N-Pyrazinoyl Substituted Amino Acids as Potential Antimycobacterial Agents—the Synthesis and Biological Evaluation of Enantiomers
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Evaluation
2.2.1. Antimycobacterial Activity Screening
pH Dependence
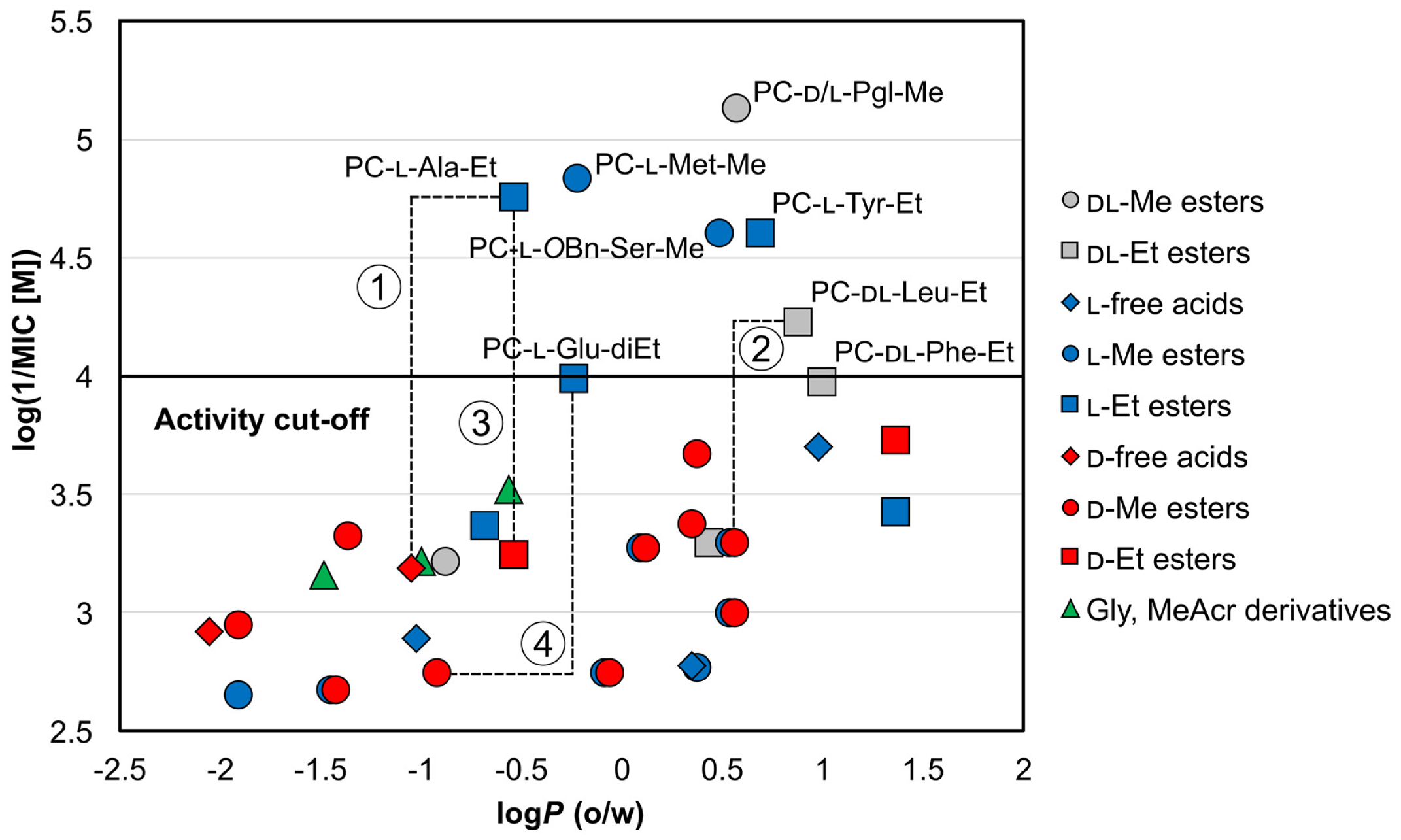
Lipophilicity and Sidechain Effects
Stereochemical Aspects of Activity
2.2.2. Antibacterial Activity Screening
2.2.3. Antifungal Activity Screening
2.2.4. Cytotoxicity Screening Results
3. Materials and Methods
3.1. Chemistry
3.1.1. General Procedure of Coupling Reaction
3.1.2. General Procedure for Esterification of Amino Acids
3.1.3. Deprotection of O-Tert-Butyl-Protected Hydroxy Amino Acids
3.1.4. General Procedure for Hydrolysis of Esters
3.1.5. Flash Chromatography
3.1.6. HPLC Analysis and Optical Rotation Measurements
3.1.7. NMR, IR Analysis, and Melting Point Measurements
3.1.8. LC-MS and GC-MS Analysis
3.2. Analytical Results
3.3. Biological Evaluation
3.3.1. Antimycobacterial Activity Screening Against Mtb H37Ra, M. smegmatis, M. aurum
3.3.2. Antibacterial and Antifungal Activity Screening
3.3.3. Cytotoxicity Screening
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- World Health Organization. Global Tuberculosis Report 2019; CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Switzerland, 17 October 2019. [Google Scholar]
- von Reyn, C.F.; Waddell, R.D.; Eaton, T.; Arbeit, R.D.; Maslow, J.N.; Barber, T.W.; Brindle, R.J.; Gilks, C.F.; Lumio, J.; Lahdevirta, J.; et al. Isolation of Mycobacterium Avium Complex from Water in the United States, Finland, Zaire, and Kenya. J. Clin. Microbiol. 1993, 31, 3227–3230. [Google Scholar] [CrossRef] [PubMed]
- Tortoli, E. Impact of Genotypic Studies on Mycobacterial Taxonomy: The New Mycobacteria of the 1990s. Clin. Microbiol. Rev. 2003, 16, 319–354. [Google Scholar] [CrossRef]
- Johnson, M.M.; Odell, J.A. Nontuberculous Mycobacterial Pulmonary Infections. J. Thorac. Dis. 2014, 6, 210–220. [Google Scholar] [CrossRef]
- Yeager, R.L.; Munroe, W.G.; Dessau, F.I. Pyrazinamide (Aldinamide) in the Treatment of Pulmonary Tuberculosis. Am. Rev. Tuberc. 1952, 65, 523–546. [Google Scholar]
- Zhang, Y.; Shi, W.; Zhang, W.; Mitchison, D. Mechanisms of Pyrazinamide Action and Resistance. Microbiol. Spectr. 2014, 2, 479–491. [Google Scholar] [CrossRef]
- Petrella, S.; Gelus-Ziental, N.; Maudry, A.; Laurans, C.; Boudjelloul, R.; Sougakoff, W. Crystal Structure of the Pyrazinamidase of Mycobacterium Tuberculosis: Insights into Natural and Acquired Resistance to Pyrazinamide. PLOS ONE 2011, 6, e15785. [Google Scholar] [CrossRef]
- Zitko, J.; Dolezal, M. Old Drugs and New Targets as an Outlook for the Treatment of Tuberculosis. Curr. Med. Chem. 2018, 25, 5142–5167. [Google Scholar] [CrossRef]
- Zimhony, O.; Vilcheze, C.; Arai, M.; Welch, J.T.; Jacobs, W.R., Jr. Pyrazinoic Acid and Its N-Propyl Ester Inhibit Fatty Acid Synthase Type I in Replicating Tubercle Bacilli. Antimicrob. Agents Chemother. 2007, 51, 752–754. [Google Scholar] [CrossRef]
- Zimhony, O.; Cox, J.S.; Welch, J.T.; Vilcheze, C.; Jacobs, W.R., Jr. Pyrazinamide Inhibits the Eukaryotic-Like Fatty Acid Synthetase I (FASI) of Mycobacterium Tuberculosis. Nat. Med. 2000, 6, 1043–1047. [Google Scholar] [CrossRef]
- Shi, W.; Zhang, X.; Jiang, X.; Yuan, H.; Lee, J.S.; Barry, C.E., 3rd; Wang, H.; Zhang, W.; Zhang, Y. Pyrazinamide Inhibits Trans-Translation in Mycobacterium Tuberculosis. Science 2011, 333, 1630–1632. [Google Scholar] [CrossRef]
