A New Family of Diverse Skin Peptides from the Microhylid Frog Genus Phrynomantis
Abstract
1. Introduction
2. Results
2.1. Skin Transcriptomes Elucidate the Diversity of Secretory Proteins in Phrynomantis
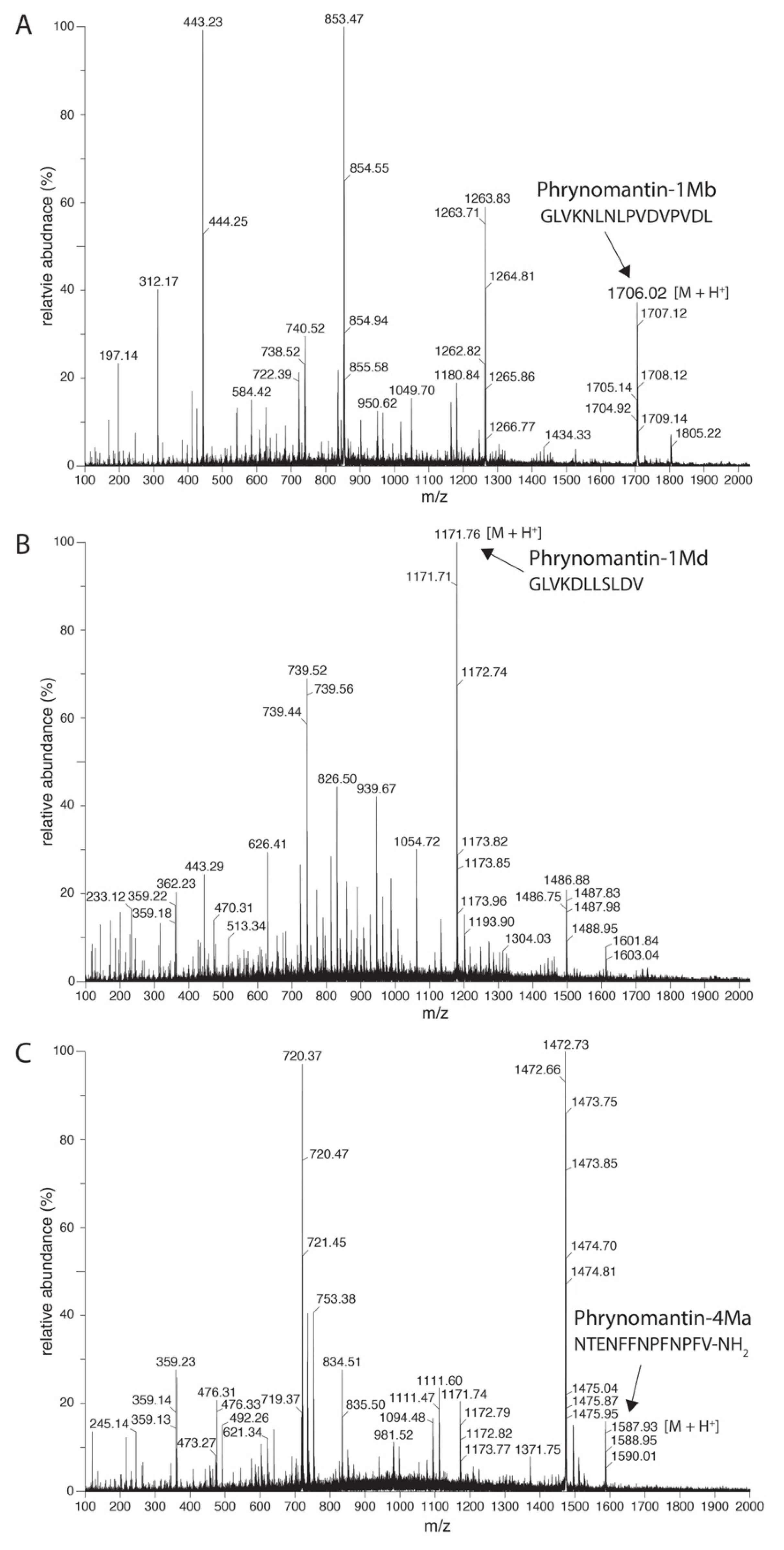
2.2. Phrynomantins: The First Microhylid Representatives of the FSAP Superfamily
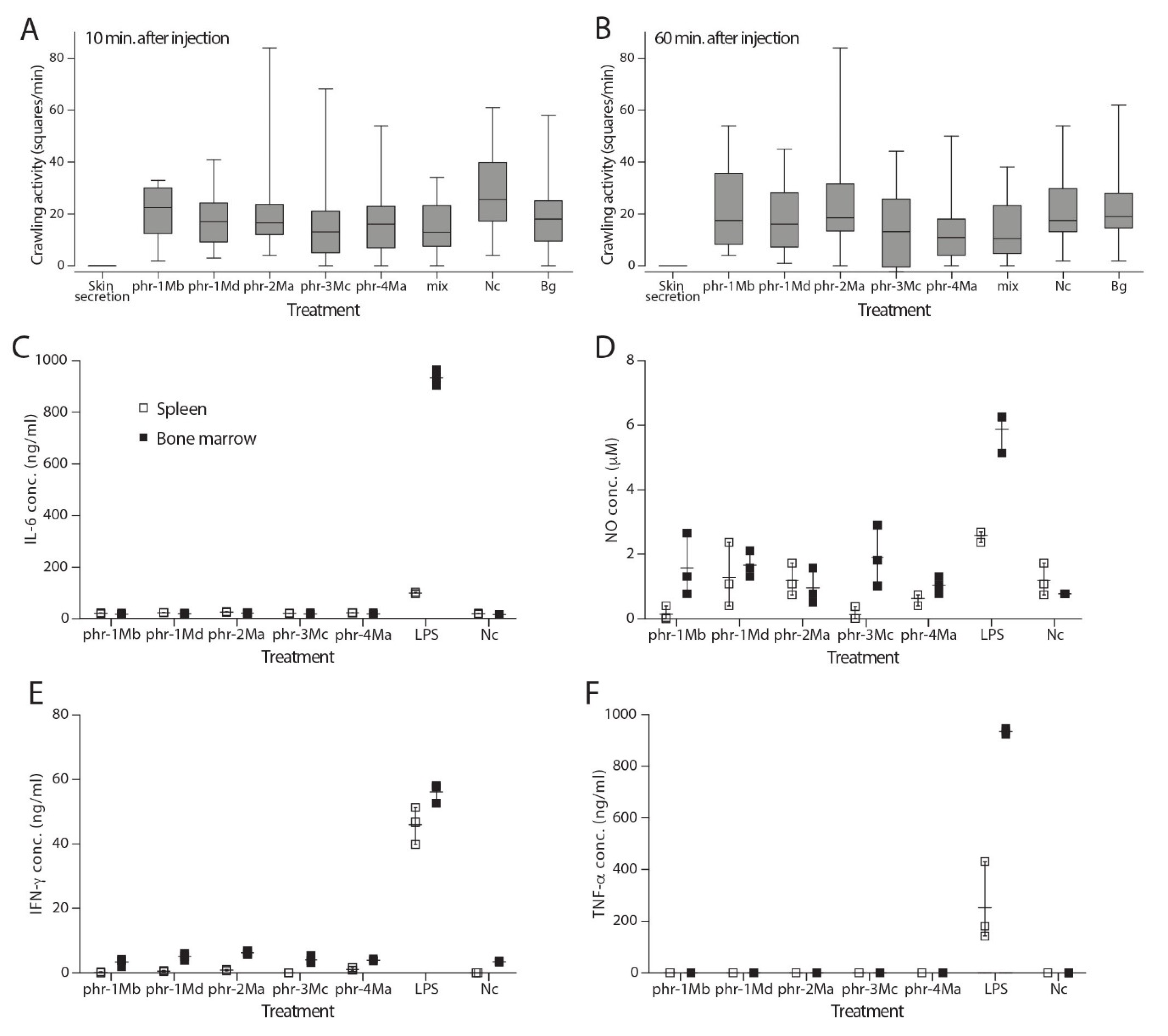
2.3. Phrynomantins Serve a yet Unknown Function
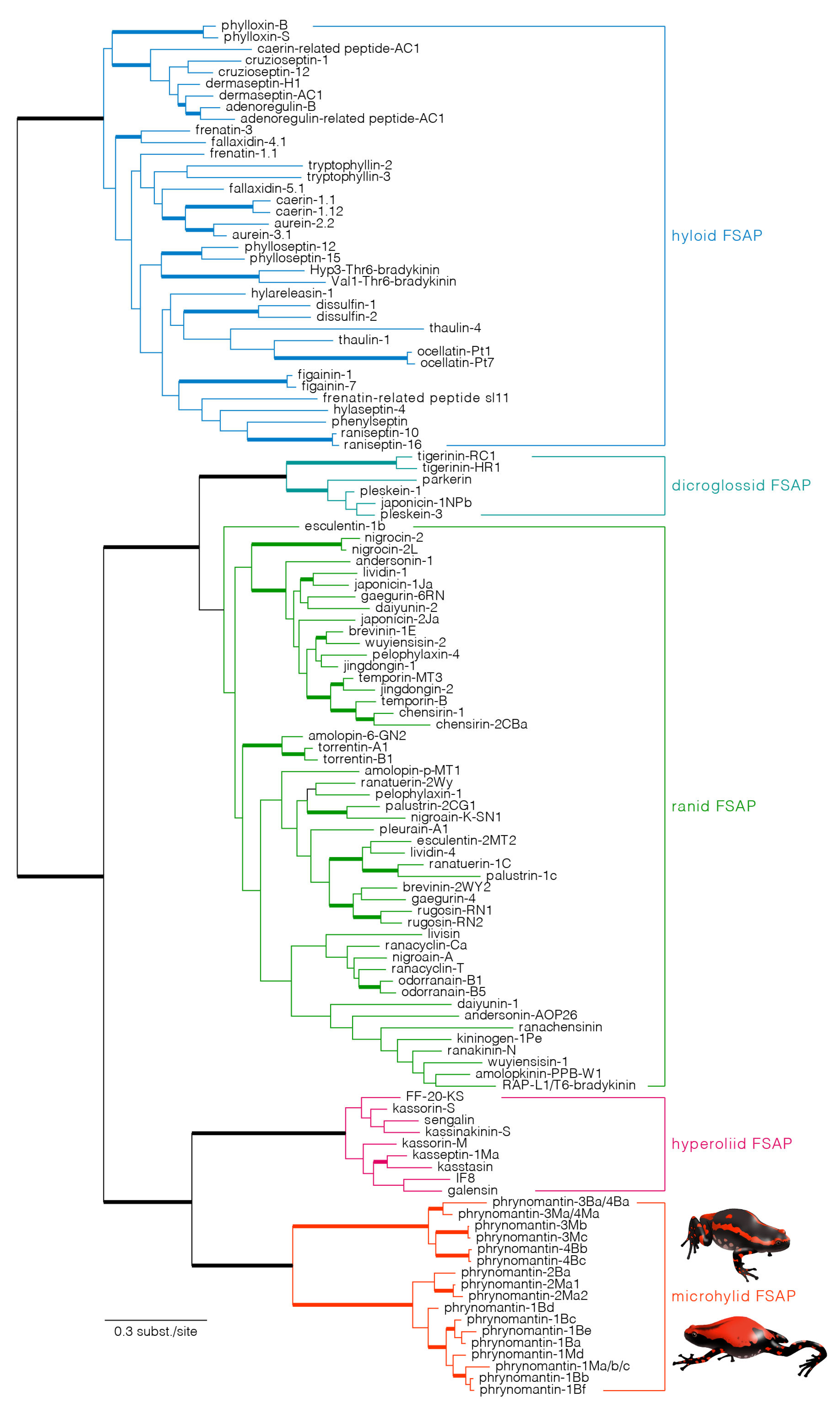
2.4. Evolutionary Origin and Diversification of Phrynomantins
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Animal Housing and Sample Collection
4.3. Transcriptome Analyses
4.4. Peptidome Analyses
4.5. Peptide Synthesis
4.6. Antimicrobial and Cytotoxicity Assays
4.7. Insecticidal Activity Experiments
4.8. Inflammation Assays
4.9. Phylogenetic Analyses
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- König, E.; Bininda-Emonds, O.R.P.; Shaw, C. The diversity and evolution of anuran skin peptides. Peptides 2015, 63, 96–117. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lai, R. The chemistry and biological activities of peptides from amphibian skin secretions. Chem. Rev. 2015, 115, 1760–1846. [Google Scholar] [CrossRef]
- Clarke, B.T. The natural history of amphibian skin secretions, their normal functioning and potential medical applications. Biol. Rev. 1997, 72, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Zou, Z.; Yang, M.; Wu, C.; Su, Y.; Tang, J.; Yang, X. Om-lv20, a novel peptide from odorous frog skin, accelerates wound healing in vitro and in vivo. Chem. Biol. Drug Des. 2018, 91, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.; Teixeira, C.; Ferraz, R.; Prudêncio, C.; Gomes, P. Wound-healing peptides for treatment of chronic diabetic foot ulcers and other infected skin injuries. Molecules 2017, 22, 1743. [Google Scholar] [CrossRef] [PubMed]
- Ojo, O.O.; Srinivasan, D.K.; Owolabi, B.O.; Vasu, S.; Conlon, J.M.; Flatt, P.R.; Abdel-Wahab, Y.H. Esculentin-2CHa-related peptides modulate islet cell function and improve glucose tolerance in mice with diet-induced obesity and insulin resistance. PLoS ONE 2015, 10, e0141549. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, D.; Ojo, O.O.; Owolabi, B.O.; Conlon, J.M.; Flatt, P.R.; Abdel-Wahab, Y.H. The frog skin host-defense peptide CPF-SE1 improves glucose tolerance, insulin sensitivity and islet function and decreases plasma lipids in high-fat fed mice. Eur. J. Pharmacol. 2015, 764, 38–47. [Google Scholar] [CrossRef]
