxide ruthenium(II) complexes of thiosemicarbazones and semicarbazone. Synthesis, characterization and biological studies. Polyhedron 2008, 27, 2743–2750. [Google Scholar] [CrossRef]
- Ghosh, B.; Adak, P.; Naskar, P.S.; Pakhira, B.; Mitra, P.; Dinda, R.; Chattopadhyay, S.K. Ruthenium(II) complexes of thiosemicarbazones: Synthesis, X-ray crystal structures, spectroscopy, electrochemistry, DFT studies and fluoride sensing properties. Inorg. Chim. Acta 2017, 459, 1–14. [Google Scholar] [CrossRef]
- Yaman, P.K.; Sen, B.; Karagoz, C.S.; Subasi, E. Half-sandwich ruthenium-arene complexes with thiophen containing thiosemicarbazones: Synthesis and structural characterization. J. Organomet. Chem. 2017, 832, 27–35. [Google Scholar] [CrossRef]
- Haribabu, J.; Sabapathi, G.; Tamizh, M.M.; Balachandran, C.; Bhuvanesh, N.S.P.; Venuvanalingam, P.; Karvembu, R. Water soluble mono- and binuclear Ru(η6-p-cymene) complexes containing indole thiosemicarbazones: Synthesis, DFT modeling, biomolecular interactions and in vitro anticancer activity through apoptosis. Organometallics 2018, 37, 1242–1257. [Google Scholar] [CrossRef]
- Muralisankar, M.; Dheepika, R.; Haribabu, J.; Balachandran, C.; Aoki, S.; Bhuvanesh, N.S.P.; Nagarajan, S. Design, synthesis, DNA/HSA binding and cytotoxic activity of half sandwich Ru(II)-arene complexes containing triarylamine-thiosemicarbazone hybrids. ACS Omega 2019, 4, 11712–11723. [Google Scholar] [CrossRef]
- Dobrova, A.; Platzer, S.; Bacher, S.; Milunovic, M.N.M.; Dobrov, A.; Spengler, G.; Enyedy, E.A.; Novitchi, G.; Arion, V.B. Structure-antiproliferative activity studies on L-proline and homoproline-4-N-pyrrolidine-3-thiosemicarbazone hydrids and their nickel(II), palladium(II) and copper(II) complexes. Dalton Trans. 2016, 45, 13427–13439. [Google Scholar] [CrossRef]
- Muralisankar, M.; Basheer, S.M.; Haribabu, J.; Bhuvanesh, N.S.P.; Karvembu, R.; Sreekanth, A. An investigation on the DNA/protein binding, DNA cleavage and in vitro anticancer properties of SNO pincer type palladium(II) complexes with N-substituted isatin thiosemicarbazone ligands. Inorg. Chim. Acta 2017, 466, 61–70. [Google Scholar] [CrossRef]
- Ramachandran, E.; Kalaivani, P.; Prabhakaran, R.; Zeller, M.; Bartlett, J.H.; Adero, P.O.; Wagner, T.R.; Natarajan, K. Synthesis, characterization, crystal structure and DNA binding studies of Pd(II) complexes containing thiosemicarbazone and triphenylphosphine/triphenylarsine. Inorg. Chim. Acta 2012, 385, 94–99. [Google Scholar] [CrossRef]
- Kalaivani, P.; Prabhakaran, R.; Kaveri, M.V.; Huang, R.; Staples, R.J.; Natarajan, K. Synthesis, spectral, X-ray crystallography, electrochemistry, DNA/protein binding and radical scavenging activity of new palladium(II) complexes containing triphenylarsine. Inorg. Chim. Acta 2013, 405, 415–526. [Google Scholar] [CrossRef]
- Ramachandran, E.; Bertani, R.; Sgarbossa, P.; Natarajan, K.; Bhuvanesh, N.S.P. Synthesis, crystal structure, DNA and protein binding studies of novel binuclear Pd(II) complex of 6-methoxy-2-oxo-1,2-dihydroquinoline-3-carbaldehyde-4(N,N)-dimethylthiosemicarbazone. J. Inorg. Biochem. 2016, 155, 1–8. [Google Scholar]