- Sun, Q.; Li, X.; Perez, L.M.; Shi, W.; Zhang, Y.; Sacchettini, J.C. The Molecular Basis of Pyrazinamide Activity on Mycobacterium Tuberculosis Pand. Nat. Commun. 2020, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Shibayama, K.; Rimbara, E.; Mori, S. Biochemical Characterization of Quinolinic Acid Phosphoribosyltransferase from Mycobacterium Tuberculosis H37rv and Inhibition of Its Activity by Pyrazinamide. PLOS ONE 2014, 9, e100062. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Cui, P.; Shi, W.; Li, Q.; Zhang, W.; Li, M.; Zhang, Y. Pyrazinoic Acid Inhibits the Bifunctional Enzyme (Rv2783) in Mycobacterium Tuberculosis by Competing with Tmrna. Pathogens 2019, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, J.; Shi, W.; Cui, P.; Zhang, J.; Cho, S.; Zhang, W.; Zhang, Y. Mutation in Clpc1 Encoding an Atp-Dependent Atpase Involved in Protein Degradation Is Associated with Pyrazinamide Resistance in Mycobacterium Tuberculosis. Emerg. Microbes Infect. 2017, 6, e8. [Google Scholar] [CrossRef]
- Sheen, P.; Requena, D.; Gushiken, E.; Gilman, R.H.; Antiparra, R.; Lucero, B.; Lizarraga, P.; Cieza, B.; Roncal, E.; Grandjean, L.; et al. A Multiple Genome Analysis of Mycobacterium Tuberculosis Reveals Specific Novel Genes and Mutations Associated with Pyrazinamide Resistance. BMC Genomics 2017, 18, 769. [Google Scholar] [CrossRef] [PubMed]
- Njire, M.; Wang, N.; Wang, B.; Tan, Y.; Cai, X.; Liu, Y.; Mugweru, J.; Guo, J.; Hameed, H.M.A.; Tan, S.; et al. Pyrazinoic Acid Inhibits a Bifunctional Enzyme in Mycobacterium Tuberculosis. Antimicrob Agents Chemother. 2017, 61, e00070–e00117. [Google Scholar] [CrossRef]
- Gopal, P.; Gruber, G.; Dartois, V.; Dick, T. Pharmacological and Molecular Mechanisms Behind the Sterilizing Activity of Pyrazinamide. Trends Pharmacol. Sci. 2019, 40, 930–940. [Google Scholar] [CrossRef]
- Via, L.E.; Savic, R.; Weiner, D.M.; Zimmerman, M.D.; Prideaux, B.; Irwin, S.M.; Lyon, E.; O’Brien, P.; Gopal, P.; Eum, S.; et al. Host-Mediated Bioactivation of Pyrazinamide: Implications for Efficacy, Resistance, and Therapeutic Alternatives. ACS Infect. Dis. 2015, 1, 203–214. [Google Scholar] [CrossRef]
- Correa, M.F.; Fernandes, J.P. Pyrazinamide and Pyrazinoic Acid Derivatives Directed to Mycobacterial Enzymes against Tuberculosis. Curr. Protein. Pept. Sci. 2016, 17, 213–219. [Google Scholar] [CrossRef]
- Cynamon, M.H.; Gimi, R.; Gyenes, F.; Sharpe, C.A.; Bergmann, K.E.; Han, H.J.; Gregor, L.B.; Rapolu, R.; Luciano, G.; Welch, J.T. Pyrazinoic Acid Esters with Broad Spectrum in Vitro Antimycobacterial Activity. J. Med. Chem. 1995, 38, 3902–3907. [Google Scholar] [CrossRef]
- Cynamon, M.H.; Klemens, S.P.; Chou, T.S.; Gimi, R.H.; Welch, J.T. Antimycobacterial Activity of a Series of Pyrazinoic Acid Esters. J. Med. Chem. 1992, 35, 1212–1215. [Google Scholar] [CrossRef]
- Bergmann, K.E.; Cynamon, M.H.; Welch, J.T. Quantitative Structure-Activity Relationships for the in Vitro Antimycobacterial Activity of Pyrazinoic Acid Esters. J. Med. Chem. 1996, 39, 3394–3400. [Google Scholar] [CrossRef] [PubMed]
- Simoes, M.F.; Valente, E.; Gomez, M.J.; Anes, E.; Constantino, L. Lipophilic Pyrazinoic Acid Amide and Ester Prodrugs Stability, Activation and Activity against M. Tuberculosis. Eur. J. Pharm. Sci. 2009, 37, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Semelkova, L.; Jandourek, O.; Konecna, K.; Paterova, P.; Navratilova, L.; Trejtnar, F.; Kubicek, V.; Kunes, J.; Dolezal, M.; Zitko, J. 3-Substituted N-Benzylpyrazine-2-Carboxamide Derivatives: Synthesis, Antimycobacterial and Antibacterial Evaluation. Molecules 2017, 22, 495. [Google Scholar] [CrossRef] [PubMed]
