Rapid Evaluation and Optimization of Medium Components Governing Tryptophan Production by Pediococcus acidilactici TP-6 Isolated from Malaysian Food via Statistical Approaches
Abstract
1. Introduction
2. Results and Discussion
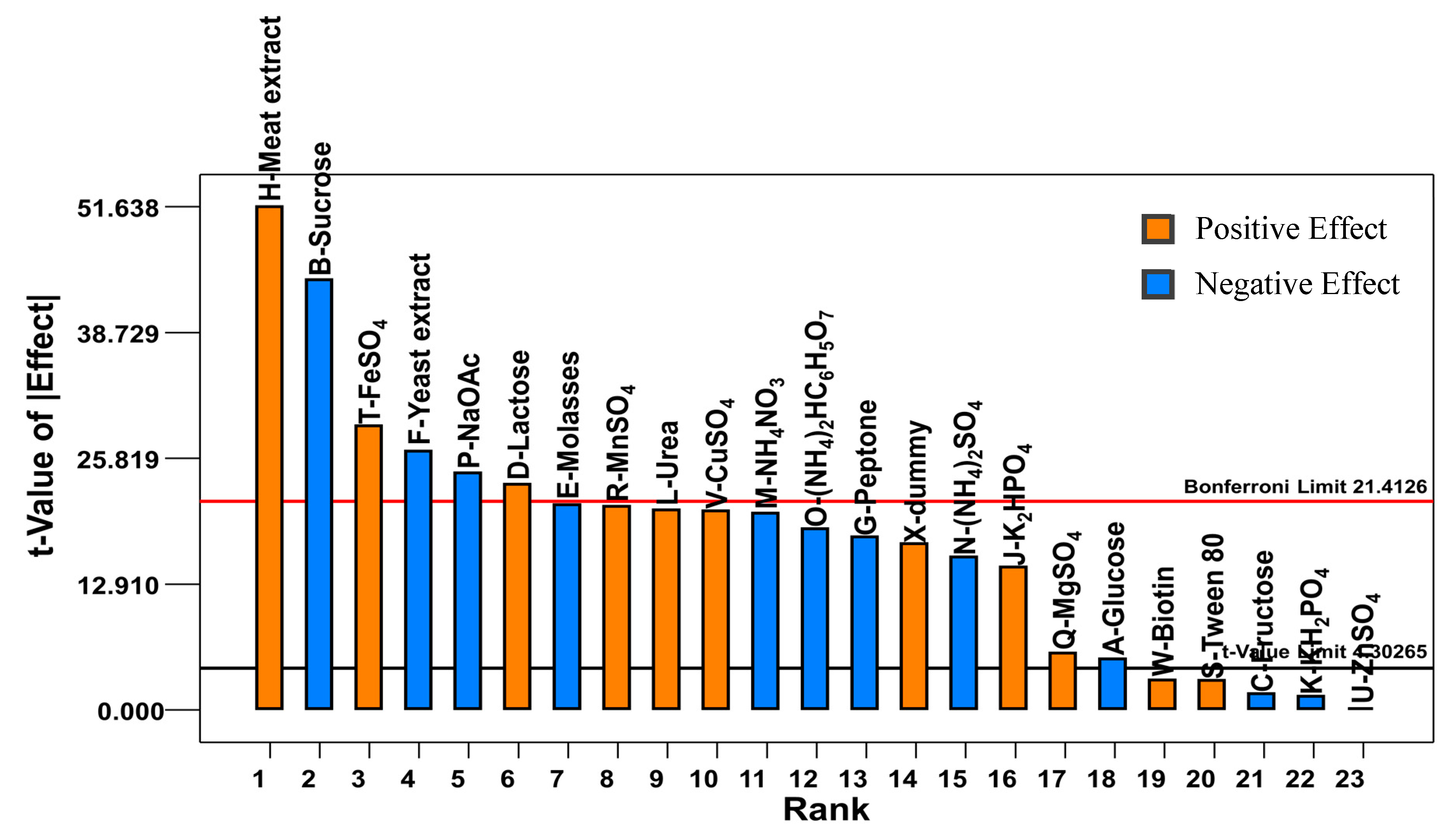
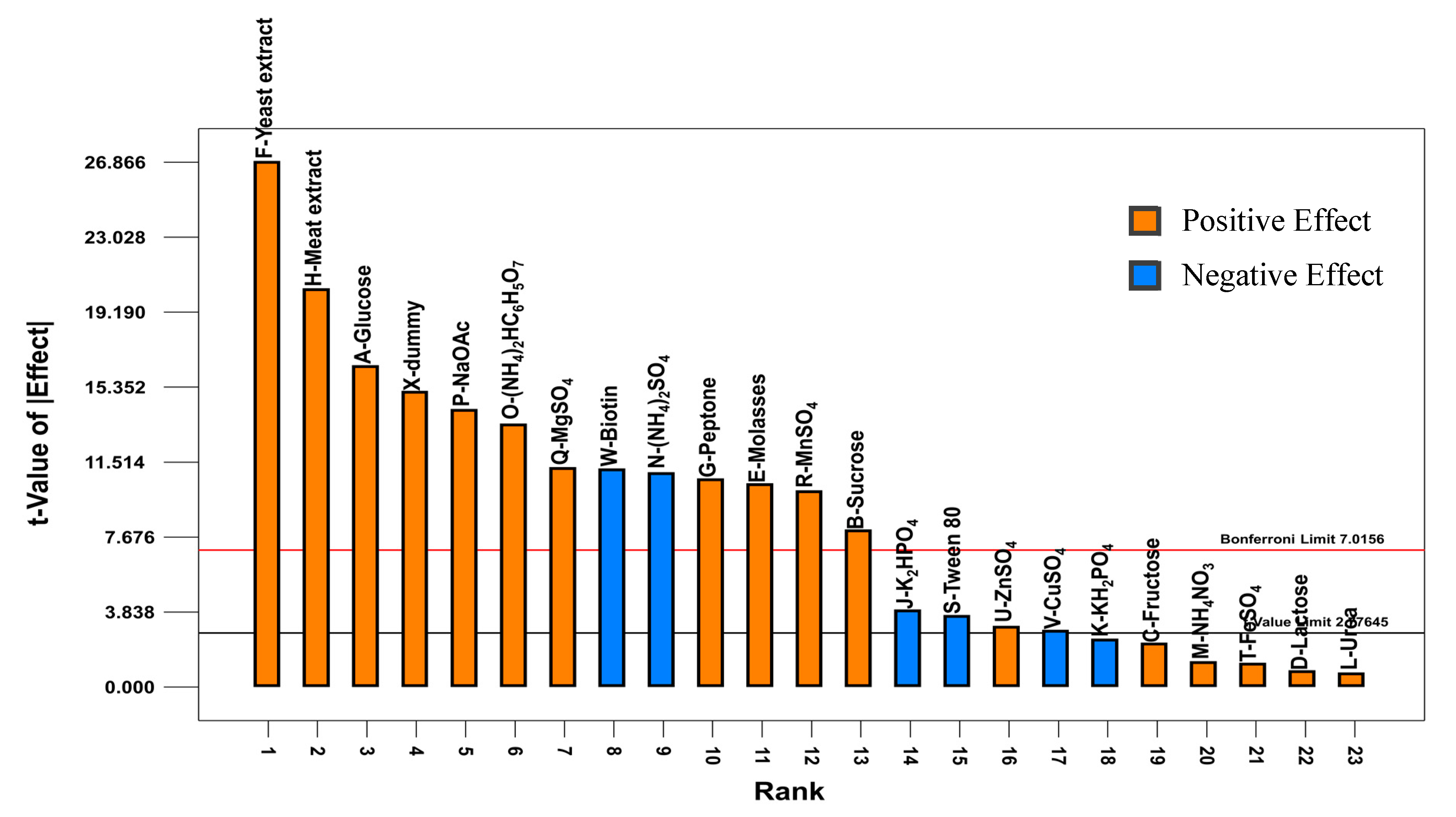
2.1. Plackett-Burman Design
1.95M − 1.51N − 1.79O − 2.34P+ 0.56Q + 2.01R + 0.29S + 2.81T + 1.97V + 0.30W + 1.64X
0.11N + 0.14O + 0.15P + 0.11Q + 0.1R − 0.037S + 0.031U − 0.029V − 0.11W + 0.16X
2.2. Steepest Ascent Method
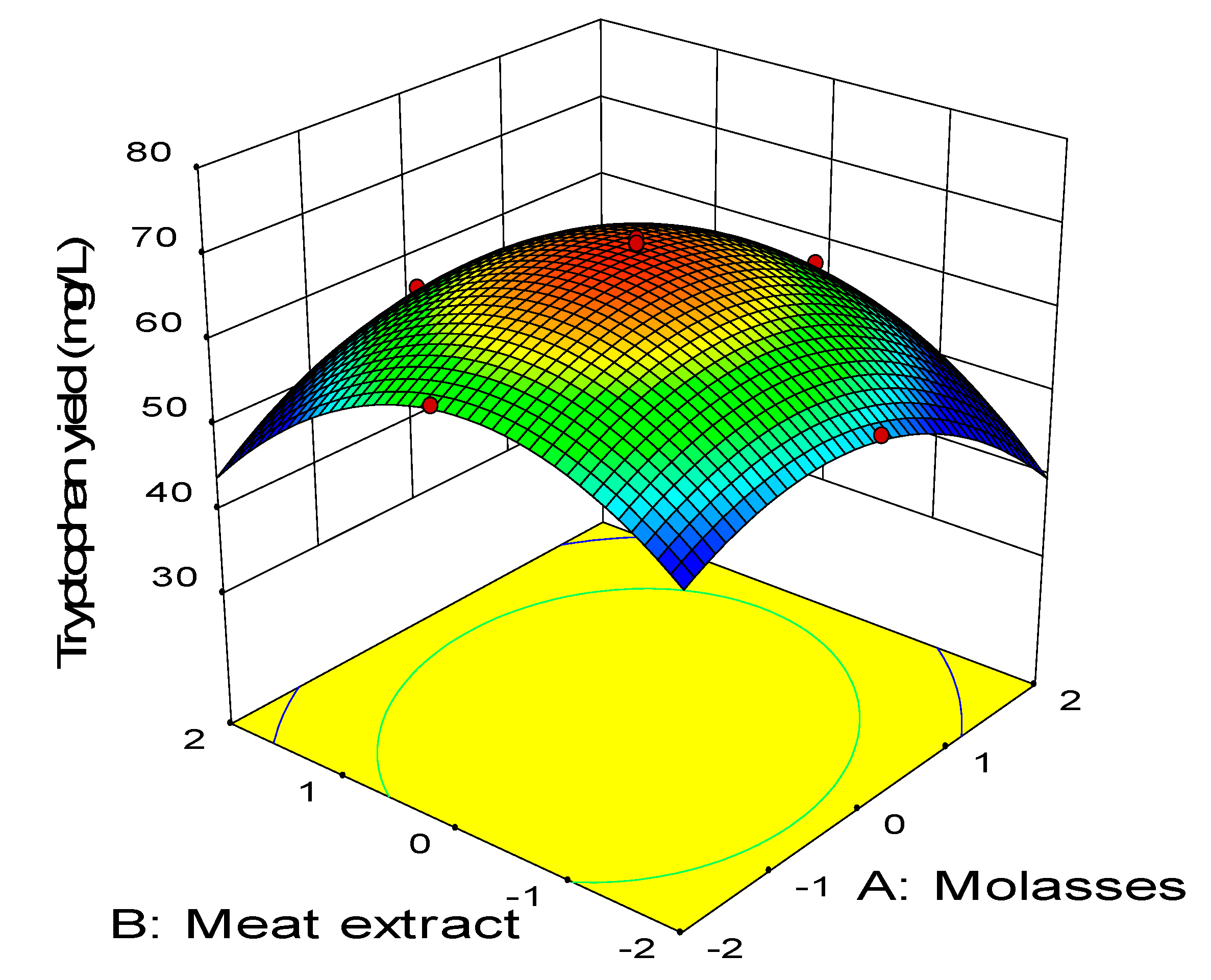
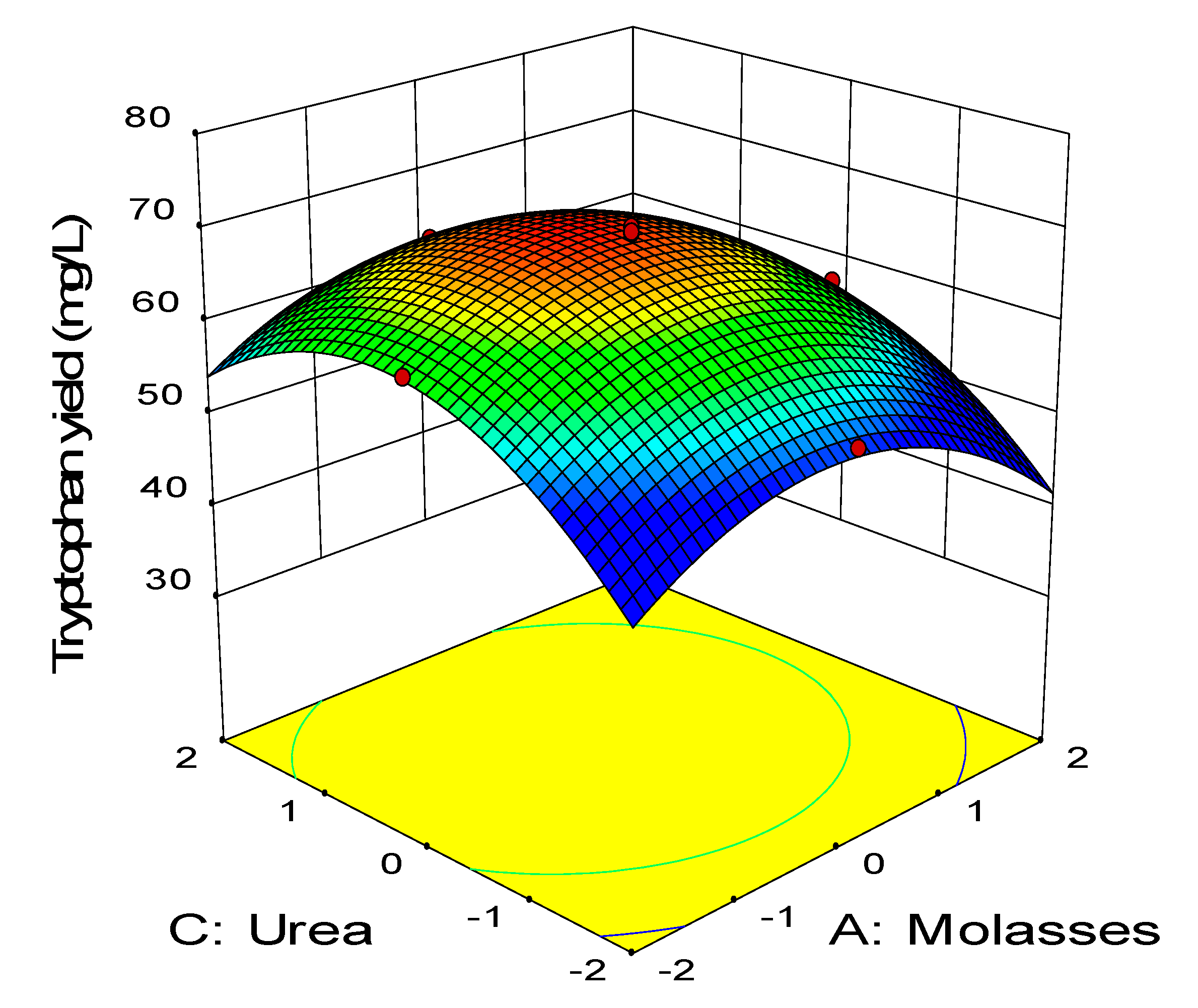
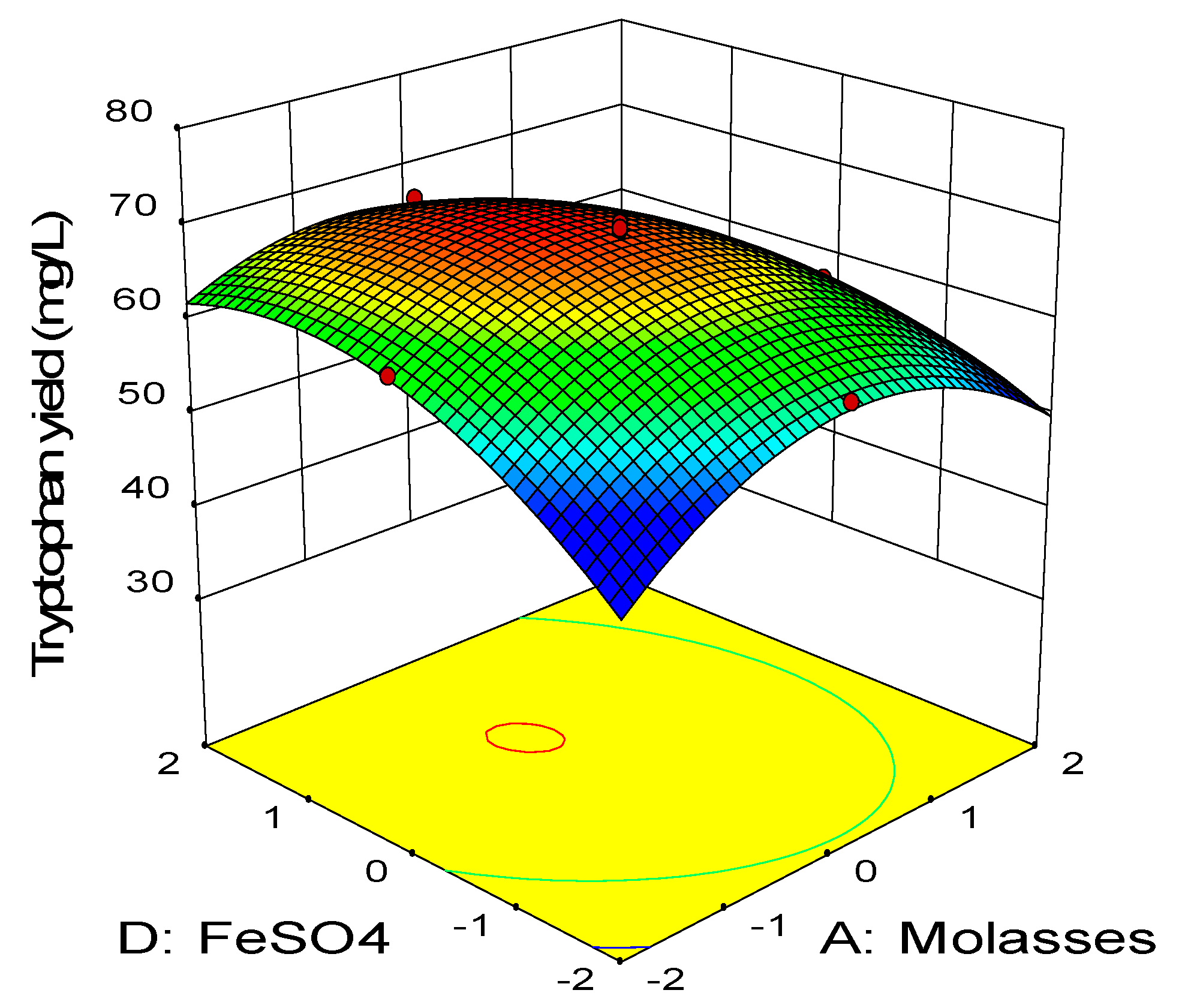
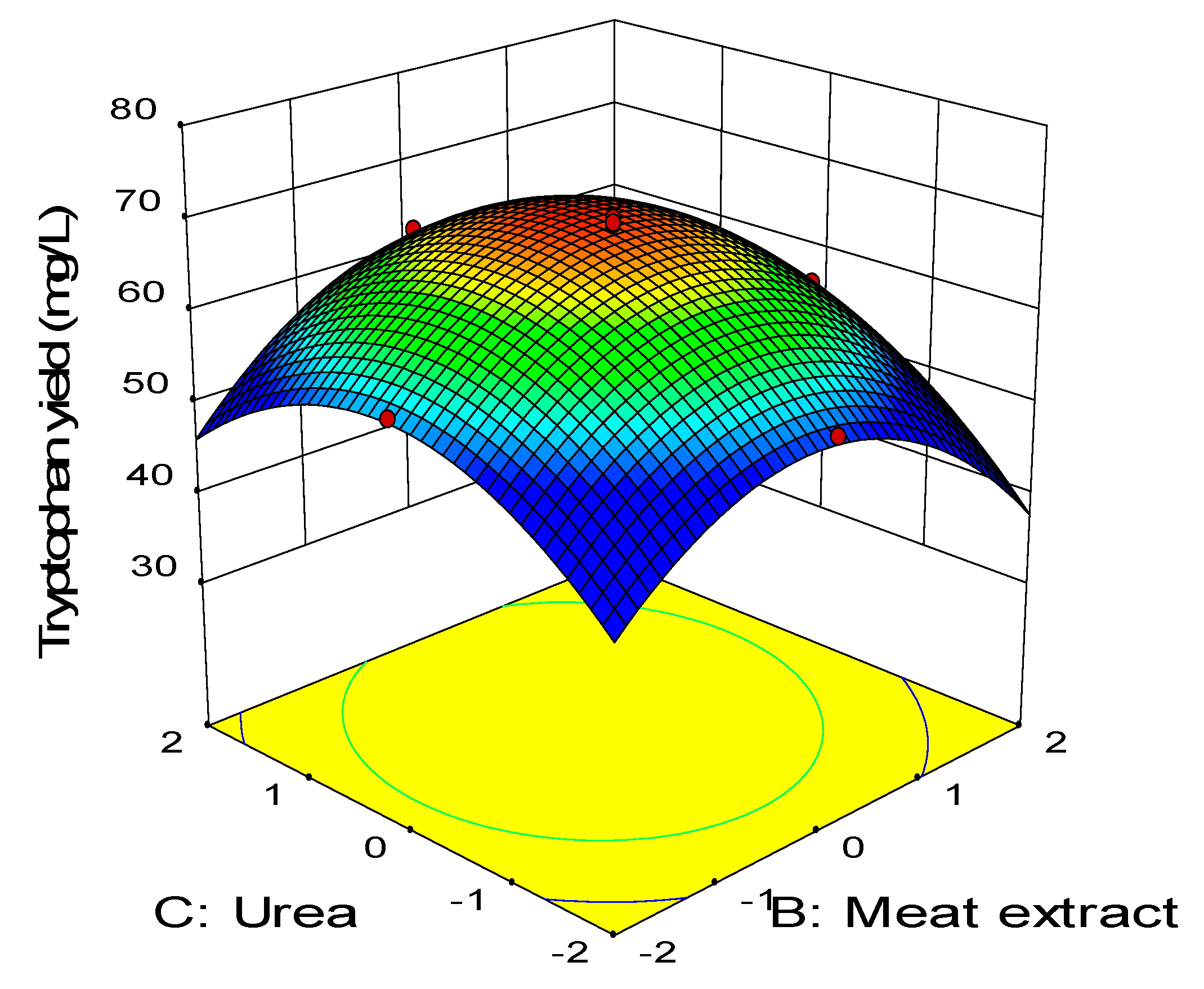
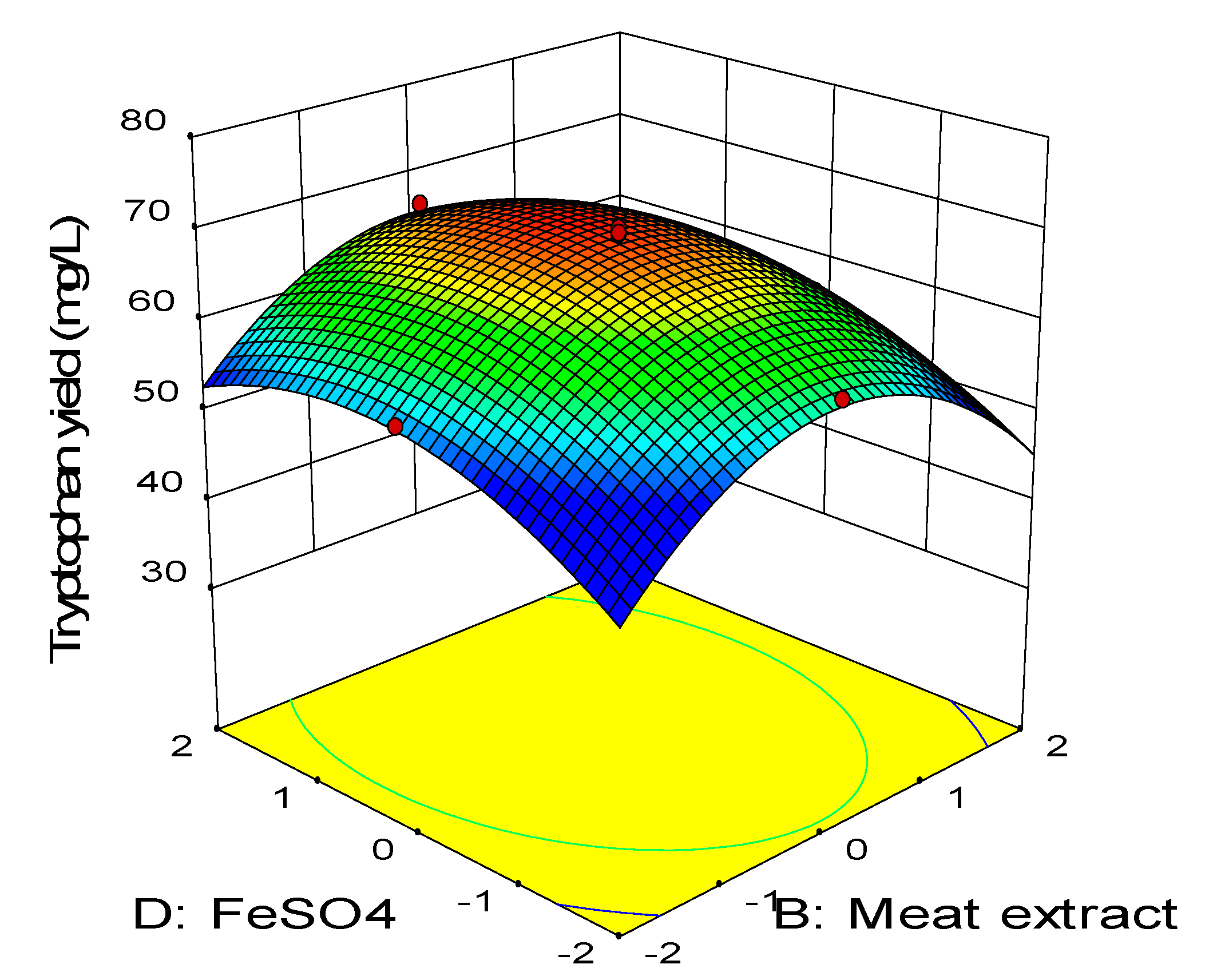
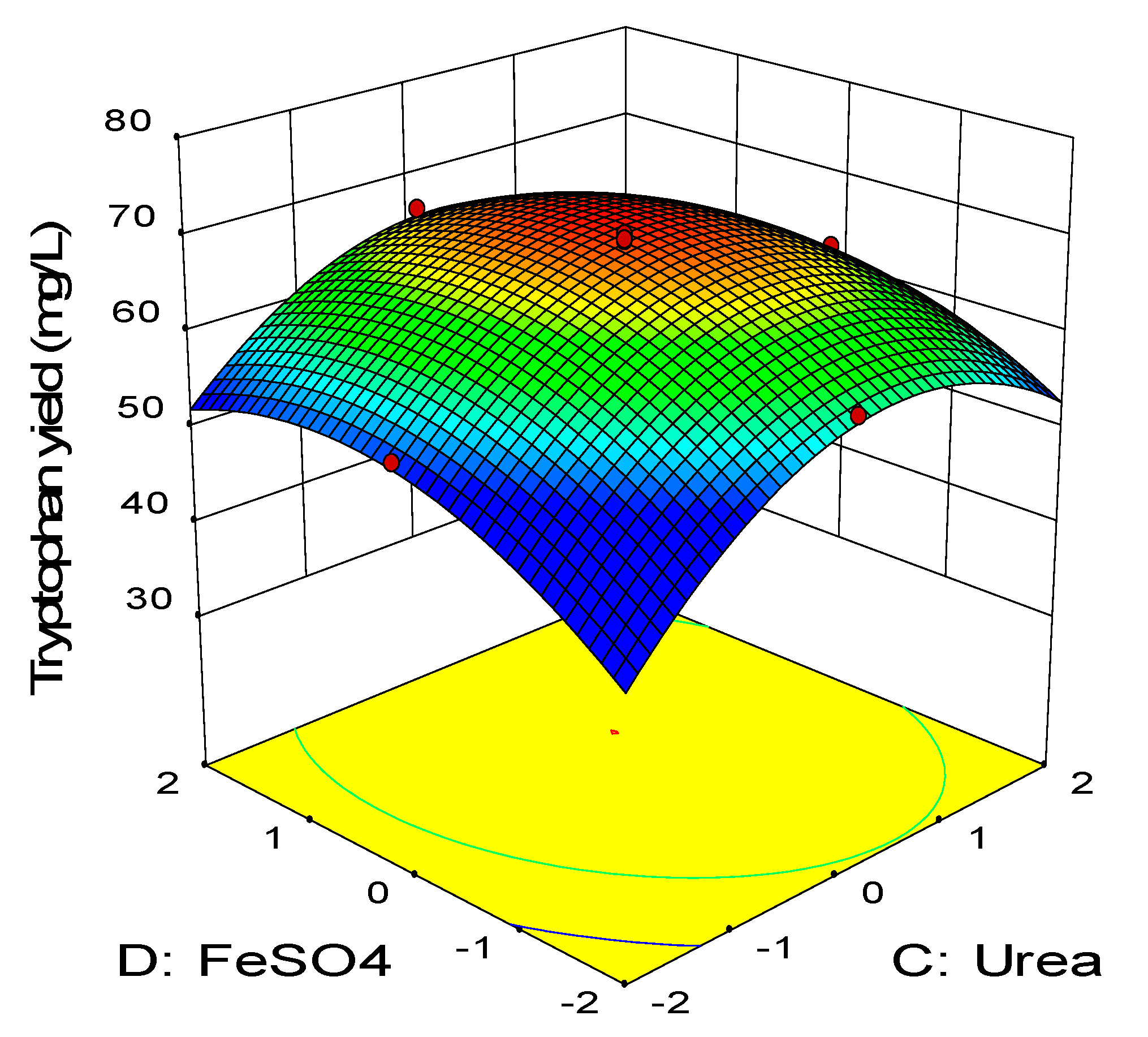
2.3. Central Composite Design
0.11BD − 0.45CD − 2.58A2 − 3.41B2 − 2.88C2 − 1.79D2
3. Materials and Methods
3.1. Inoculum Preparation
3.2. Experimental Design
3.3. Plackett-Burman Design
3.4. Steepest Ascent Method
3.5. Central Composite Design
3.6. Production of Tryptophan
3.7. Analytical Methods
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| LAB | Lactic acid bacteria |
| RSM | Response surface methodology |
| PBD | Plackett-Burman Design |
| CCD | Central Composite Design |
| PLP | Pyridoxal phosphate |
| MRS | deMan, Rogosa and Sharpe |
| SEM | Standard error of mean |
| HPLC | High performance liquid chromatography |
| OPA | o-phthalaldehyde |
| FMOC | 9-fluorenylmethyl chloroformate |
References
- Panda, B.P.; Ali, M.; Javed, S. Fermentation process optimization. Res. J. Microbiol. 2007, 2, 201–208. [Google Scholar]
- Weuster-Botz, D. Experimental design for fermentation media development: Statistical design or global random search. J. Biosci. Bioeng. 2000, 90, 473–483. [Google Scholar] [CrossRef]