- Oliveira, A.A.; Perdigao, G.M.C.; Rodrigues, L.E.; da Silva, J.G.; Souza-Fagundes, E.; Takahashi, J.A.; Rocha, W.R.; Beraldo, H. Cytotoxic and antimicrobial effects of indium(III) complexes with 2-acetylpyridine-derived thiosemicarbazones. Dalton Trans. 2017, 46, 918–932. [Google Scholar] [CrossRef]
- Arce, E.R.; Machado, I.; Rodriguez, B.; Lapier, M.; Zuniga, M.C.; Maya, J.D.; Azar, C.O.; Otero, L.; Gambino, D. Rhenium(I) tricarbonyl compounds of bioactive thiosemicarbazones: Synthesis, characterization and activity against Trypanosoma cruzi. J. Inorg. Biochem. 2017, 170, 125–133. [Google Scholar] [CrossRef]
- Nguyen, T.B.Y.; Pham, C.T.; Trieu, T.N.; Abram, U.; Nguyen, H.H. Syntheses, structures and biological evaluation of some transition metal complexes with a tetradentate benzamidine/thiosemicarbazone ligand. Polyhedron 2015, 96, 66–70. [Google Scholar] [CrossRef]
- Matesanz, A.I.; Albacete, P.; Souza, P. Synthesis and characterization of a new bioactive mono(thiosemicarbazone) ligand based on 3,5-diacetyl-1,2,4-triazol-diketone and its palladium and platinum complexes. Polyhedron 2016, 109, 161–165. [Google Scholar] [CrossRef]
- Rettondin, A.R.; Carneira, Z.A.; Goncalves, A.C.R.; Ferreira, V.F.; Oliveira, C.G.; Lima, A.N.; Oliveira, R.J.; de Albuquerque, S.; Deflon, V.M.; Maia, P.I.S. Gold(III) complexes with ONS-tridentate thosemicarbazones: Toward selective trypanocidal drugs. Eur. J. Med. Chem. 2016, 120, 217–226. [Google Scholar] [CrossRef]
- Bedier, R.A.; Yousef, T.A.; Abu El-Reash, G.M.; El-Gemmal, O.A. Synthesis, structural and optical band gap and biological studies on iron(III), nickel(II), zinc(II) and mercury(II) complexes of benzyl α-monoxime pyridyl thiosemicarbazone. J. Mol. Struct. 2017, 1139, 436–446. [Google Scholar] [CrossRef]
- Sharma, D.; Jasinski, J.P.; Smolinski, V.A.; Kaur, M.; Paul, K.; Sharma, R. Synthesis and structure of complexes (NiII, AgI) of substituted benzaldehyde thiosemicarbazones and antitubercular activity of NiII complex. Inorg. Chim. Acta 2020, 499, 119187–119194. [Google Scholar] [CrossRef]
- Medici, S.; Peana, M.; Nurchi, V.M.; Lachowicz, J.I.; Crisponi, G.; Zoroddu, M.A. Noble metals in medicine: Latest advances. Coord. Chem. Rev. 2015, 284, 329–350. [Google Scholar] [CrossRef]
- Serda, M.; Kalinowski, D.S.; Rasko, N.; Potuckova, E.; Mrozek-Wilczkiewicz, A.; Musiol, R.; Malecki, J.G.; Sajewicz, M.; Ratuszna, A.; Muchowicz, A.; et al. Exploring the anti-cancer activity of novel thiosemicarbazones generated through the combination of retro-fragments: Dissection of critical structure-activity relationships. PLoS ONE 2014, 9, e110291. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Li, J.; Fang, Y.; Chang, B.; Meng, Q.; Li, M.; Wang, C.; Zhu, X. Synthesis, characterization and bioactivities of a new covalent copper compound derived from {P2Mo5O23}6- and thiosemicarbazones. Bioorg. Med. Chem. Lett. 2010, 30, 126781–126785. [Google Scholar] [CrossRef] [PubMed]