- Conlon, J.M. The contribution of skin antimicrobial peptides to the system of innate immunity in anurans. Cell Tissue Res. 2011, 343, 201–212. [Google Scholar] [CrossRef]
- Conlon, J.M. The therapeutic potential of antimicrobial peptides from frog skin. Rev. Med. Microbiol. 2004, 15, 17–25. [Google Scholar] [CrossRef]
- Conlon, J.M.; Guilhaudis, L.; Leprince, J.; Coquet, L.; Mangoni, M.L.; Attoub, S.; Jouenne, T.; King, J.D. Peptidomic analysis of skin secretions of the Mexican burrowing toad Rhinophrynus dorsalis (Rhinophrynidae): Insight into the origin of host-defense peptides within the Pipidae and characterization of a proline-arginine-rich peptide. Peptides 2017, 97, 22–28. [Google Scholar] [CrossRef]
- Nicolas, P.; El Amri, C. The dermaseptin superfamily: A gene-based combinatorial library of antimicrobial peptides. Biochim. Biophys. Acta 2009, 1788, 1537–1550. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, P.; Vanhoye, D.; Amiche, M. Molecular strategies in biological evolution of antimicrobial peptides. Peptides 2003, 24, 1669–1680. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Kolodziejek, J.; Nowotny, N. Antimicrobial peptides from ranid frogs: Taxonomic and phylogenetic markers and a potential source of new therapeutic agents. Biochim. Biophys. Acta 2004, 1696, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Vanhoye, D.; Bruston, F.; Nicolas, P.; Amiche, M. Antimicrobial peptides from hylid and ranin frogs originated from a 150-million-year-old ancestral precursor with a conserved signal peptide but a hypermutable antimicrobial domain. Eur. J. Biochem. 2003, 270, 2068–2081. [Google Scholar] [CrossRef] [PubMed]
- König, E.; Bininda-Emonds, O.R.P. Evidence for convergent evolution in the antimicrobial peptide system in anuran amphibians. Peptides 2011, 32, 20–25. [Google Scholar] [CrossRef]
- Khamis, A.M.; Essack, M.; Gao, X.; Bajic, V.B. Distinct profiling of antimicrobial peptide families. Bioinformatics 2015, 31, 849–856. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Peloso, P.; Paraense, M.; Goeldi, E.; Richards, S.J.; Rodrigues, M.T. The impact of anchored phylogenomics and taxon sampling on phylogenetic inference in narrow-mouthed frogs (Anura, Microhylidae). Cladistics 2016, 32, 113–140. [Google Scholar] [CrossRef]
