Identification of ε-Poly-L-lysine as an Antimicrobial Product from an Epichloë Endophyte and Isolation of Fungal ε-PL Synthetase Gene
Abstract
1. Introduction
2. Results
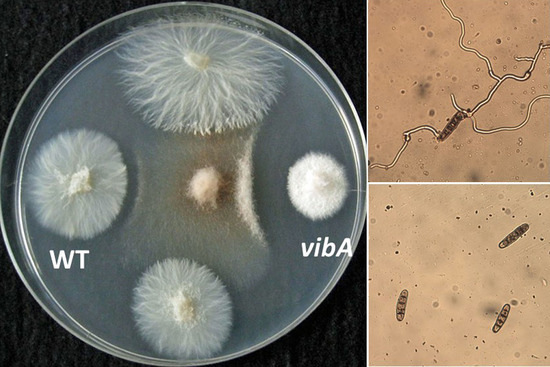
2.1. Chemical Identification of an Antifungal Substance Produced by a VibA-Overexpressed Strain of E. festucae E437
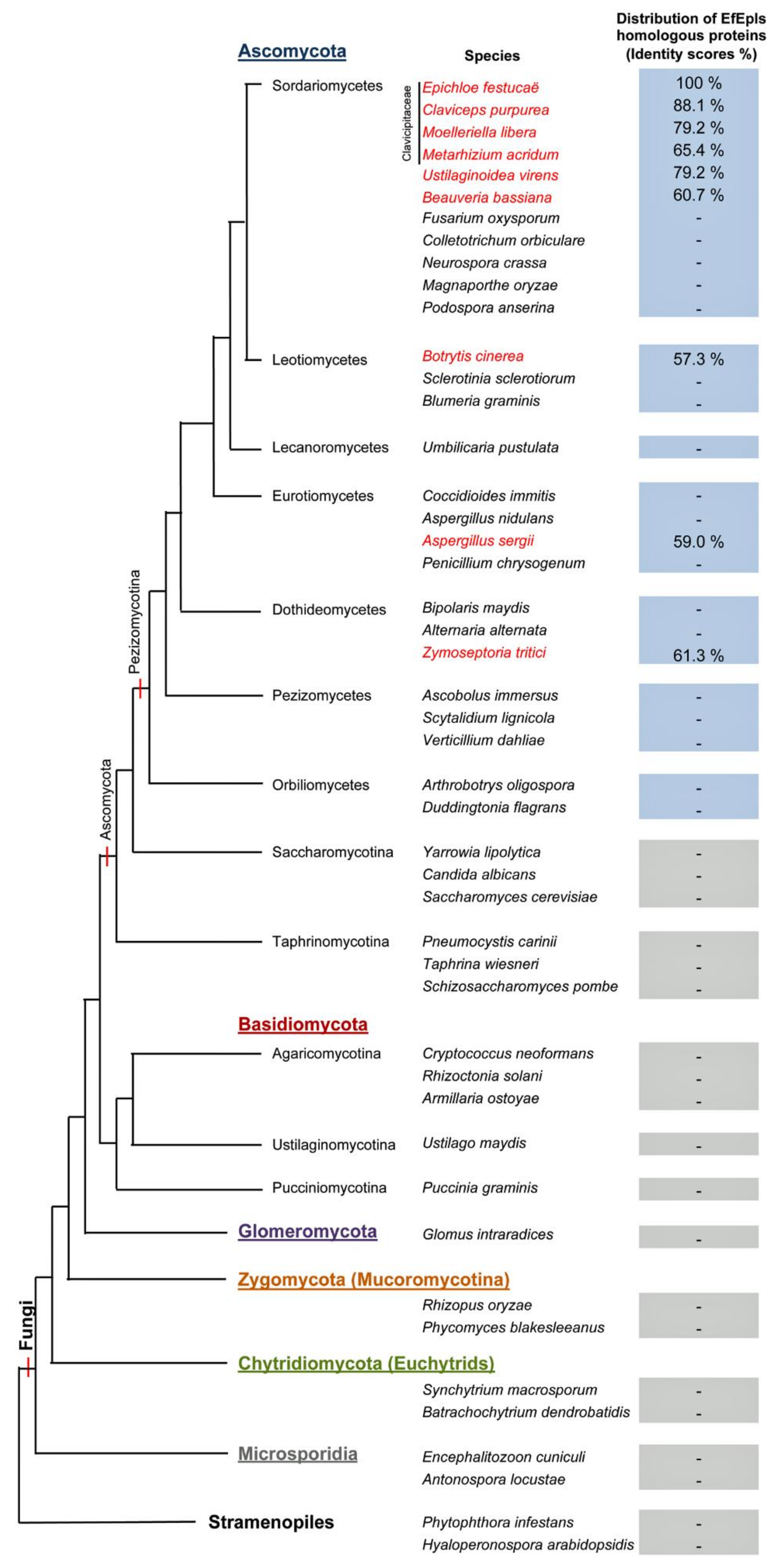
2.2. Identification of ε-PL Biosynthetic Gene Epls from E. festucae
2.3. Overexpression of Epls Gene in E. festucae Strains
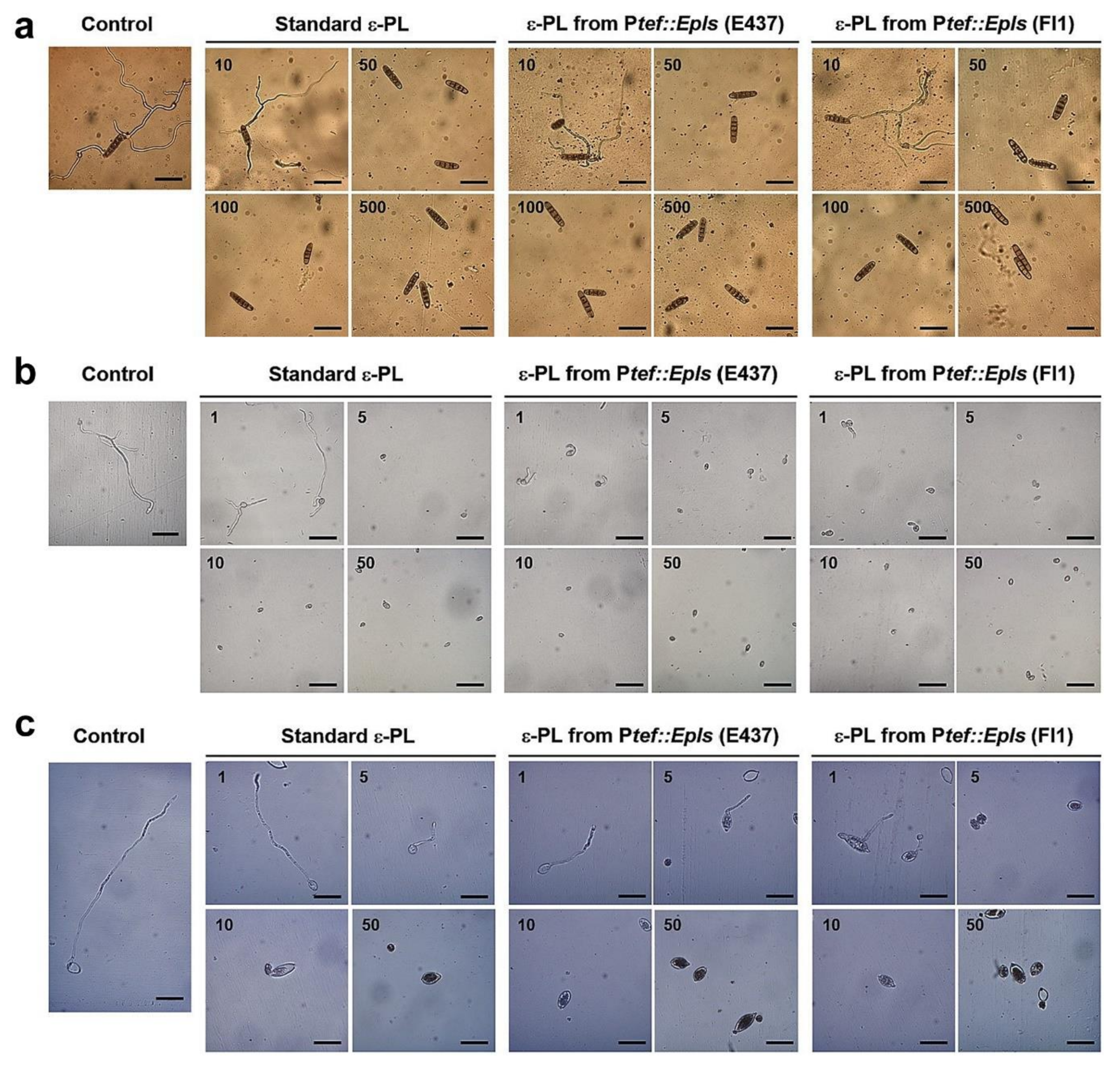
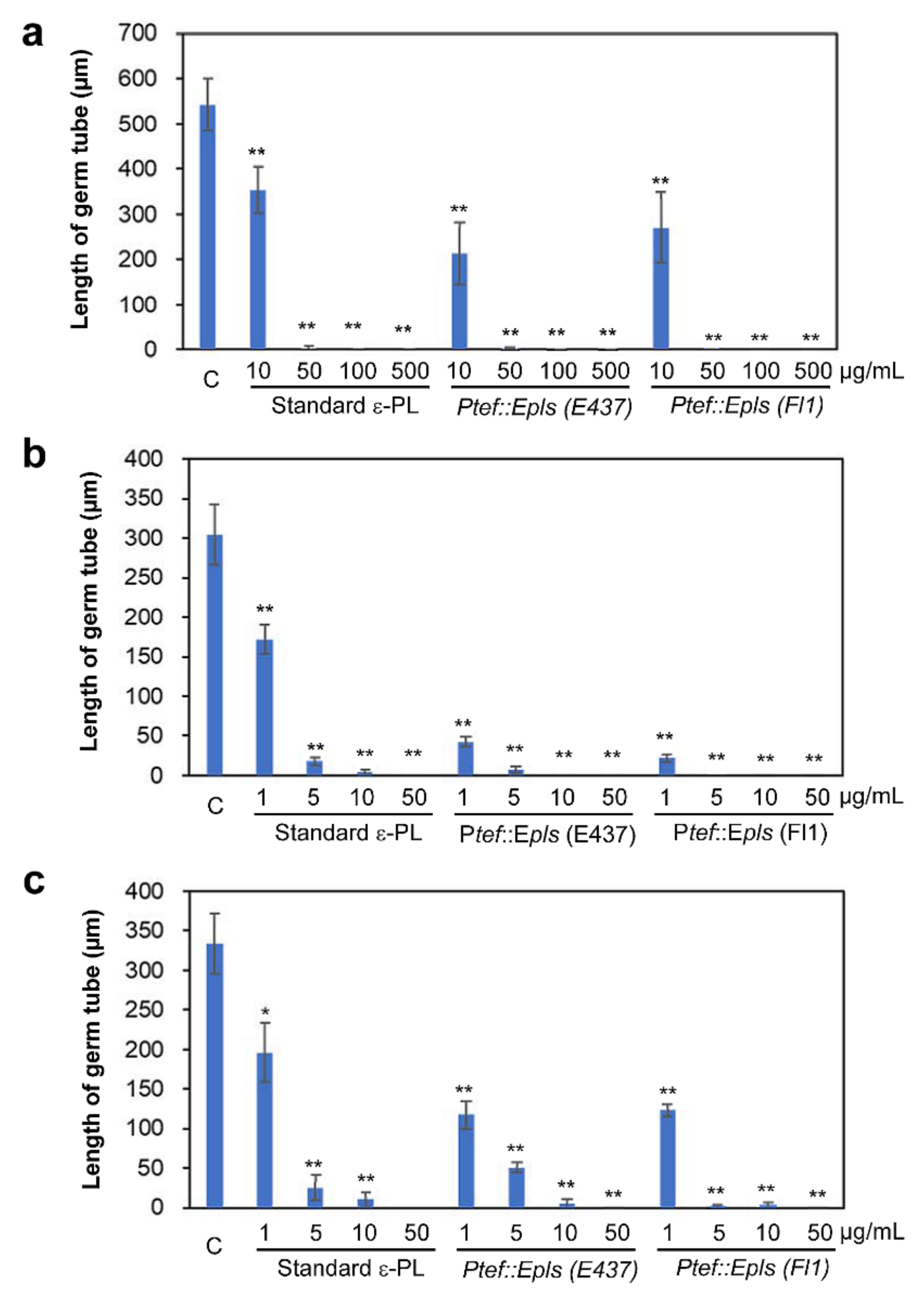
2.4. Inhibitory Activity of ε-PL against Spore Germination of Fungal and Oomycete Pathogens
2.5. Antifungal Activity of ε-PL
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Spectroscopic Analyses
4.3. Vector Construction and Transformation of E. festucae for Overexpression of Epls Gene
4.4. Purification and Structure Determination of ε-PL
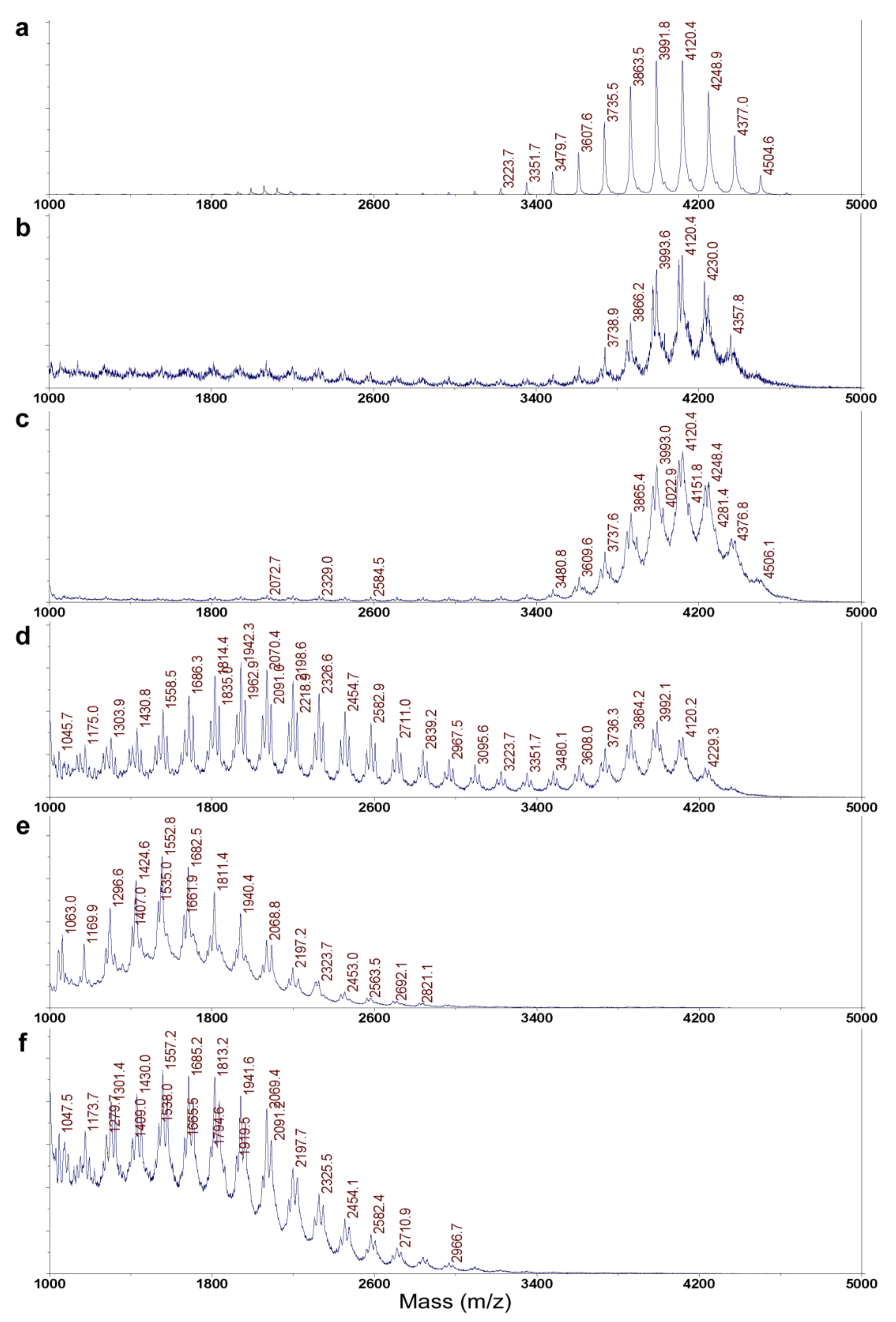
4.5. MALDI-TOF MS Analysis of ε-PL
4.6. Absolute Configuration
4.7. Spore Germination Assay
4.8. Cell Wall Staining
4.9. Antifungal Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hawsworth, D.L. The fungal dimension of biodiversity: Magnitude, significance, and conservation. Mycol. Res. 1991, 95, 641–655. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compante, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Shiba, T.; Sugawara, K. Resistance to the rice leaf bug, Trigonotylus caelestialium, is conferred by Neotyphodium endophyte infection of perennial ryegrass, Lolium perenne. Entomol. Exp. Appl. 2005, 115, 387–392. [Google Scholar] [CrossRef]