- Zitko, J.; Jand’ourek, O.; Paterova, P.; Navratilova, L.; Kunes, J.; Vinsova, J.; Dolezal, M. Design, Synthesis and Antimycobacterial Activity of Hybrid Molecules Combining Pyrazinamide with a 4-Phenylthiazol-2-Amine Scaffold. Medchemcomm 2018, 9, 685–696. [Google Scholar] [CrossRef]
- Vale, N.; Ferreira, A.; Matos, J.; Fresco, P.; Gouveia, M.J. Amino Acids in the Development of Prodrugs. Molecules 2018, 23, 2318. [Google Scholar] [CrossRef]
- Ibrahim, M.A.; Panda, S.S.; Birs, A.S.; Serrano, J.C.; Gonzalez, C.F.; Alamry, K.A.; Katritzky, A.R. Synthesis and Antibacterial Evaluation of Amino Acid-Antibiotic Conjugates. Bioorg. Med. Chem. Lett. 2014, 24, 1856–1861. [Google Scholar] [CrossRef]
- Pochopin, N.L.; Charman, W.N.; Stella, V.J. Amino-Acid Derivatives of Dapsone as Water-Soluble Prodrugs. Int. J. Pharm. 1995, 121, 157–167. [Google Scholar] [CrossRef]
- Kushner, S.; Dalalian, H.; Sanjurjo, J.L.; Bach, F.L.; Safir, S.R.; Smith, V.K.; Williams, J.H. Experimental Chemotherapy of Tuberculosis. Ii. The Synthesis of Pyrazinamides and Related Compounds1. J. Am. Chem. Soc. 1952, 74, 3617–3621. [Google Scholar] [CrossRef]
- Badie, M.F.; Azab, M.S. Synthesis and Antimicrobial Activity of Some Pyrazine- and Disubstituted Pyrazine-Amino Acid Derivatives. Alex. J. Pharm. Sci. 1991, 5, 176–180. [Google Scholar]
- Pinheiro, A.C.; Kaiser, C.R.; Lourenco, M.C.S.; de Souzaa, M.V.N.; Wardell, S.M.S.V.; Wardell, J.L. Synthesis and in Vitro Activity Towards Mycobacterium Tuberculosis of l-Serinyl Ester and Amino Derivatives of Pyrazinoic Acid. Chemin- 2007, 2007, 180–184. [Google Scholar] [CrossRef]
- Panda, S.S.; Girgis, A.S.; Mishra, B.B.; Elagawany, M.; Devarapalli, V.; Littlefield, W.F.; Samir, A.; Fayad, W.; Fawzy, N.G.; Srour, A.M.; et al. Synthesis, Computational Studies, Antimycobacterial and Antibacterial Properties of Pyrazinoic Acid-Isoniazid Hybrid Conjugates. RSC Advances 2019, 9, 20450–20462. [Google Scholar] [CrossRef]
- Makino, E.; Iwasaki, N.; Yagi, N.; Ohashi, T.; Kato, H.; Ito, Y.; Azuma, H. Studies on Antiallergic Agents. I. Synthesis and Antiallergic Activity of Novel Pyrazine Derivatives. Chem. Pharm. Bull. (Tokyo) 1990, 38, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Himaja, M.; Venkataramana, M.; Shaifali, M.; Kilaru, J.P.; Ranjitha, A.; Saisaraswathi, V.; Asif, K. Ultrasound-Mediated Synthesis Pyrazine-2-Carboxylamino Acids and Dipeptides as Potent Insecticidal and Anthelmintic Agents. Int. J. Res. Ayurveda Pharm. 2010, 1, 180–185. [Google Scholar]
- Moreira, W.; Santhanakrishnan, S.; Ngan, G.J.Y.; Low, C.B.; Sangthongpitag, K.; Poulsen, A.; Dymock, B.W.; Dick, T. Towards Selective Mycobacterial Clpp1p2 Inhibitors with Reduced Activity against the Human Proteasome. Antimicrob. Agents Chemother. 2017, 61, e02307–e02316. [Google Scholar] [CrossRef]
- Larsen, E.M.; Johnson, R.J. Microbial Esterases and Ester Prodrugs: An Unlikely Marriage for Combating Antibiotic Resistance. Drug Dev. Res. 2019, 80, 33–47. [Google Scholar] [CrossRef]
- Li, J.; Sha, Y. A Convenient Synthesis of Amino Acid Methyl Esters. Molecules 2008, 13, 1111–1119. [Google Scholar] [CrossRef]
- Amblard, M.; Fehrentz, J.A.; Martinez, J.; Subra, G. Methods and Protocols of Modern Solid Phase Peptide Synthesis. Mol. Biotechnol. 2006, 33, 239–254. [Google Scholar] [CrossRef]