- de Sá, R.O.; Streicher, J.W.; Sekonyela, R.; Forlani, M.C.; Loader, S.P.; Greenbaum, E.; Richards, S.; Haddad, C.F. Molecular phylogeny of microhylid frogs (Anura: Microhylidae) with emphasis on relationships among New World genera. BMC Evol. Biol. 2012, 12, 241. [Google Scholar] [CrossRef]
- Rödel, M.-O.; Brede, C.; Hirschfeld, M.; Schmitt, T.; Favreau, P.; Stocklin, R.; Wunder, C.; Mebs, D. Chemical Camouflage---A Frog’s Strategy to Co-Exist with Aggressive Ants. PLoS ONE 2012, 8, e81950. [Google Scholar]
- Cavalcante, I.D.; Antoniazzi, M.M.; Jared, C.; Pires Jr, O.R.; Sciani, J.M.; Pimenta, D.C. Venomics analyses of the skin secretion of Dermatonotus muelleri: Preliminary proteomic and metabolomic profiling. Toxicon 2017, 130, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Kim, J.B. A protease inhibitor of the Kunitz family from skin secretions of the tornato frog, Dyscophus guineti (Microhylidae). Biochem. Biophys. Res. Commun. 2000, 279, 961–964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, M.; Wei, S. Isolation and characterization of a trypsin inhibitor from the skin secretions of Kaloula pulchra hainana. Toxicon 2010, 56, 502–507. [Google Scholar] [CrossRef]
- Jaeger, R.G. Toxic reaction to skin secretions of the frog, Phrynomerus bifasciatus. Copeia 1971, 1, 160–161. [Google Scholar] [CrossRef]
- Passmore, N.I.; Carruthers, V.C. South. african frogs, 1st ed.; Witwatersrand University Pres: Johannesburg, South Africa, 1979; pp. 1–270. [Google Scholar]
- Braun, U. Associations between Anurans and Ants in a West African Savanna (Anura: Microhylidae, Hyperoliidae and Hymenoptera: Formicidae). Biotropica 1999, 31, 178–183. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Helle, K.B. The granin family of uniquely acidic proteins of the diffuse neuroendocrine system: Comparative and functional aspects. Biol. Rev. 2004, 79, 769–794. [Google Scholar] [CrossRef]
- McGuffin, L.J.; Bryson, K.; Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef]
- Pantanowitz, L.; Naude, T.W.; Leisewitz, A. Noxious toads and frogs of South Africa. J. South. Afr. Stud. 1998, 88, 1408–1414. [Google Scholar]
- Stijlemans, B.; Leng, L.; Brys, L.; Sparkes, A.; Vansintjan, L.; Caljon, G.; Raes, G.; Van Den Abbeele, J.; Van Ginderachter, J.A.; Beschin, A.; et al. MIF contributes to Trypanosoma brucei associated immunopathogenicity development. PLoS Pathog. 2014, 10, e1004414. [Google Scholar] [CrossRef] [PubMed]