- Bilyi, J.K.; Harmer, J.R.; Bernhardt, P.V. Formation and reactivity of copper acetylacetone bis(thiosemicarbazone) complexes. Eur. J. Inorg. Chem. 2018, 4731–4741. [Google Scholar]
- Mahendiran, D.; Amuthakala, S.; Bhuvanesh, N.S.P.; Kumar, R.S.; Rahiman, A.K. Copper complexes as prospective anticancer agents: In vitro and in vivo evaluation, selective targeting of cancer cells by DNA damage and S phase arrest. RSC Adv. 2018, 8, 16973–16990. [Google Scholar] [CrossRef]
- Anjum, R.; Palanimuthu, D.; Kalinowski, D.S.; Lewis, W.; Park, K.C.; Kovacevic, Z.; Khan, I.U.; Richardson, D.R. Synthesis, characterization and in vitro anticancer activity of copper and zinc bis(thiosemicarbazone) complexes. Inorg. Chem. 2019, 58, 13709–13723. [Google Scholar] [CrossRef]
- Brown, O.C.; Torres, J.B.; Holt, K.B.; Blower, P.J.; Went, M.J. Copper complexes with dissimmetrically substituted bis(thiosemicarbazone) ligands as a basis for PET radiopharmaceuticals: Control of redox potential and lipophilicity. Dalton Trans. 2017, 46, 14612–14630. [Google Scholar] [CrossRef]
- Zhang, Z.; Gou, Y.; Wang, J.; Yang, K.; Qi, J.; Zhou, Z.; Liang, S.; Liang, H.; Yang, F. Four copper(II) compounds synthesized by anion regulation: Structure, anticancer function and anticancer mechanism. Eur. J. Med. Chem. 2016, 121, 399–409. [Google Scholar] [CrossRef]
- Ramachandran, E.; Senthil Raja, D.; Bhuvanesh, N.P.S.; Natarajan, K. Mixed ligand palladium(II) complexes of 6-methoxy-2-oxo-1,2-dihydroquinoline-3-carbaldehyde 4N-substituted thiosemicarbazones with triphenylphosphine co-ligand: Synthesis, crystal structure and biological properties. Dalton Trans. 2012, 41, 13308–13323. [Google Scholar] [CrossRef]
- Senthil Raja, D.; Bhuvanesh, N.S.P.; Natarajan, K. Biological evaluation of a novel water soluble sulphur bridged binuclear copper(II) thiosemicarbazone complex. Eur. J. Med. Chem. 2011, 46, 4584–4594. [Google Scholar] [CrossRef]
- Ramachandran, E.; Senthil Raja, D.; Mike, J.L.; Wagner, T.R.; Zeller, M.; Natarajan, K. Evaluation on the role of terminal N-substitution in 6-methoxy-2-oxo-1,2-dihydroquinoline-3-carbaldehyde thiosemicarbazones on the biological properties of new water soluble nickel(II) complexes. RSC Adv. 2012, 2, 8515–8525. [Google Scholar] [CrossRef]
- Senthil Raja, D.; Paramaguru, G.; Bhuvanesh, N.S.P.; Reibenspies, J.H.; Renganathan, R.; Natarajan, K. Effect of terminal N-substitution in 2-oxo-1,2-dihydroquinoline-3-carbaldehyde thiosemicarbazones on the mode of coordination, structure, interaction with protein, radical scavenging and cytotoxic activiity of copper(II) complexes. Dalton Trans. 2011, 40, 4548–4559. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, Y.; Liang, H.; Wang, H.; Hu, K.; Liu, X.; Yi, X.; Peng, Y. Synthesis and antioxidant activities of 2-oxo-quinoline-3-carbaldehyde Schiff-base derivatives. Bioorg. Med. Chem. Lett. 2013, 23, 107–111. [Google Scholar] [CrossRef]