- Elibol, M. Optimization of medium composition for actinorhodin production by Streptomyces coelicolor A3 with response surface methodology. Process. Biochem. 2004, 39, 1057–1062. [Google Scholar] [CrossRef]
- Djekrif-Dakhmouche, S.; Gheribi-Aoulmi, Z.; Meraihi, Z.; Bennamoun, L. Application of a statistical design to the optimization of culture medium for α-amylase production by Aspergillus niger ATCC 16404 grown on orange waste powder. J. Food. Eng. 2006, 73, 190–197. [Google Scholar] [CrossRef]
- Chen, H.; Xu, X.Q.; Zhu, Y. Optimization of hydroxyl radical scavenging activity of exo-polysaccharides from Inonotus obliquus in submerged fermentation using response surface methodology. J. Microbiol. Biotechnol. 2010, 20, 835–843. [Google Scholar]
- Vohra, A.; Satyanarayana, T. Statistical optimization of the medium components by response surface methodology to enhance phytase production by Pichia anomala. Process. Biochem. 2002, 37, 999–1004. [Google Scholar] [CrossRef]
- Montserrat, S.; Iñaki, R.; François, O.; Francesc, G.; Carles, C. Application of factorial design to the optimization of medium composition in batch cultures of Streptomyces lividans TK21 producing a hybrid antibiotic. Biotechnol. Lett. 1993, 15, 559–564. [Google Scholar] [CrossRef]
- Faghfuri, E.; Fooladi, J.; Moosavi-Nejad, S.Z. L-tryptophan production by whole cells of Escherichia coli based on Iranian sugar beet molasses. Jundishapur J. Microbiol. 2013, 6, 1–5. [Google Scholar] [CrossRef]
- Cheng, L.K.; Wang, J.; Xu, Q.Y.; Xie, X.X.; Zhang, Y.J.; Zhao, C.G.; Chen, N. Effect of feeding strategy on L-tryptophan production by recombinant Escherichia coli. Ann. Microbiol. 2012, 62, 1625–1634. [Google Scholar] [CrossRef]