- Tanaka, A.; Tapper, B.A.; Popay, A.; Parker, E.J.; Scott, B. A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol. Microbiol. 2005, 57, 1036–1050. [Google Scholar] [CrossRef] [PubMed]
- Bonos, S.A.; Wilson, M.M.; Meyer, W.A.; Funk, C.R. Suppression of red thread in fine fescues through endophyte-mediated resistance. Appl. Turfgrass. Sci. 2005, 2, 105–112. [Google Scholar] [CrossRef]
- Clarke, B.B.; White, J.F., Jr.; Hurley, R.H.; Torres, M.S.; Sun, S.; Huff, D.R. Endophyte-mediated suppression of dollar spot disease in fine fescues. Plant Dis. 2006, 90, 994–998. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, D.P.; Belesky, D.P. Adaptations of endophyte-infected cool-season grasses to environmental stresses: Mechanisms of drought and mineral stress tolerance. Crop Sci. 2000, 40, 923–940. [Google Scholar] [CrossRef]
- Bastias, D.A.; Martínez-Ghersa, M.A.; Ballaré, C.L.; Gundel, P.E. Epichloë fungal endophytes and plant defenses: Not just alkaloids. Trends Plant Sci. 2017, 22, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L.; Florea, S.; Pan, J.; Nagabhyru, P.; Bec, S.; Calie, P.J. The epichloae: Alkaloid diversity and roles in symbiosis with grasses. Curr. Opin. Plant Biol. 2013, 16, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Yue, Q.; Miller, C.J.; White, J.F., Jr.; Rihardson, M.D. Isolation and characterization of fungal inhibitors from Epichloë festucae. J. Agric. Food Chem. 2000, 48, 4687–4692. [Google Scholar] [CrossRef]