- Van Bocxlaer, I.; Roelants, K.; Biju, S.D.; Nagaraju, J.; Bossuyt, F. Late-Cretaceous vicariance in Gondwanan amphibians. PLoS ONE 2006, 1, e74. [Google Scholar] [CrossRef] [PubMed]
- Pyron, A.R.; Wiens, J.J. A large-scale phylogeny of Amphibia including over 2800 species and a revised classification of extant frogs, salamanders and caecilians. Mol. Phylogenet. Evol. 2011, 61, 543–583. [Google Scholar] [CrossRef] [PubMed]
- Roelants, K.; Gower, D.J.; Wilkinson, M.; Loader, S.P.; Biju, S.D.; Guillaume, K.; Moriau, L.; Bossuyt, F. Global patterns of diversification in the history of modern amphibians. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Mignogna, G.; Pascarella, S.; Wechselberger, C.; Hinterleitner, C.; Mollay, C.; Amiconi, G.; Barra, D.; Kreil, G. BSTI, a trypsin inhibitor from skin secretions of Bombina bombina related to protease inhibitors of nematodes. Protein Sci. 1996, 5, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, Y.; Wei, L.; Ye, H.; Liu, H.; Wang, L.; Liu, R.; Li, D.; Lai, R. Snake venom-like waprin from the frog of Ceratophrys calcarata contains antimicrobial function. Gene 2013, 514, 99–104. [Google Scholar] [CrossRef]
- Proaño-Bolaños, C.; Li, R.; Zhou, M.; Wang, L.; Xi, X.; Tapia, E.E.; Coloma, L.A.; Chen, T.; Shaw, C. Novel Kazal-type proteinase inhibitors from the skin secretion of the Splendid leaf frog, Cruziohyla calcarifer. EuPA Open Proteom. 2017, 15, 1–13. [Google Scholar] [CrossRef]
- Wu, J.; Liu, H.; Yang, H.; Yu, H.; You, D.; Ma, Y.; Ye, H.; Lai, R. Proteomic analysis of skin defensive factors of tree frog Hyla simplex. J. Proteome Res. 2011, 10, 4230–4240. [Google Scholar] [CrossRef]
- Mu, L.; Zhou, L.; Yang, J.; Zhuang, L.; Tang, J.; Liu, T.; Wu, J.; Yang, H. The first identified cathelicidin from tree frogs possesses anti-inflammatory and partial LPS neutralization activities. Amino Acids 2017, 49, 1571–1585. [Google Scholar] [CrossRef]
- Meng, P.; Yang, S.; Shen, C.; Jiang, K.; Rong, M.; Lai, R. The First Salamander Defensin Antimicrobial Peptide. PLoS ONE 2013, 8, E83044. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Jin, Y.; Lee, W.-H.; Zhang, Y. Purification of a lysozyme from skin secretions of Bufo andrewsi. Comp. Biochem. Physiol. 2006, 142C, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.; Muello, K.; Victor, M.; Elsbach, P. The role of lipopolysaccharides in the action of the bactericidal/permeability-increasing neutrophil protein on the bacterial envelope. J. Immunol. 1984, 132, 3109–3115. [Google Scholar] [PubMed]