- Hill, T.A.; Lohman, R.J.; Hoang, H.N.; Nielsen, D.S.; Scully, C.C.; Kok, W.M.; Liu, L.; Lucke, A.J.; Stoermer, M.J.; Schroeder, C.I.; et al. Cyclic Penta- and Hexaleucine Peptides without N-Methylation Are Orally Absorbed. ACS Med. Chem. Lett. 2014, 5, 1148–1151. [Google Scholar] [CrossRef]
- Yamada, S.; Hongo, C.; Yoshioka, R.; Chibata, I. Method for the Racemization of Optically-Active Amino-Acids. J. Org. Chem. 1983, 48, 843–846. [Google Scholar] [CrossRef]
- Paul, R.; Anderson, G.W. N,N‘-Carbonyldiimidazole, a New Peptide Forming Reagent. J. Am. Chem. Soc. 1960, 82, 4596–4600. [Google Scholar] [CrossRef]
- E-Abadelah, M.M.; Sabri, S.S.; Jarrar, A.A.; Zarga, M.H.A. Chiroptical Properties of N-(2-Pyrazinoyl)-A-Amino-Esters, -Aziridines, and Related Compounds. J. Chem. Soc. Perkin Trans. 1 1979, 2881–2885. [Google Scholar] [CrossRef]
- Popovic, S.; Bieraugel, H.; Detz, R.J.; Kluwer, A.M.; Koole, J.A.; Streefkerk, D.E.; Hiemstra, H.; van Maarseveen, J.H. Epimerization-Free C-Terminal Peptide Activation. Chemistry 2013, 19, 16934–16937. [Google Scholar] [CrossRef] [PubMed]
- Franzblau, S.G.; Witzig, R.S.; McLaughlin, J.C.; Torres, P.; Madico, G.; Hernandez, A.; Degnan, M.T.; Cook, M.B.; Quenzer, V.K.; Ferguson, R.M.; et al. Rapid, Low-Technology Mic Determination with Clinical Mycobacterium Tuberculosis Isolates by Using the Microplate Alamar Blue Assay. J. Clin. Microbiol 1998, 36, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Namouchi, A.; Cimino, M.; Favre-Rochex, S.; Charles, P.; Gicquel, B. Phenotypic and Genomic Comparison of Mycobacterium Aurum and Surrogate Model Species to Mycobacterium Tuberculosis: Implications for Drug Discovery. BMC Genomics 2017, 18, 530. [Google Scholar] [CrossRef][Green Version]
- Chaturvedi, V.; Dwivedi, N.; Tripathi, R.P.; Sinha, S. Evaluation of Mycobacterium Smegmatis as a Possible Surrogate Screen for Selecting Molecules Active against Multi-Drug Resistant Mycobacterium Tuberculosis. J. Gen. Appl. Microbiol. 2007, 53, 333–337. [Google Scholar] [CrossRef]
- Heinrichs, M.T.; May, R.J.; Heider, F.; Reimers, T.; SK, B.S.; Peloquin, C.A.; Derendorf, H. Mycobacterium Tuberculosis Strains H37ra and H37rv Have Equivalent Minimum Inhibitory Concentrations to Most Antituberculosis Drugs. Int. J. Mycobacteriol. 2018, 7, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Toida, I.; Watanabe, N.; Ura, T. In Vitro Antimycobacterial Activities of Pyrazinamide Analogs. Antimicrob. Agents Chemother. 1995, 39, 2088–2091. [Google Scholar] [CrossRef][Green Version]
- Vandal, O.H.; Nathan, C.F.; Ehrt, S. Acid Resistance in Mycobacterium Tuberculosis. J. Bacteriol. 2009, 191, 4714–4721. [Google Scholar] [CrossRef]
- den Hertog, A.L.; Menting, S.; Pfeltz, R.; Warns, M.; Siddiqi, S.H.; Anthony, R.M. Pyrazinamide Is Active against Mycobacterium Tuberculosis Cultures at Neutral Ph and Low Temperature. Antimicrob. Agents Chemother. 2016, 60, 4956–4960. [Google Scholar] [CrossRef][Green Version]
- Bansa-Mutalik, R.; Nikaido, H. Mycobacterial Outer Membrane Is a Lipid Bilayer and the Inner Membrane Is Unusually Rich in Diacyl Phosphatidylinositol Dimannosides. Proc. Natl. Acad. Sci. USA 2014, 111, 4958–4963. [Google Scholar] [CrossRef]
- Chen, H.; Nyantakyi, S.A.; Li, M.; Gopal, P.; Aziz, D.B.; Yang, T.; Moreira, W.; Gengenbacher, M.; Dick, T.; Go, M.L. The Mycobacterial Membrane: A Novel Target Space for Anti-Tubercular Drugs. Front. Microbiol. 2018, 9, 1627. [Google Scholar] [CrossRef] [PubMed]
- Molecular Operating Environment (MOE), Chemical Computing Group ULC, 1010 Sherbrooke St. West, Suite #910: Montreal, QC, Canada, 2019.