- Singh, M.K.; Chandra, A.; Singh, B.; Singh, R.H. Synthesis of diastereomeric 2,4-disubstituted pyrano[2, 3-b]quinolines from 3-formyl-2-quinolines through O-C bond formation via intramolecular electrophilic cyclization. Tetrahedron Lett. 2007, 48, 5987–5990. [Google Scholar] [CrossRef]
- Turkkan, E.; Sayin, U.; Erbilen, N.; Pehlivanoglu, S.; Erdogan, G.; Tasdemir, H.U.; Saf, A.O.; Guler, L.; Akgemci, E.G. Anticancer, antimicrobial, spectra, voltammetric and DFT studies with Cu(II) complexes of 2-hydroxy-5-methoxyacetophenone thiosemicarbazone and its N(4)-substituted derivatives. J. Organomet. Chem. 2017, 831, 23–35. [Google Scholar] [CrossRef]
- Khalilian, H.; Mirzaei, S.; Taherpour, A. Comprehensive insights into the structure and coordination behaviour of thiosemicarbazone ligands: A computational assessment of the E-Z interconversion mechanism during coordination. New J. Chem. 2015, 39, 9313–9324. [Google Scholar] [CrossRef]
- Sripathi, M.P.; Berely, S.; Venkata Ramana Reddy, C. Computational studies of 4-formylpyridinethiosemicarbazone and structural and biological studies of its Ni(II) and Cu)II) complexes. Heteroat. Chem. 2019, 2019, 3507837. [Google Scholar] [CrossRef]
- Zou, B.Q.; Lu, X.; Qin, Q.P.; Bai, Y.X.; Zhang, Y.; Wang, M.; Liu, Y.C.; Chen, Z.F.; Liang, H. Three novel transition metal complexes of 6-methyl-2-oxo-quinoline-3-carbaldehyde thiosemicarbazone: Synthesis, crystal structure, cytotoxicity, and mechanism of action. RSC Adv. 2017, 7, 17923–17933. [Google Scholar] [CrossRef]
- Ramachandran, E.; Senthil Raja, D.; Rath, N.P.; Natarajan, K. Role of substitution at terminal nitrogen of 2-oxo-quinoline-3-carbaldehyde thiosemicarbazones on the coordination behavior and structure and biological properties of their palladium(II) complexes. Inorg. Chem. 2013, 52, 1504–1514. [Google Scholar] [CrossRef]
- Pahontu, E.; Julea, F.; Chumakov, Y.; Petrenco, P.; Rosu, T.; Gulea, A. Synthesis, characterization, crystal structure and antiproliferative activity studies of Cu(II), Ni(II) and Co(II) complexes with 4-benzoyl-5-pyrazolones derived compounds. J. Organomet. Chem. 2017, 836–837, 44–55. [Google Scholar] [CrossRef]
- Schlosser, G.; Stefanescu, R.; Przybylsk, M.; Murariu, M.; Hudecz, F.; Drochioiu, G. Copper-Induced Oligomerization of Peptides: A Model Study. Eur. J. Mass Spectrom. 2007, 13, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Tom, L.; Aiswarya, N.; Sreejith, S.S.; Prathapachandra Kurup, M.R. Self-organized three dimensional architectures based on non-covalent interactions in square planar Cu(II) thiosemicarbazone: Solvent mediated crystallization and EPR based correlation study. Inorg. Chim. Acta 2018, 473, 223–235. [Google Scholar] [CrossRef]
- Ilies, D.; Pahontu, E.; Shova, S.; Georgescu, R.; Stanica, N.; Olar, R.; Gulea, A.; Rosu, T. Synthesis, characterization, crystal structure and antimicrobial activity of copper(II) complexes with a thiosemicarbazone derived from 3-formyl-6-methylchromone. Polyhedron 2014, 81, 123–131. [Google Scholar] [CrossRef]