- Seto, Y.; Takahashi, K.; Matsuura, H.; Kogami, Y.; Yada, H.; Yoshihara, T.; Nabeta, K. Novel cyclic peptide, epichlicin, from the endophytic fungus, Epichloe typhina. Biosci. Biotechnol. Biochem. 2007, 71, 1470–1475. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Song, Q.Y.; Nan, Z.B.; Gao, K.; Song, H.; Tian, P.; Zhang, X.X.; Li, C.J.; Xu, W.B.; Li, X.Z. Antifungal, phytotoxic, and cytotoxic activities of metabolites from Epichloë bromicola, a fungus obtained from Elymus tangutorum Grass. J. Agric. Food Chem. 2015, 63, 8787–8792. [Google Scholar] [CrossRef] [PubMed]
- Niones, J.T.; Takemoto, D. An isolate of Epichloë festucae, an endophytic fungus of temperate grasses, shows growth inhibitory activity to selective grass pathogens. J. Gen. Plant Pathol. 2014, 80, 337–347. [Google Scholar] [CrossRef]
- Niones, J.T.; Takemoto, D. VibA, a homologue of a transcription factor for fungal heterokaryon incompatibility, is involved in antifungal compound production in the plant symbiotic fungus Epichloë festucae. Eukaryot. Cell 2015, 14, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Q.; Glass, N.L. Identification of vib-1, a locus involved in vegetative incompatibility mediated by het-c in Neurospora crassa. Genetics 2002, 162, 89–101. [Google Scholar]
- Xiang, Q.; Glass, N.L. The control of mating type heterokaryon incompatibility by vib-1, a locus involved in het-c heterokaryon incompatibility in Neurospora crassa. Fungal Genet. Biol. 2004, 41, 1063–1076. [Google Scholar] [CrossRef]
- Hutchison, E.A.; Glass, N.L. Meiotic regulators Ndt80 and ime2 have different roles in Saccharomyces and Neurospora. Genetics 2010, 185, 1271–1282. [Google Scholar] [CrossRef]