- Durban, J.; Juarez, P.; Angulo, Y.; Lomonte, B.; Flores-Diaz, M.; Alape-Girón, A.; Sasa, M.; Sanz, L.; Gutiérrez, J.M.; Dopazo, J.; et al. Profiling the venom gland transcriptomes of Costa Rican snakes by 454 pyrosequencing. BMC Genomics 2011, 12, 259. [Google Scholar] [CrossRef]
- Tan, C.H.; Tan, K.Y.; Fung, S.Y.; Tan, N.H. Venom-gland transcriptome and venom proteome of the Malaysian king cobra (Ophiophagus hannah). BMC Genomics. 2015, 16, 687. [Google Scholar] [CrossRef]
- Harris, F.; Dennison, S.; Phoenix, D. Anionic Antimicrobial Peptides from Eukaryotic Organisms. Curr. Protein Pept. Sci. 2009, 10, 585–606. [Google Scholar] [CrossRef]
- Siano, A.; Humpola, M.; de Oliveira, E.; Albericio, F.; Simonetta, A.; Lajmanovich, R.; Tonarelli, G.G. Leptodactylus latrans amphibian skin secretions as a novel source for the isolation of antibacterial peptides. Molecules 2018, 23, 2943. [Google Scholar] [CrossRef]
- Li, S.; Hao, L.; Bao, W.; Zhang, P.; Su, D.; Cheng, Y.; Nie, L.; Wang, G.; Hou, F.; Yang, Y. A novel short anionic antibacterial peptide isolated from the skin of Xenopus laevis with broad antibacterial activity and inhibitory activity against breast cancer cell. Arch. Microbiol. 2016, 198, 473–482. [Google Scholar] [CrossRef]
- Daly, J.W. The chemistry of poisons in amphibian skin. Proc. Natl. Acad. Sci. U.S.A. 1995, 92, 9–13. [Google Scholar] [CrossRef]
- Erspamer, V. Biogenic amines and active polypeptides of the amphibian skin. Ann. Rev. Pharmacol. 1971, 11, 327–350. [Google Scholar] [CrossRef]
- Matsuura, H.N.; Fett-Neto, A.G. Plant alkaloids: Main features, toxicity and mechanisms of action. In Plant Toxins, 1st ed.; Gopalakrishnakone, P., Carlini, C.R., Ligabue-Braun, R., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 243–261. [Google Scholar]
- Glendinning, J.I. How do predators cope with chemically defended foods? Biol. Bull. 2007, 213, 252–266. [Google Scholar] [CrossRef] [PubMed]
- Rowland, H.M.; Ruxton, G.D.; Skelhorn, J. Bitter taste enhances predatory biases against aggregations of prey with warning coloration. Behav. Ecol. 2013, 24, 942–948. [Google Scholar] [CrossRef]