- Ilies, D.; Pahontu, E.; Shova, S.; Gulea, A.; Rosu, T. Synthesis, characterization and crystal structures of nickel(II), palladium(II), and copper(II) complexes with 2-furaldehyde-4-phenylthiosemicarbazone. Polyhedron 2013, 51, 307–315. [Google Scholar] [CrossRef]
- Yokoi, H.; Addison, A.W. Spectroscopic and redox properties of pseudotetrahedral copper(II) complexes. Their relation to copper proteins. Inorg. Chem. 1977, 16, 1341–1349. [Google Scholar] [CrossRef]
- Cowley, A.R.; Dilworth, J.R.; Donnelly, P.S.; White, J.M. Copper Complexes of Thiosemicarbazone-Pyridylhydrazine (THYNIC) Hybrid Ligands: A New Versatile Potential Bifunctional Chelator for Copper Radiopharmaceuticals. Inorg. Chem. 2006, 45, 496–498. [Google Scholar] [CrossRef] [PubMed]
- Dearling, J.L.J.; Lewis, J.S.; Mullen, G.E.D.; Welch, M.J.; Blower, P.J. Copper bis(thiosemicarbazone) complexes as hypoxia imaging agents: Structure-activity relationships. J. Biol. Inorg. Chem. 2002, 7, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, E.; Gandin, V.; Bertani, R.; Sgarbossa, P.; Natarayan, K.; Bhuvanesh, N.S.P.; Venzo, A.; Zoleo, A.; Glisenti, A.; Dolmella, A.; et al. Synthesis, characterization and cytotoxic activity of novel copper(II) complexes with aroylhydrazone derivatives of 2-oxo-1,2-dihydrobenzo[h]quinoline-3-carbaldehyde. J. Inorg. Biochem. 2018, 182, 18–28. [Google Scholar] [CrossRef]
- Marverti, G.; Andrews, P.A.; Piccinini, G.; Ghiaroni, S.; Barbieri, D.; Moruzzi, M.S. Modulation of cis-diamminedichloroplatinum(II) accumulation and cytotoxicity by spermine in sensitive and resistant human ovarian carcinoma cells. Eur. J. Cancer 1997, 33, 669–675. [Google Scholar] [CrossRef]
- Andrews, P.A.; Murphy, M.P.; Howell, S.B. Differential potentiation of alkylating and platinating agent cytotoxicity in human ovarian carcinoma cells by glutathione depletion. Cancer Res. 1985, 45, 6250–6253. [Google Scholar] [PubMed]
- Scanlon, K.J.; Kashani-Sabet, M.; Tone, T.; Funato, T. Cisplatin resistance in human cancers. Pharmacol. Ther. 1991, 52, 385–406. [Google Scholar] [CrossRef]
- Marzano, C.; Gandin, V.; Folda, A.; Scutari, G.; Bindoli, A.; Rigobello, M.P. Inhibition of thioredoxin reductase by auranofin induces apoptosis in cisplatin-resistant human ovarian cancer cells. Free Radic. Biol. Med. 2007, 42, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Ekblad, L.; Kjellström, J.; Johnsson, A. Reduced drug accumulation is more important in acquired resistance against oxaliplatin than against cisplatin in isogenic colon cancer cells. Anticancer Drugs 2010, 21, 523–531. [Google Scholar] [CrossRef]
- Kankotia, S.; Stacpoole, P.W. Dichloroacetate and cancer: New home for an orphan drug? Biochim. Biophys. Acta 2014, 1846, 617–629. [Google Scholar] [CrossRef]
- Bisceglie, F.; Orsoni, N.; Pioli, M.; Bonati, B.; Tarasconi, P.; Rivetti, C.; Amidani, D.; Montalbano, S.; Buschini, A.; Pelosi, G. Cytotoxicity activity of copper(II), nickel(II) and platinum (II) thiosemicarbazone derivatives: Interaction with DNA and the H2A histone opeptide. Metallomics 2019, 11, 1729–1742. [Google Scholar] [CrossRef]