- Marfey, P. Determination of D-amino acids. II. use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 1984, 49, 591–596. [Google Scholar] [CrossRef]
- Yamanaka, K.; Maruyama, C.; Takagi, H.; Hamano, Y. ε-poly-L-lysine dispersity is controlled by a highly unusual nonribosomal peptide synthetase. Nat. Chem. Biol. 2008, 4, 766–772. [Google Scholar] [CrossRef]
- Hamano, Y.; Kito, N.; Kita, A.; Imokawa, Y.; Yamanaka, K.; Maruyama, C.; Katano, H. ε-Poly-l-lysine peptide chain length regulated by the linkers connecting the transmembrane domains of ε-poly-l-lysine synthetase. Appl. Environ. Microbiol. 2014, 80, 4993–5000. [Google Scholar] [CrossRef]
- Challis, G.L.; Ravel, J.; Townsend, C.A. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 2000, 7, 211–224. [Google Scholar] [CrossRef]
- Green, K.A.; Eaton, C.J.; Savoian, M.S.; Scott, B. A homologue of the fungal tetraspanin Pls1 is required for Epichloë festucae expressorium formation and establishment of a mutualistic interaction with Lolium perenne. Mol. Plant Pathol. 2019, 20, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Kito, N.; Maruyama, C.; Yamanaka, K.; Imokawa, Y.; Utagawa, T.; Hamano, Y. Mutational analysis of the three tandem domains of ε-poly-L-lysine synthetase catalyzing the L-lysine polymerization reaction. J. Biosci. Bioeng. 2013, 115, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Vanden Wymelenberg, A.J.; Cullen, D.; Spear, R.N.; Schoenike, B.; Andrews, J.H. Expression of green fluorescent protein in Aureobasidium pullulans and quantification of the fungus on leaf surfaces. Biotechniques 1997, 23, 686–690. [Google Scholar] [CrossRef] [PubMed]