- Hagino, H.; Nakayama, K. L-tryptophan production by analog-resistant mutants derived from a phenylalanine and tyrosine double auxotroph of Corynebacterium glutamicum. Agric. Biol. Chem. 1975, 39, 343–349. [Google Scholar]
- Toe, C.J.; Foo, H.L.; Loh, T.C.; Rosfarizan, M.; Raha, A.R.; Zulkifli, I. Extracellular proteolytic activity and amino acid production by lactic acid bacteria Isolated from Malaysian foods. Int. J. Mol. Sci. 2019, 20, 1777. [Google Scholar] [CrossRef] [PubMed]
- Norfarina, M.N.; Mohd Shamzi, M.; Loh, T.C.; Foo, H.L.; Raha, A.R.; Tan, J.S.; Rosfarizan, M. Comparative analyses on medium optimization using one-factor-at-a-time, response surface methodology and artificial neural network for lysine–methionine biosynthesis by Pediococcus pentosaceus RF-1. Biotechnol. Biotechnol. Equip. 2017, 31, 935–947. [Google Scholar]
- Izuddin, W.I.; Loh, T.C.; Foo, H.L.; Samsudin, A.A.; Humam, A.M. Postbiotic, L. Plantarum RG14 improves ruminal epithelium growth, immune status and upregulates the intestinal barrier function in post-weaning lambs. Sci. Rep. 2019, 9, 9938. [Google Scholar] [CrossRef] [PubMed]
- Abdulla, N.R.; Mohd Zamri, A.N.; Sabow, A.B.; Kareem, K.Y.; Nurhazirah, S.; Foo, H.L.; Awis, Q.S.; Loh, T.C. Physico-Chemical properties of breast muscle in broiler chickens fed probiotics, antibiotics or antibiotic–probiotic mix. J. Appl. Anim. Res. 2017, 45, 64–70. [Google Scholar] [CrossRef]
- Kareem, K.Y.; Loh, T.C.; Foo, H.L.; Asmara, S.A.; Akit, H. Influence of postbiotic RG14 and inulin combination on cecal microbiota, organic acid concentration and cytokine expression in broiler chickens. Poult. Sci. 2016, 96, 966–975. [Google Scholar] [CrossRef]