- Öhrvik, H.; Aaseth, J.; Horn, N. Orchestration of dynamic copper navigation—New and missing pieces. Metallomics 2017, 9, 1204–1229. [Google Scholar] [CrossRef]
- Bax, A.; Subramanian, S. Sensitivity-enhanced two-dimensional heteronuclear shift correlation NMR spectroscopy. J. Magn. Reson. 1986, 67, 565–569. [Google Scholar] [CrossRef]
- Bax, A.; Summers, M.F. 1H and 13C assignments from sensitivity-enhanced detection of heteronuclear multiple-bond connectivity by 2D multiple quantum NMR. J. Am. Chem. Soc. 1986, 108, 2093–2094. [Google Scholar] [CrossRef]
- Otting, G.; Wuthrich, K. Efficient purging scheme for proton-detected heteronuclear two-dimensional NMR. J. Magn. Reson. 1988, 76, 569–574. [Google Scholar] [CrossRef]
- Friedrich, J.; Eder, W.; Castaneda, J.; Doss, M.; Huber, E.; Ebner, R.; Kunz-Schughart, L.A. A reliable tool to determine cell viability in complex 3-d culture: The acid phosphatase assay. J. Biomol. Screen. 2007, 12, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Gabr, M.T.; El-Gohary, N.S.; El-Bendary, E.R.; El-Kerdawy, M.M.; Ni, N. Isatin-β-thiosemicarbazones: Microwave-assisted synthesis, antitumor activity and structure activity relationship. Eur. J. Med. Chem. 2017, 128, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Bisceglie, F.; Tavone, M.; Mussi, F.; Azzoni, S.; Montalbano, S.; Franzoni, S.; Tarasconi, P.; Buschini, A.; Pelosi, G. Effects of polar substituents on the biological activity of thiosemicarbazone metal complexes. J. Inorg. Biochem. 2018, 179, 60–70. [Google Scholar] [CrossRef]
- Kalinovski, D.S.; Stefani, C.; Toyakuni, S.; Ganz, T.; Anderson, G.J.; Subramanian, N.V.; Trinder, D.; Olynyk, J.K.; Chua, A.; Jansson, P.J.; et al. Redox cycling metals: Pedaling their roles in metabolism and their use in the development of novel therapeutics. Biochim. Biophys. Acta 2016, 1863, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Rogolino, D.; Gatti, A.; Carcelli, M.; Pelosi, G.; Bisceglie, F.; Restive, F.M.; Degola, F.; Buschini, A.; Montalbano, S.; Feretti, D.; et al. Thiosemicarbazone scaffold for the design of antifungal and antiaflatoxigenic agents: Evaluation of ligands and related copper complexes. Sci. Rep. 2017, 7, 11214–11225. [Google Scholar] [CrossRef] [PubMed]
- Marzano, C.; Sbovata, S.M.; Gandin, V.; Colavito, D.; Del Giudice, E.; Michelin, R.A.; Venzo, A.; Seraglia, R.; Benetollo, F.; Schiavon, M.; et al. A New Class of Antitumor trans-Amine-Amidine-Pt(II) Cationic Complexes: Influence of Chemical Structure and Solvent on in Vitro and in Vivo Tumor Cell Proliferation. J. Med. Chem. 2010, 53, 6210–6227. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.S.; Yu, P.; Hu, J.N.; Liu, Y.; Li, Z.W.; Qian, Y.; Wamg, Y.; Gou, Y.; Yang, F. Mitochondria-localizing N-heterocyclic thiosemicarbazone copper complexes with good cytotoxicity and high antimetastatic activity. Eur. J. Med. Chem. 2019, 164, 654–664. [Google Scholar] [CrossRef]