- Bankar, S.B.; Singhal, R.S. Panorama of poly-ε-lysine. RSC Adv. 2013, 3, 8586–8603. [Google Scholar] [CrossRef]
- El-Sersy, N.A.; Abdelwahab, A.E.; Abouelkhiir, S.S.; Abou-Zeid, D.M.; Sabry, S.A. Antibacterial and anticancer activity of ε-poly-L-lysine (ε-PL) produced by a marine Bacillus subtilis sp. J. Basic Microbiol. 2012, 52, 513–522. [Google Scholar] [CrossRef]
- Chheda, A.H.; Vernekar, M.R. Enhancement of ε-poly-L-lysine (ε-PL) production by a novel producer Bacillus cereus using metabolic precursors and glucose feeding. 3 Biotech 2015, 5, 839–846. [Google Scholar] [CrossRef]
- Szókán, G.; Almás, M.; Krizsán, K.; Khlafulla, A.R.; Tyihák, E.; Szende, B. Structure determination and synthesis of lysine isopeptides influencing on cell proliferation. Biopolymers 1997, 42, 305–318. [Google Scholar]
- Nishikawa, M.; Ogawa, K. Distribution of microbes producing antimicrobial ε-poly-L-lysine polymers in soil microflora determined by a novel method. Appl. Environ. Microbiol. 2002, 68, 3575–3581. [Google Scholar] [CrossRef]
- Dementhon, K.; Iyer, G.; Glass, N.L. VIB-1 is required for expression of genes necessary for programmed cell death in Neurospora crassa. Eukaryot Cell. 2006, 5, 2161–2173. [Google Scholar] [CrossRef]
- Katz, M.E.; Gray, K.-A.; Cheetham, B.F. The Aspergillus nidulans xprG (phoG) gene encodes a putative transcriptional activator involved in the response to nutrient limitation. Fungal Genet. Biol. 2006, 43, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Wang, R.; Xu, Z.; Xu, Z.; Li, S.; Wang, M.; Feng, X.; Xu, H. Discovery of a short chain ε-poly-L-lysine and its highly efficient production via synthetase swap strategy. J. Agric. Food Chem. 2019, 67, 1453–1462. [Google Scholar] [CrossRef] [PubMed]