- Palos, I.; Luna-Herrera, J.; Lara-Ramirez, E.E.; Loera-Piedra, A.; Fernandez-Ramirez, E.; Aguilera-Arreola, M.G.; Paz-Gonzalez, A.D.; Monge, A.; Wan, B.; Franzblau, S.; et al. Anti-Mycobacterium Tuberculosis Activity of Esters of Quinoxaline 1,4-Di-N-Oxide. Molecules 2018, 23, 1453. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Tsujimoto, M. Cloning and Expression of Dipeptidase from Acinetobacter Calcoaceticus Atcc 23055. J. Biochem. 1995, 118, 555–561. [Google Scholar] [CrossRef]
- Reichau, S.; Blackmore, N.J.; Jiao, W.; Parker, E.J. Probing the Sophisticated Synergistic Allosteric Regulation of Aromatic Amino Acid Biosynthesis in Mycobacterium Tuberculosis Using -Amino Acids. PLOS ONE 2016, 11, e0152723. [Google Scholar] [CrossRef]
- European Committee for Antimicrobial Susceptibility Testing (Eucast) of the European Society for Clinical Microbiology and Infectious Diseases (Escmid). Eucast Discussion Document E. Dis 5.1: Determination of Minimum Inhibitory Concentrations (Mics) of Antibacterial Agents by Broth Dilution. Clin. Microbiol. Infec. 2003, 9, 1–7. Available online: http://www.eucast.org/documents/publications_in_journals/ (accessed on 11 December 2019).
- Eucast Definitive Document E.Def 7.3.1. Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Yeasts. 2017. Available online: http://www.eucast.org/astoffungi/methodsinantifungalsusceptibilitytesting/susceptibility_testing_of_yeasts/ (accessed on 11 December 2019).
- Eucast Definitive Document E.Def 9.3.1. Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Conidia Forming Moulds. 2017. Available online: http://www.eucast.org/astoffungi/methodsinantifungalsusceptibilitytesting/susceptibility_testing_of_moulds/ (accessed on 11 December 2019).
- Bagla, V.P.; McGaw, L.J.; Elgorashi, E.E.; Eloff, J.N. Antimicrobial Activity, Toxicity and Selectivity Index of Two Biflavonoids and a Flavone Isolated from Podocarpus Henkelii (Podocarpaceae) Leaves. BMC Complement. Altern. Med. 2014, 14, 383. [Google Scholar] [CrossRef]
- Shih, T.Y.; Pai, C.Y.; Yang, P.; Chang, W.L.; Wang, N.C.; Hu, O.Y. A Novel Mechanism Underlies the Hepatotoxicity of Pyrazinamide. Antimicrob. Agents Chemother. 2013, 57, 1685–1690. [Google Scholar] [CrossRef]
- Kočevar, M.; Polanc, S.; Verček, B.; Tišler, M. Syntheses of Some N-(Pyrazinecarbonyl) Amino Acids and Peptides. Recl. Trav. Chim. Pays.-Bas 1988, 107, 366–369. [Google Scholar] [CrossRef]
- Kakemi, K.; Arta, T.; Kitazawa, S.; Kiyotaki, T. Studies on the Synthesis of Pyrazinoic Acid Derivatives. Ii. Derivatives of 3-Aminopyrazinoic Acid. Yakugaku Zasshi 1961, 81, 1650–1653. [Google Scholar] [CrossRef]
- Naredla, R.R.; Dash, B.P.; Klumpp, D.A. Preparation of Pyrazine Carboxamides: A Reaction Involving N-Heterocyclic Carbene (Nhc) Intermediates. Org. Lett. 2013, 15, 4806–4809. [Google Scholar] [CrossRef] [PubMed]
- Cynamon, M.H.; Speirs, R.J.; Welch, J.T. In Vitro Antimycobacterial Activity of 5-Chloropyrazinamide. Antimicrob. Agents Chemother. 1998, 42, 462–463. [Google Scholar] [PubMed]
- Fieweger, R.A.; Wilburn, K.M.; VanderVen, B.C. Comparing the Metabolic Capabilities of Bacteria in the Mycobacterium Tuberculosis Complex. Microorganisms 2019, 7, 177. [Google Scholar] [CrossRef] [PubMed]