- Biswas, N.; Saha, S.; Khanra, S.; Sarkar, A.; Mandal, D.P.; Bhattacharjee, S.; Chaudhuri, A.; Chakraborty, S.; Choudhury, C.R. Example of two novel thiocyanato bridged copper(II) complexes derived from substituted thiosemicarbazone ligand: Structural elucidation, DNA/albumin binding, biological profile analysis, and molecular docking study. J. Biomol. Struct. Dyn. 2019, 37, 2801–2822. [Google Scholar] [CrossRef]
- Kaushal, M.; Lobana, T.S.; Nim, L.; Bala, R.; Aropra, D.S.; Garcia-Santos, I.; Duff, C.E.; Jasinski, J.P. Synthesis of 2-acetylpyridine-N-substituted thiosemicarbazonates of copper(II) with high antimicrobial activity against methillicin resistant S.aureus, K.pneumoniae 1 and C.albicans. New J. Chem. 2019, 43, 11727–11742. [Google Scholar] [CrossRef]
- Rajendran, N.; Periyasamy, A.; Kamatchi, N.; Solomon, V. Biological evaluation of copper(II) complexes on N(4)-substituted thiosemicarbazide derivatives, and diimine co-ligands using DNA interaction, antibacterial and in vitro cytotoxicity. J. Coord. Chem. 2019, 72, 1937–1956. [Google Scholar] [CrossRef]
- Kalaiarasi, G.; Rajkumar, S.R.J.; Dharani, S.; Matecki, J.G.; Prabhakaran, R. An investigation on 3-acetyl-7-methoxy-coumarin Schiff bases and their Ru(II) metallates with potent antiproliferative activity and enhanced LDH and NO release. RSC Adv. 2018, 8, 1539–1561. [Google Scholar] [CrossRef]

| Assignment | gzz | Azz | gxx | gyy | Axx | Ayy |
|---|---|---|---|---|---|---|
| 1a | 2.388 | 137 × 10−4 | 2.07 | 2.08 | 3.3 × 10−4 | 3.3 × 10−4 |
| 1b | 2.260 | 173 × 10−4 | 2.07 | 2.10 | 5.0 × 10−4 | 5.0 × 10−4 |
| 3a | 2.385 | 147 × 10−4 | 2.01 | 2.08 | 4.0 × 10−4 | 4.0 × 10−4 |
| 3b | 2.350 | 182 × 10−4 | 2.07 | 2.10 | 5.0 × 10−4 | 5.0 × 10−4 |
| Compound | Cathodic Peak Potential | Anodic Peak Potential 3 | ||
|---|---|---|---|---|
| Lred | Ired 2 | A | Iox | |
| H2L1 H2L2 H2L3 | −1.56 | |||
| 1 | −1.59 | −0.26 | −0.05 | 0.36 |
| 2 | −1.56 | −0.32 | −0.05 | 0.27 |
| 3 | −1.56 | −0.31 | −0.07 | 0.24 |
| 2 | 1a * | 1b * | |
|---|---|---|---|
| Cu1–O1 | 1.948(3) | 1.973(3) | 1.946(3) |
| Cu1–S1 | 2.266(2) | 2.289(2) | 2.278(1) |
| Cu1–N2 | 1.977(3) | 1.975(3) | 1.969(3) |
| Cu1–Ow1 | 1.928(3) | 1.961(2) | 1.943(3) |
| Cu1–Ow2 | 2.226(4) | 2.254(2) | |
| Cu1–ON1 | 2.695(4) | ||
| Cu1–ON2 | 2.525(4) | ||
| N2–Cu1–Ow2 | 96.6(1) | 100.7(1) | 88.5(3) |
| Ow1–Cu1–Ow2 | 90.9(2) | 97.0(1)) | 82.2(2) |
| Ow1–Cu1–ON1 | 113.2(2) | ||
| N2–Cu1–Ow1 | 172.4(1) | 169.2(1) | 170.6(1) |
| ON1–Cu1–ON2 | 158.1(2) |
| 3a(Cu1) * | 3b(Cu2) * | |
|---|---|---|
| Cu–O1 | 1.930(3) | 1.929(3) |
| Cu–S1 | 2.263(1) | 2.258(1) |
| Cu–N2 | 1.986(3) | 1.977(3) |
| Cu–Om1 | 1.989(3) | |
| Cu–Ow1 | 1.931(3) | |
| Cu–ON1 | 2.318(3) | 2.388(3) |
| N2–Cu–O1 | 90.8(1) | 91.3(1) |
| N2–Cu–S1 | 87.0(1) | 87.4(1) |
| N2–Cu–ON1 | 122.3(1) | 86.4(1) |
| ON1–Cu–Om1 | 80.1(1) | |
| ON1–Cu–Ow1 | 91.7(1) | |
| S1–Cu–ON1 | 90.86(8) | 99.93(8) |
| N2–Cu–Om1 | 157.6(1) | |
| N2–Cu–Ow1 | 178.0(1) | |
| S1–Cu–O1 | 177.16(9) | 168.12(9) |
| IC50(µM) ± S.D. | |||||
|---|---|---|---|---|---|