- Hirohara, H.; Saimura, M.; Takehara, M.; Miyamoto, M.; Ikezaki, A. Substantially monodispersed poly(ε-L-lysine)s frequently occurred in newly isolated strains of Streptomyces sp. Appl Microbiol Biotechnol. 2007, 76, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Hirohara, H.; Takehara, M.; Saimura, M.; Ikezaki, A.; Miyamoto, M. Biosynthesis of poly(ε-L-lysine)s in two newly isolated strains of Streptomyces sp. Appl. Microbiol. Biotechnol. 2006, 73, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, X.; Wu, G.; Li, S.; Zeng, X.; Ren, X.; Tang, L.; Mao, Z. Enhanced ε-poly-L-lysine production by inducing double antibiotic-resistant mutations in Streptomyces albulus. Bioprocess Biosyst. Eng. 2017, 40, 271–283. [Google Scholar] [CrossRef]
- Zhou, Y.P.; Ren, X.D.; Wang, L.; Chen, X.S.; Mao, Z.G.; Tang, L. Enhancement of ε-poly-lysine production in ε-poly-lysine-tolerant Streptomyces sp. by genome shuffling. Bioprocess Biosyst Eng. 2015, 38, 1705–1713. [Google Scholar] [CrossRef]
- Olsen, I.; Jantzen, E. Sphingolipids in bacteria and fungi. Anaerobe 2001, 7, 103–112. [Google Scholar] [CrossRef]
- Mélida, H.; Sandoval-Sierra, J.V.; Diéguez-Uribeondo, J.; Bulone, V. Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryot. Cell. 2013, 12, 194–203. [Google Scholar] [CrossRef]