- Agapova, A.; Serafini, A.; Petridis, M.; Hunt, D.M.; Garza-Garcia, A.; Sohaskey, C.D.; de Carvalho, L.P.S. Flexible Nitrogen Utilisation by the Metabolic Generalist Pathogen Mycobacterium Tuberculosis. eLife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Naffin-Olivos, J.L.; Daab, A.; White, A.; Goldfarb, N.E.; Milne, A.C.; Liu, D.; Baikovitz, J.; Dunn, B.M.; Rengarajan, J.; Petsko, G.A.; et al. Structure Determination of Mycobacterium Tuberculosis Serine Protease Hip1 (Rv2224c). Biochemistry 2017, 56, 2304–2314. [Google Scholar] [CrossRef] [PubMed]
- Akopian, T.; Kandror, O.; Tsu, C.; Lai, J.H.; Wu, W.; Liu, Y.; Zhao, P.; Park, A.; Wolf, L.; Dick, L.R.; et al. Cleavage Specificity of Mycobacterium Tuberculosis Clpp1p2 Protease and Identification of Novel Peptide Substrates and Boronate Inhibitors with Anti-Bacterial Activity. J. Biol. Chem. 2015, 290, 11008–11020. [Google Scholar] [CrossRef]
- Soni, D.K.; Dubey, S.K.; Bhatnagar, R. Atp-Binding Cassette (Abc) Import Systems of Mycobacterium Tuberculosis: Target for Drug and Vaccine Development. Emerg. Microbes Infect. 2020, 9, 207–220. [Google Scholar] [CrossRef]
- Dasgupta, A.; Sureka, K.; Mitra, D.; Saha, B.; Sanyal, S.; Das, A.K.; Chakrabarti, P.; Jackson, M.; Gicquel, B.; Kundu, M.; et al. An Oligopeptide Transporter of Mycobacterium Tuberculosis Regulates Cytokine Release and Apoptosis of Infected Macrophages. PLOS ONE 2010, 5, e12225. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the corresponding authors. |
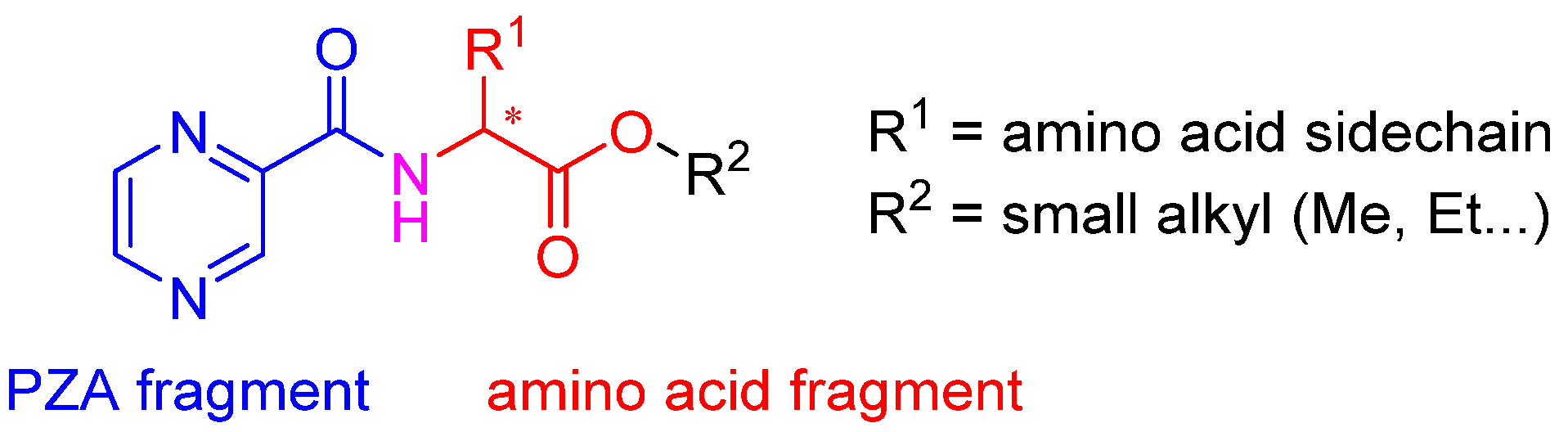

| Code | Structure |
|---|---|
| PC-d-Ala | 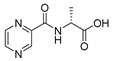 |
| PC-l-Ala | 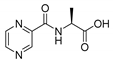 |
| PC-dl-Ala-Me | 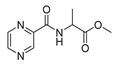 |
| PC-d-Ala-Et | 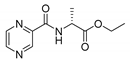 |
| PC-l-Ala-Et | 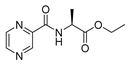 |
| PC-d-Asp-diMe | 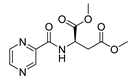 |
| PC-l-Asp-diEt | 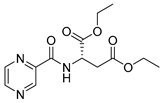 |
| PC-l-SBn-Cys | 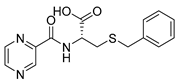 |
| PC-d-Glu-diMe | 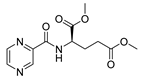 |
| PC-l-Glu-diEt |  |
| PC-Gly | 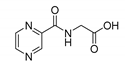 |
| PC-Gly-Et |  |
| PC-d-Ile-Me | 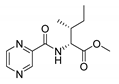 |
| PC-l-Ile-Me |  |
| PC-dl-Leu-Et | 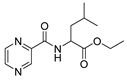 |
| PC-d-Leu-Me | 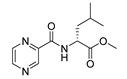 |
| PC-l-Leu-Me | 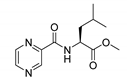 |
| PC-MeAcr |  |
| PC-l-Met-Me | 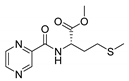 |
| PC-d/l-Pgl-Me |  |
| PC-dl-Phe-Et |  |
| PC-d-Ser | 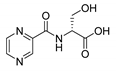 |
| PC-d-Ser-Me | 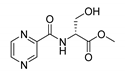 |
| PC-l-Ser-Me |  |
| PC-l-OBn-Ser | 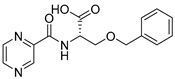 |
| PC-l-OBn-Ser-Me | 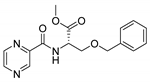 |
| PC-d-OtBu-Ser-Me | 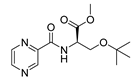 |
| PC-l-OtBu-Ser-Me | 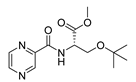 |
| PC-d-Thr-Me |  |
| PC-l-Thr-Me | 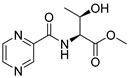 |
| PC-d-OtBu-Thr-Me |  |
| PC-l-OtBu-Thr-Me |  |
| PC-d-Trp-Et | 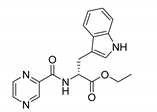 |
| PC-l-Trp-Et |  |
| PC-d-Tyr-Me | 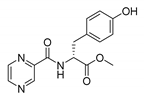 |
| PC-l-Tyr-Et | 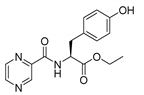 |
| PC-dl-Val-Et | 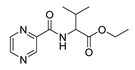 |
| PC-d-Val-Me |  |
| PC-l-Val-Me |  |
| Code | Mtb H37Ra | M. smegmatis | M. aurum |
|---|---|---|---|
| PC-d-Ala | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-l-Ala | ≥ 250 | ≥ 250 | ≥ 250 |