- Kareem, K.Y.; Loh, T.C.; Foo, H.L.; Asmara, S.A.; Akit, H.; Abdulla, N.R.; Ooi, M.F. Carcass, meat and bone quality of broiler chickens fed with postbiotic and prebiotic combinations. Int. J. Probiotzics Prebiotics 2015, 10, 23. [Google Scholar]
- Leuchtenberger, W.; Huthmacher, K.; Drauz, K. Biotechnological production of amino acids and derivatives: Current status and prospects. Appl. Microbiol. Biot. 2005, 69, 1–8. [Google Scholar] [CrossRef]
- Rosebrough, R.W. Crude protein and supplemental dietary tryptophan effects on growth and tissue neurotransmitter levels in the broiler chicken. Br. J. Nutr. 1996, 76, 87–96. [Google Scholar] [CrossRef]
- Moshirfar, A.; Kamara, K.; Castonguay, T.W. Intragastrically administered tryptophan blocks gluconeogenesis in 48-hr starved rats. J. Nutr. Biochem. 1996, 7, 567–570. [Google Scholar] [CrossRef]
- Iwuji, T.C.; Akinmutimi, A.H.; Ogbuewu, I.P.; Etuk, I.F.; Odoemelam, V.U. Roles of tryptophan in monogastric nutrition: A review. Adv. Agric. Sci. Eng. Res. 2014, 4, 1544–1556. [Google Scholar]
- Duarte, K.F.; Junqueira, O.M.; Filardi, R.D.S.; Siqueira, J.C.D.; Puzotti, M.M.; Garcia, E.A.; Molino, A.D.B.; Laurentiz, A.C.D. Digestible tryptophan requirements for broilers from 22 to 42 days old. R. Bras. Zootec. 2013, 42, 728–733. [Google Scholar] [CrossRef]
- Mateus, D.M.R.; Alves, S.S.; Da Fonseca, M.M.R. Kinetics of L-tryptophan production from indole and L-serine catalyzed by whole cells with tryptophanase activity. J. Biosci. Bioeng. 2004, 97, 289–293. [Google Scholar] [CrossRef]
- Turner, E.H.; Loftis, J.M.; Blackwell, A.D. Serotonin a la carte: Supplementation with the serotonin precursor 5-hydroxytryptophan. Pharmacol. Ther. 2006, 109, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Tarek, M.; Hesham, H.M. Screening of potential infants’ Lactobacilli isolates for amino acids production. Afr. J. Microbiol. Res. 2010, 4, 226–232. [Google Scholar]