- Yoshida, T. Biochemistry and enzymology of poly-ε-l-lysine degradation. In Amino-Acid Homopolymers Occurring in Nature; Hamano, Y., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 15, pp. 45–59. [Google Scholar]
- Byrd, A.D.; Schardl, C.L.; Songlin, P.J.; Mogen, K.L.; Siegel, M.R. The β-tubulin gene of Epichloë typhina from perennial ryegrass (Lolium perenne). Curr. Genet. 1990, 18, 347–354. [Google Scholar] [CrossRef]
- Takemoto, D.; Tanaka, A.; Scott, B. A p67Phox-like regulator is recruited to control hyphal branching in a fungal-grass mutualistic symbiosis. Plant Cell. 2006, 18, 2807–2821. [Google Scholar] [CrossRef]
- Young, C.A.; Bryant, M.K.; Christensen, M.J.; Tapper, B.A.; Bryan, G.T.; Scott, B. Molecular cloning and genetic analysis of a symbiosis-expressed gene cluster for lolitrem biosynthesis from a mutualistic endophyte of perennial ryegrass. Mol. Gen. Genomics. 2005, 274, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Johnson, R.; Scott, B. Integrative transformation of the myco-toxin-producing fungus, Penicillium paxilli. Curr. Genet. 1994, 25, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Shibata, Y.; Kawakita, K.; Takemoto, D. Age-related resistance of Nicotiana benthamiana against hemibiotrophic pathogen Phytophthora infestans requires both ethylene- and salicylic acid-mediated signaling pathways. Mol. Plant Microbe Interact. 2010, 23, 1130–1142. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |


| Position | δC (ppm), Type | δH (ppm), Mult. (J, Hz) | HMBC Correlations (to C) |
|---|---|---|---|
| α | 53.3, CH | 3.90, t (6.8) | γ, β, C=O |
| β | 30.5, CH2 | 1.85, m | α, C=O |
| γ | 21.7, CH2 | 1.37, quint (7.3) | β, ε ε |