| PC-dl-Ala-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-d-Ala-Et | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-l-Ala-Et | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-d-Asp-diMe | ≥ 125 | ≥ 125 | ≥ 125 |
| PC-l-Asp-diEt | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-l-SBn-Cys | ≥ 250 | ≥ 250 | ≥ 250 |
| PC-d-Glu-diMe | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-l-Glu-diEt | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-Gly | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-Gly-Et | ≥ 500 | 250 | ≥ 500 |
| PC-d-Ile-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-l-Ile-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-dl-Leu-Et | 250 | 250 | 250 |
| PC-d-Leu-Me | ≥ 500 | 250 | ≥ 500 |
| PC-l-Leu-Me | 250 | 250 | ≥ 500 |
| PC-MeAcr | 15.625 | 31.25 | 62.5 |
| PC-l-Met-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-d/l-Pgl-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-dl-Phe-Et | 250 | 250 | ≥ 500 |
| PC-d-Ser | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-d-Ser-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-l-Ser-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-l-OBn-Ser | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-l-OBn-Ser-Me | 250 | ≥ 500 | ≥ 500 |
| PC-d-OtBu-Ser-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-l-OtBu-Ser-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-d-Thr-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-l-Thr-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-d-OtBu-Thr-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-l-OtBu-Thr-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-d-Trp-Et | 125 | 250 | 250 |
| PC-l-Trp-Et | 125 | 250 | 125 |
| PC-d-Tyr-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-l-Tyr-Et | 500 | ≥ 500 | ≥ 500 |
| PC-dl-Val-Et | ≥ 500 | 250 | ≥ 500 |
| PC-d-Val-Me | ≥ 500 | ≥ 500 | ≥ 500 |
| PC-l-Val-Me | ≥ 500 | 250 | ≥ 500 |
| PZA | ≥ 500 | ≥ 500 | ≥ 500 |
| INH | 0.125–0.25 | 7.81–15.625 | 1.95–3.91 |
| RIF | 0.0039–0.0078 | 12.5–25 | 0.39–0.78 |
| CPX | 0.125–0.25 | 0.0625–0.125 | 0.0078–0.0156 |
| Code | Mtb H37Ra | Mtb H37Ra | M. smegmatis | M. aurum | logP ** |
|---|---|---|---|---|---|
| µg/mL | µM * | µg/mL | µg/mL | ||
| PC-d-Ala | ≥ 125 | ≥ 640.4 | ≥ 125 | 62.5 | −1.0155 |
| PC-l-Ala | ≥ 250 | ≥ 1280.9 | 125 | 62.5 | −1.0155 |
| PC-dl-Ala-Me | ≥ 125 | ≥ 597.5 | ≥ 125 | ≥ 125 | −0.8725 |
| PC-d-Ala-Et | ≥ 125 | ≥ 560.0 | ≥ 125 | ≥ 125 | −0.5315 |
| PC-l-Ala-Et | 3.91 | 17.5 | ≥ 125 | ≥ 125 | −0.5315 |
| PC-d-Asp-diMe | ≥ 125 | ≥ 467.7 | ≥ 125 | ≥ 125 | −1.3565 |
| PC-l-Asp-diEt | ≥ 125 | ≥ 423.3 | ≥ 125 | ≥ 125 | −0.6745 |
| PC-l-SBn-Cys | 62.5 | 196.9 | ≥ 250 | 125 | 0.9855 |
| PC-d-Glu-diMe | ≥ 500 | ≥ 1777.7 | ≥ 500 | ≥ 500 | −0.9145 |
| PC-l-Glu-diEt | 31.25 | 101.0 | ≥ 500 | ≥ 500 | −0.2325 |
| PC-Gly | ≥ 125 | ≥ 690.0 | ≥ 125 | ≥ 125 | −1.4775 |
| PC-Gly-Et | ≥ 125 | ≥ 597.5 | ≥ 125 | ≥ 125 | −0.9935 |
| PC-d-Ile-Me | 250 | 994.9 | 250 | 250 | 0.5415 |
| PC-l-Ile-Me | 250 | 994.9 | ≥ 500 | ≥ 500 | 0.5415 |
| PC-dl-Leu-Et | 15.625 | 58.9 | ≥ 125 | ≥ 125 | 0.8825 |
| PC-d-Leu-Me | ≥ 125 | ≥ 497.4 | ≥ 125 | ≥ 125 | 0.5415 |
| PC-l-Leu-Me | ≥ 125 | ≥ 497.4 | ≥ 125 | ≥ 125 | 0.5415 |
| PC-MeAcr | 62.5 | 301.7 | ≥ 125 | 62.5 | −0.5555 |
| PC-l-Met-Me | < 3.91 | < 14.5 | ≥ 500 | 62.5 | −0.2175 |
| PC-d/l-Pgl-Me | < 1.95 | < 7.3 | ≥ 250 | 31.25 | 0.5745 |
| PC-dl-Phe-Et | 31.25 | 104.4 | ≥ 125 | 62.5 | 1.0035 |