- Lim, Y.H.; Foo, H.L.; Loh, T.C.; Rosfarizan, M.; Norhani, A. Comparative studies of versatile extracellular proteolytic activities of lactic acid bacteria and their potential for extracellular amino acid productions as feed supplements. J. Anim. Sci. Biotechnol. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Miller, J.N. Experimental design and optimisation (4): Plackett–Burman designs. Anal. Methods 2013, 5, 1901–1903. [Google Scholar]
- Li, Y.; Liu, Z.; Zhao, H.; Xu, Y.; Cui, F. Statistical optimization of xylanase production from new isolated Penicillium oxalicum ZH-30 in submerged fermentation. Biochem. Eng. J. 2007, 34, 82–86. [Google Scholar] [CrossRef]
- Pirie, P.; Naeimpoor, F.; Hejazi, P. A microcosm study on P-Nitrophenol biodegradation in soil slurry by Alcaligenes faecalis: Plackett-Burman design. Iran. J. Chem. Eng. 2011, 8, 57–68. [Google Scholar]
- Voet, D.; Voet, J.G. Biochemistry, 3rd ed.; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2004. [Google Scholar]
- Tavakkoli, M.; Hamidi-Esfahani, Z.; Azizi, M.H. Optimization of Corynebacterium glutamicum glutamic acid production by response surface methodology. Food Bioproc. Tech. 2012, 5, 92–99. [Google Scholar] [CrossRef]
- Li, J.; Ma, C.; Ma, Y.; Li, Y.; Zhou, W.; Xu, P. Medium optimization by combination of response surface methodology and desirability function: An application in glutamine production. Appl. Microbiol. Biotechnol. 2007, 74, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, P.; Heinzle, E.; Wittmann, C. Influence of glucose, fructose and sucrose as carbon sources on kinetics and stoichiometry of lysine production by Corynebacterium glutamicum. J. Ind. Microbiol. Biotechnol. 