- de Magalhaes, M.T.; Barbosa, E.A.; Prates, M.V.; Verly, R.M.; Munhoz, V.H.O.; de Araujo, I.E.; Bloch Jr, C. Conformational and functional effects induced by D-and L-amino acid epimerization on a single gene encoded peptide from the skin secretion of Hypsiboas punctatus. PLoS ONE 2013, 8, e59255. [Google Scholar] [CrossRef]
- Ishibashi, N.; Sadamori, K.; Yamamoto, O.; Kanehisa, H.; Kouge, K.; Kikuchi, E.; Okai, H.; Fukui, S. Bitterness of phenylalanine-and tyrosine-containing peptides. Agricult. Biol. Chem. 1987, 51, 3309–3313. [Google Scholar] [CrossRef]
- Chen, T.; Reid, C.N.; Walker, B.; Zhou, M.; Shaw, C. Kassinakinin S: A novel histamine-releasing heptadecapeptide from frog (Kassina senegalensis) skin secretion. Biochem. Biophys. Res. Commun. 2005, 337, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Van der Meijden, A.; Vences, M.; Hoegg, S.; Boistel, R.; Channing, A.; Meyer, A. Nuclear gene phylogeny of narrow-mouthed toads (Family: Microhylidae) and a discussion of competing hypotheses concerning their biogeographical origins. Mol. Phylogenet. Evol. 2007, 44, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Kurabayashi, A.; Matsui, M.; Belabut, D.M.; Yong, H.-S.; Ahmad, N.; Sudin, A.; Kuramoto, M.; Hamidy, A.; Sumida, M. From Antarctica or Asia? New colonization scenario for Australian-New Guinean narrow mouth toads suggested from the findings on a mysterious genus Gastrophrynoides. BMC Evol. Biol. 2011, 11, 175. [Google Scholar] [CrossRef]
- Tennessen, J.A.; Woodhams, D.C.; Chaurand, P.; Reinert, L.K.; Billheimer, D.; Shyr, Y.; Caprioli, R.M.; Blouin, M.S.; Rollins-Smith, L.A. Variations in the expressed antimicrobial peptide repertoire of northern leopard frog (Rana pipiens) populations suggest intraspecies differences in resistance to pathogens. Dev. Comp. Immunol. 2009, 33, 1247–1257. [Google Scholar] [CrossRef]
- Samgina, T.Y.; Artemenko, K.A.; Bergquist, J.; Trebse, P.; Torkar, G.; Tolpina, M.D.; Lebedev, A.T. Differentiation of frogs from two populations belonging to the Pelophylax esculentus complex by LC-MS/MS comparison of their skin peptidomes. Anal. Bioanal. Chem. 2017, 409, 1951–1961. [Google Scholar] [CrossRef]
- Magnoni, M.L.; Miele, R.; Renda, T.G.; Barra, D.; Simmaco, M. The synthesis of antimicrobial peptides in the skin of Rana esculenta is stimulated by microorganisms. FASEB J. 2001, 8, 1431–1432. [Google Scholar]