| Compounds | HCT-15 | A549 | BxPC3 | PSN-1 | A375 |
| 1 | 8.4 ± 1.7 | 6.8 ± 1.3 | 7.8 ± 0.5 | 9.2 ± 1.6 | 0.6 ± 0.2 |
| H2L1 | >25 | >50 | 16.8 ± 2.6 | 18.3 ± 2.8 | 17.5 ± 3.2 |
| 2 | 2.6 ± 1.2 | 5.6 ± 0.5 | 0.1 ± 0.08 | 4.6 ± 2.0 | 0.027 ± 0.005 |
| H2L2 | >25 | >50 | >25 | >100 | >25 |
| 3 | 1.7 ± 1.5 | 3.2 ± 0.6 | 0.3 ± 0.2 | 5.3 ± 0.3 | 0.031 ± 0.001 |
| H2L3 | 23.4 ± 1.0 | >25 | >25 | >100 | >100 |
| cisplatin | 15.5 ± 3.8 | 6.9 ± 2.1 | 13.9 ± 5.9 | 13.5 ± 4.1 | 1.3 ± 0.6 |
| IC50(µM) ± S.D. | ||||||
|---|---|---|---|---|---|---|
| Compounds | 2008 | C13* | R.F. | LoVo | LoVo-OXP | R.F. |
| 1 | 9.7 ± 3.3 | 6.7 ± 1.2 | 0.7 | 0.7 ± 0.07 | 1.2 ± 0.6 | 1.8 |
| 2 | 11.2 ± 2.0 | 8.5 ± 3.5 | 0.8 | 1.9 ± 0.6 | 2.0 ± 0.04 | 1.1 |
| 3 | 2.8 ± 0.9 | 0.9 ± 0.7 | 0.3 | 0.7 ± 0.5 | 1.1 ± 0.1 | 1.6 |
| cisplatin | 2.1 ± 0.6 | 26.9 ± 4.3 | 12.8 | |||
| oxaliplatin | 3.1 ± 0.3 | 53.4 ± 0.8 | 17.2 | |||
| Compounds | HEK293 IC50(µM) ± S.D. | SI A375 |
|---|---|---|
| 1 | 3.9 ± 3.1 | 6.5 |
| 2 | 1.3 ± 0.8 | 41.9 |
| 3 | 1.1 ± 0.2 | 47.8 |
| cisplatin | 21.6 ± 3.5 | 16.6 |
| IC50(µM) ± S.D. | ||
|---|---|---|
| Compounds | HCT-15 | A375 |
| 1 | 14.9 ± 2.3 | 18.6 ± 3.9 |
| 2 | 6.50 ± 0.80 | 7.6 ± 1.7 |
| 3 | 16.0 ± 2.8 | 11.3 ± 2.5 |
| cisplatin | 54.75 ± 1.26 | 32.8 ± 6.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramachandran, E.; Gandin, V.; Bertani, R.; Sgarbossa, P.; Natarajan, K.; Bhuvanesh, N.S.P.; Venzo, A.; Zoleo, A.; Mozzon, M.; Dolmella, A.; et al. Synthesis, Characterization and Biological Activity of Novel Cu(II) Complexes of 6-Methyl-2-Oxo-1,2-Dihydroquinoline-3-Carbaldehyde-4n-Substituted Thiosemicarbazones. Molecules 2020, 25, 1868. https://doi.org/10.3390/molecules25081868
Ramachandran E, Gandin V, Bertani R, Sgarbossa P, Natarajan K, Bhuvanesh NSP, Venzo A, Zoleo A, Mozzon M, Dolmella A, et al. Synthesis, Characterization and Biological Activity of Novel Cu(II) Complexes of 6-Methyl-2-Oxo-1,2-Dihydroquinoline-3-Carbaldehyde-4n-Substituted Thiosemicarbazones. Molecules. 2020; 25(8):1868. https://doi.org/10.3390/molecules25081868
Chicago/Turabian StyleRamachandran, Eswaran, Valentina Gandin, Roberta Bertani, Paolo Sgarbossa, Karuppannan Natarajan, Nattamai S. P. Bhuvanesh, Alfonso Venzo, Alfonso Zoleo, Mirto Mozzon, Alessandro Dolmella, and et al. 2020. "Synthesis, Characterization and Biological Activity of Novel Cu(II) Complexes of 6-Methyl-2-Oxo-1,2-Dihydroquinoline-3-Carbaldehyde-4n-Substituted Thiosemicarbazones" Molecules 25, no. 8: 1868. https://doi.org/10.3390/molecules25081868
APA StyleRamachandran, E., Gandin, V., Bertani, R., Sgarbossa, P., Natarajan, K., Bhuvanesh, N. S. P., Venzo, A., Zoleo, A., Mozzon, M., Dolmella, A., Albinati, A., Castellano, C., Reis Conceição, N., C. Guedes da Silva, M. F., & Marzano, C. (2020). Synthesis, Characterization and Biological Activity of Novel Cu(II) Complexes of 6-Methyl-2-Oxo-1,2-Dihydroquinoline-3-Carbaldehyde-4n-Substituted Thiosemicarbazones. Molecules, 25(8), 1868. https://doi.org/10.3390/molecules25081868