2002, 28, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Lee, H.W.; Park, J.H.; Ahn, J.O.; Jung, J.K.; Hwang, Y.I. Improved L-threonine production of Escherichia coli mutant by optimization of culture conditions. J. Biosci. Bioeng. 2006, 101, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Ikeda, M. Development of an industrially stable process for L-threonine fermentation by an L-methionine-auxotrophic mutant of Escherichia coli. J. Biosci. Bioeng. 2000, 89, 87–89. [Google Scholar] [CrossRef]
- Chen, N.; Huang, J.; Feng, Z.B.; Yu, L.; Xu, Q.Y.; Wen, T.Y. Optimization of fermentation conditions for the biosynthesis of L-threonine by Escherichia coli. Appl. Biochem. Biotechnol. 2009, 158, 595–604. [Google Scholar] [CrossRef]
- Wang, J.; Cheng, L.K.; Chen, N. High-level production of L-threonine by recombinant Escherichia coli with combined feeding strategies. Biotechnol. Biotechnol. Equip. 2014, 28, 495–501. [Google Scholar] [CrossRef]
- Roy, D.K.; Chatterjee, S.P. Production of glutamic acid by Arthrobacter globiformis: Influence of cultural conditions. Folia Microbiol. 1989, 34, 11–24. [Google Scholar] [CrossRef]
- Coruzzi, G.; Last, R.; Dudareva, N.; Amrhein, N. Amino Acids. In Biochemistry and Molecular Biology of Plants; Buchanan, B.B., Gruissem, W., Jones, R.L., Eds.; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 289–336. [Google Scholar]
- Alexander, J.C.; Beckneh, C.; Elvehjem, C.A. The alanine, cystine, glycine and serine content of meat. J. Nutr. 1953, 51, 319–328. [Google Scholar] [CrossRef]
- Powers, H.J. Riboflavin (vitamin B-2) and health. Am. J. Clin. Nutr. 2003, 77, 1352–1360. [Google Scholar] [CrossRef]