- Rollins-Smith, L.A. The role of amphibian antimicrobial peptides in protection of amphibians from pathogens linked to global amphibian declines. Biochim. Biophys. Acta 2009, 1788, 1593–1599. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-Seq quantification. Nat. Biotechnol. 2016, 34, 525. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H. Predicting secretory proteins with SignalP. Methods Mol. Biol. 2017, 1611, 59–73. [Google Scholar] [PubMed]
- Desbois, A.P.; Coote, P.J. Utility of greater wax moth larva (Galleria mellonella) for evaluating the toxicity and efficacy of new antimicrobial agents. Adv. Appl. Microbiol. 2012, 78, 25–53. [Google Scholar] [PubMed]
- Megaw, J.; Thompson, T.P.; Lafferty, R.A.; Gilmore, B.F. Galleria mellonella as a novel in vivo model for assessment of the toxicity of 1-alkyl-3-methylimidazolium chloride ionic liquids. Chemosphere 2015, 139, 197–201. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |

| Phrynomantis bifasciatus | Phrynomantis microps | |||
|---|---|---|---|---|
| Number of Transcript Contigs | ∑TPM 1 | Number of Transcript Contigs | ∑TPM 1 | |
| Frog Skin Active Peptide (FSAP) superfamily | 10 | 13,437.3 | 7 | 12,636.8 |
| Antimicrobial peptides/protein families | ||||
| Bactericidal permeability-increasing protein (BPI) | 48 | 992.4 | 38 | 479.6 |
| Beta-defensin | 1 | 1.7 | 1 | 3.5 |
| Cathelicidin | 6 | 5.1 | 5 | 4.8 |
| C-type lysozyme | 9 | 127.4 | 6 | 578.9 |
| G-type lysozyme | 12 | 5.6 | 2 | 5.0 |
| Serine protease inhibitor families | ||||
| Kazal-like | 1 | 408.4 | 4 | 534.7 |
| Kunitz/Bovine pancreatic trypsin inhibitor (BPTI) | 20 | 941.0 | 15 | 423.4 |
| Serpin-type | 60 | 773.2 | 60 | 2119.8 |
| Trypsin inhibitor-like (TIL) | 39 | 10,424.7 | 45 | 4252.5 |
| Whey acidic protein (WAP) | 26 | 506.0 | 20 | 825.2 |
| Hormone/neuropeptide-like peptide families | ||||
| Angiotensin | 1 | 1.9 | 0 | 0 |
| Bradykinin (kininogen) | 2 | 2.9 | 1 | 7.2 |
| Gastrin | 1 | 0.2 | 0 | 0 |
| Glucagon | 1 | 1.0 | 0 | 0 |
| Natriuretic peptide | 2 | 2.2 | 1 | 3.3 |
| Neurotensin/Neuromedin N | 1 | 0.7 | 0 | 0 |
| Secretin | 1 | 5.0 | 1 | 1.4 |
| Secretogranin | 11 | 35.6 | 8 | 70.0 |
| Tachykinin | 0 | 0 | 2 | 7.1 |
| Peptide | Sequence 1 | Length | MW (Da) 3 | Net charge 4 | GRAVY 5 | Helicity (%) 6 |
|---|---|---|---|---|---|---|
| phrynomantin-1Ba | GLVTNLLSSVR | 11 | 1158.4 | 1 | 0.84 | 72.7 |