| δ | 28.0, CH2 | 1.55, quint (7.3) | β, ε |
| ε | 39.1, CH2 | 3.24, m | C=O |
| C=O | 169.6, C | - |
| Strains | Production (mg/L) a | Chain length, DP b |
|---|---|---|
| E437 (wild type) | 10.4 | 28–34 (32) |
| Ptef::VibA (E437) | 38.6 | 28–34 (32) |
| Ptef::Epls (E437) | 69.9 | 8–25 (15) and 26–33 (31) |
| Fl1 (wild type) | trace | - |
| Ptef::VibA (Fl1) | 2.7 | 8–18 (12) |
| Ptef::Epls (Fl1) | 13.9 | 8–20 (12) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Purev, E.; Kondo, T.; Takemoto, D.; Niones, J.T.; Ojika, M. Identification of ε-Poly-L-lysine as an Antimicrobial Product from an Epichloë Endophyte and Isolation of Fungal ε-PL Synthetase Gene. Molecules 2020, 25, 1032. https://doi.org/10.3390/molecules25051032
Purev E, Kondo T, Takemoto D, Niones JT, Ojika M. Identification of ε-Poly-L-lysine as an Antimicrobial Product from an Epichloë Endophyte and Isolation of Fungal ε-PL Synthetase Gene. Molecules. 2020; 25(5):1032. https://doi.org/10.3390/molecules25051032
Chicago/Turabian StylePurev, Enkhee, Tatsuhiko Kondo, Daigo Takemoto, Jennifer T. Niones, and Makoto Ojika. 2020. "Identification of ε-Poly-L-lysine as an Antimicrobial Product from an Epichloë Endophyte and Isolation of Fungal ε-PL Synthetase Gene" Molecules 25, no. 5: 1032. https://doi.org/10.3390/molecules25051032
APA StylePurev, E., Kondo, T., Takemoto, D., Niones, J. T., & Ojika, M. (2020). Identification of ε-Poly-L-lysine as an Antimicrobial Product from an Epichloë Endophyte and Isolation of Fungal ε-PL Synthetase Gene. Molecules, 25(5), 1032. https://doi.org/10.3390/molecules25051032