- Osborne, A.; Thorneley, R.N.; Abell, C.; Bornemann, S. Studies with substrate and cofactor analogues provide evidence for a radical mechanism in the chorismate synthase reaction. J. Biol. Chem. 2000, 275, 35825–35830. [Google Scholar] [CrossRef]
- Lin, X.; Xu, S.; Yang, Y.; Wu, J.; Wang, H.; Shen, H.; Wang, H. Purification and characterization of anthranilate synthase component I (TrpE) from Mycobacterium tuberculosis H37Rv. Protein Expr. Purif. 2009, 64, 8–15. [Google Scholar] [CrossRef]
- Davati, N.; Hamidi Esfahani, Z.; Shojaosadati, S.A. Optimization of medium composition for microbial production of glutamic acid from Date fruit wastes using fractional factorial method. Iran. J. Food Sci. Technol. 2010, 7, 61–67. [Google Scholar]
- Nampoothiri, K.M.; Pandey, A. Solid state fermentation for L-glutamic acid production using Brevibacterium sp. Biotechnol. Lett. 1996, 18, 199–204. [Google Scholar] [CrossRef]
- Zareian, M.; Ebrahimpour, A.; Bakar, F.A.; Mohamed, A.K.S.; Forghani, B.; Ab-Kadir, M.S.B.; Saari, N. A glutamic acid-producing lactic acid bacteria isolated from Malaysian fermented foods. Int. J. Mol. Sci. 2012, 13, 5482–5497. [Google Scholar] [CrossRef]
- Ikeda, M.; Katsumata, R. Metabolic engineering to produce tyrosine or phenylalanine in a tryptophan-producing Corynebacterium glutamicum strain. Appl. Environ. Microbiol. 1992, 58, 781–785. [Google Scholar] [CrossRef]
- Rodwell, V.; Bender, D.; Botham, K.M.; Kennelly, P.J.; Weil, P.A. Harper’s Illustrated Biochemistry, 30th ed.; McGraw-Hill Education: New York, NY, USA, 2015. [Google Scholar]
- Zalkin, H.; Kling, D. Anthranilate synthetase. Purification and properties of component I from Salmonella typhimurium. Biochemistry 1968, 7, 3566–3573. [Google Scholar] [CrossRef] [PubMed]