| PC-d-Ser | ≥ 250 | ≥ 1183.8 | 125 | 62.5 | −2.0505 |
| PC-d-Ser-Me | 250 | 1110.1 | ≥ 500 | 250 | −1.9075 |
| PC-l-Ser-Me | ≥ 500 | ≥ 2220.2 | ≥ 500 | ≥ 500 | −1.9075 |
| PC-l-OBn-Ser | ≥ 500 | ≥ 1659.5 | 250 | 62.5 | 0.3495 |
| PC-l-OBn-Ser-Me | 7.81 | 24.8 | ≥ 500 | 62.5 | 0.4925 |
| PC-d-OtBu-Ser-Me | ≥ 500 | ≥ 1777.4 | ≥ 500 | ≥ 500 | −0.0855 |
| PC-l-OtBu-Ser-Me | ≥ 500 | ≥ 1777.4 | 250 | 62.5 | −0.0855 |
| PC-d-Thr-Me | ≥ 500 | ≥ 2090.0 | ≥ 500 | ≥ 500 | −1.4455 |
| PC-l-Thr-Me | ≥ 500 | ≥ 2090.0 | ≥ 500 | ≥ 500 | −1.4455 |
| PC-d-OtBu-Thr-Me | 62.5 | 211.6 | ≥ 500 | ≥ 500 | 0.3765 |
| PC-l-OtBu-Thr-Me | ≥ 500 | ≥ 1693.0 | ≥ 500 | ≥ 500 | 0.3765 |
| PC-d-Trp-Et | 62.5 | 184.7 | 250 | 62.5 | 1.3695 |
| PC-l-Trp-Et | 125 | 369.4 | 250 | 62.5 | 1.3695 |
| PC-d-Tyr-Me | 125 | 414.9 | ≥ 500 | ≥ 250 | 0.3545 |
| PC-l-Tyr-Et | 7.81 | 24.8 | ≥ 250 | ≥ 250 | 0.6955 |
| PC-dl-Val-Et | ≥ 125 | ≥ 497.4 | ≥ 125 | ≥ 125 | 0.4405 |
| PC-d-Val-Me | ≥ 125 | ≥ 526.9 | ≥ 125 | ≥ 125 | 0.0995 |
| PC-l-Val-Me | ≥ 125 | ≥ 526.9 | ≥ 125 | ≥ 125 | 0.0995 |
| PZA | < 3.91 | < 31.8 | ≥ 500 | 250 | −1.4595 |
| INH | 0.125–0.25 | 0.91–1.82 | 7.81–15.625 | 3.91–7.81 | −0.7970 |
| RIF | 0.0016–0.0062 | 0.0019–0.0075 | 6.25–12.5 | 0.39–1.56 | 4.5560 |
| CPX | 0.25 | 0.75 | 0.0625–0.25 | 0.0625 | 1.0370 |
| Tested compound | Tested range (μM) | HepG2 IC50 (µM) | Mtb pH 6 MIC (µM) | SI pH 6 |
|---|---|---|---|---|
| PC-l-Ala | 1–1000 | > 250* | ≥ 1281 | ≥ 0.2 |
| PC-l-Ala-Et | 1–1000 | > 1000 | 18 | > 57.1 |
| PC-dl-Leu-Et | 1–1000 | 788.2 | 59 | 13.4 |
| PC-MeAcr | 1–500 | 704.2 ** | 302 | 1.7 |
| PC-l-Met-Me | 1–1000 | > 100 * | 15 | 6.9 |
| PC-d/l-Pgl-Me | 1–1000 | > 1000 | < 7 | > 68.5 |
| PC-l-OBn-Ser-Me | 1–1000 | 742.3 | 25 | 30.0 |
| PC-l-OtBu-Thr-Me | 1–500 | > 500 | 1693 | 0.3 |
| PC-l-Trp-Et | 1–500 | 146.6 | 125 | 1.2 |
| PC-l-Tyr-Et | 1–1000 | 177.1 | 25 | 7.2 |
| PZA | 9619.1 *** | 31.8 | 302.9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juhás, M.; Kučerová, L.; Horáček, O.; Janďourek, O.; Kubíček, V.; Konečná, K.; Kučera, R.; Bárta, P.; Janoušek, J.; Paterová, P.; et al. N-Pyrazinoyl Substituted Amino Acids as Potential Antimycobacterial Agents—the Synthesis and Biological Evaluation of Enantiomers. Molecules 2020, 25, 1518. https://doi.org/10.3390/molecules25071518
Juhás M, Kučerová L, Horáček O, Janďourek O, Kubíček V, Konečná K, Kučera R, Bárta P, Janoušek J, Paterová P, et al. N-Pyrazinoyl Substituted Amino Acids as Potential Antimycobacterial Agents—the Synthesis and Biological Evaluation of Enantiomers. Molecules. 2020; 25(7):1518. https://doi.org/10.3390/molecules25071518
Chicago/Turabian StyleJuhás, Martin, Lucie Kučerová, Ondřej Horáček, Ondřej Janďourek, Vladimír Kubíček, Klára Konečná, Radim Kučera, Pavel Bárta, Jiří Janoušek, Pavla Paterová, and et al. 2020. "N-Pyrazinoyl Substituted Amino Acids as Potential Antimycobacterial Agents—the Synthesis and Biological Evaluation of Enantiomers" Molecules 25, no. 7: 1518. https://doi.org/10.3390/molecules25071518
APA StyleJuhás, M., Kučerová, L., Horáček, O., Janďourek, O., Kubíček, V., Konečná, K., Kučera, R., Bárta, P., Janoušek, J., Paterová, P., Kuneš, J., Doležal, M., & Zitko, J. (2020). N-Pyrazinoyl Substituted Amino Acids as Potential Antimycobacterial Agents—the Synthesis and Biological Evaluation of Enantiomers. Molecules, 25(7), 1518. https://doi.org/10.3390/molecules25071518