| phrynomantin-1Bb | GLVPDLDLPVDL | 12 | 1265.5 | −3 | 0.79 | 0 |
| phrynomantin-1Bc | GIVNNLLSTVL | 11 | 1142.4 | 0 | 1.40 | 81.8 |
| phrynomantin-1Bd | GPVFDYLSQVYPVR | 14 | 1639.9 | 0 | 0.05 | 50 |
| phrynomantin-1Be | GLVKDILSLDVL | 12 | 1284.5 | −1 | 1.33 | 66.7 |
| phrynomantin-1Bf | GLVPDLDLPVDLPV | 14 | 1461.7 | −3 | 0.86 | 0 |
| phrynomantin-1Ma | GLVKNQDLPVDLAAVF | 16 | 1699.0 | −1 | 0.66 | 12.5 |
| phrynomantin-1Mb | GLVKNLNLPVDVPVDL 2 | 16 | 1705.0 | −1 | 0.66 | 0 |
| phrynomantin-1Mc | GLVKDLNLPVDVPVDLPV | 18 | 1902.2 | −2 | 0.73 | 0 |
| phrynomantin-1Md | GLVKDLLSLDV2 | 11 | 1171.4 | −1 | 1.05 | 63.6 |
| phrynomantin-2Ba | DYEAVSL | 7 | 795.8 | −2 | 0.10 | 0 |
| phrynomantin-2Ma | DYEPASL | 7 | 793.8 | −2 | −0.73 | 0 |
| phrynomantin-3Ba | SEWPPVRGDNGEYDVEL | 17 | 1962.0 | −4 | −1.19 | 0 |
| phrynomantin-3Ma | SEWPPVRGDAGEYDVEL | 17 | 1919.0 | −4 | −0.88 | 0 |
| phrynomantin-3Mb | AEWRLL | 6 | 786.9 | 0 | 0.08 | 0 |
| phrynomantin-3Mc | AEWRLLKNa | 8 | 1028.2 | 2 | −0.86 | 0 |
| phrynomantin-4Ba | NAASRLYTPYSPTK | 14 | 1568.7 | 2 | −0.95 | 0 |
| phrynomantin-4Bb | NARNFNSFDPFNTSD | 15 | 1745.8 | −1 | −1.28 | 0 |
| phrynomantin-4Ma | NTENFFNPFNPFV2 | 13 | 1586.7 | −1 | −0.46 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raaymakers, C.; Stijlemans, B.; Martin, C.; Zaman, S.; Ballet, S.; Martel, A.; Pasmans, F.; Roelants, K. A New Family of Diverse Skin Peptides from the Microhylid Frog Genus Phrynomantis. Molecules 2020, 25, 912. https://doi.org/10.3390/molecules25040912
Raaymakers C, Stijlemans B, Martin C, Zaman S, Ballet S, Martel A, Pasmans F, Roelants K. A New Family of Diverse Skin Peptides from the Microhylid Frog Genus Phrynomantis. Molecules. 2020; 25(4):912. https://doi.org/10.3390/molecules25040912
Chicago/Turabian StyleRaaymakers, Constantijn, Benoit Stijlemans, Charlotte Martin, Shabnam Zaman, Steven Ballet, An Martel, Frank Pasmans, and Kim Roelants. 2020. "A New Family of Diverse Skin Peptides from the Microhylid Frog Genus Phrynomantis" Molecules 25, no. 4: 912. https://doi.org/10.3390/molecules25040912
APA StyleRaaymakers, C., Stijlemans, B., Martin, C., Zaman, S., Ballet, S., Martel, A., Pasmans, F., & Roelants, K. (2020). A New Family of Diverse Skin Peptides from the Microhylid Frog Genus Phrynomantis. Molecules, 25(4), 912. https://doi.org/10.3390/molecules25040912