- Hertel, S.C.; Hieke, M.; Gröger, D. Anthranilate synthase from Ruta graveolens: Partial purification and properties. Biochem. Physiol. Pflanz. 1991, 187, 121–129. [Google Scholar] [CrossRef]
- Widholm, J. Anthranilate synthetase from 5-methyltryptophan-susceptible and-resistant cultured Daucus carota cells. Biochim. Biophys. Acta. 1972, 279, 48–57. [Google Scholar] [CrossRef]
- Momose, H.; Takagi, T. Glutamic acid production in biotin-rich media by temperature-sensitive mutants of Brevibacterium lactofermentum, a novel fermentation process. Agric. Biol. Chem. 1978, 42, 1911–1917. [Google Scholar] [CrossRef][Green Version]
- Todorov, S.D.; Dicks, L.M. Bacteriocin production by Lactobacillus pentosus ST712BZ isolated from boza. Braz. J. Microbiol. 2007, 38, 166–172. [Google Scholar] [CrossRef]
- Saraniya, A.; Jeevaratnam, K. Optimization of nutritional and non-nutritional factors involved for production of antimicrobial compounds from Lactobacillus pentosus SJ65 using response surface methodology. Braz. J. Microbiol. 2014, 45, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.H.; Salam, A.I. Current limitations and challenges with lactic acid bacteria: A review. Food Nutr. Sci. 2013, 4, 73–87. [Google Scholar]
- de Carvalho, A.A.T.; Mantovani, H.C.; Paiva, A.D.; De Melo, M.R. The effect of carbon and nitrogen sources on bovicin HC5 production by Streptococcus bovis HC5. J. Appl. Microbiol. 2009, 107, 339–347. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oh, H.; Wee, Y.J.; Yun, J.S.; Han, S.H.; Jung, S.; Ryu, H.W. Lactic acid production from agricultural resources as cheap raw materials. Bioresour. Technol. 2005, 96, 1492–1498. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, I.A.; Loh, T.C.; Foo, H.L.; Lau, W.H.; Awis, Q.S. Biodegradation of palm kernel cake by cellulolytic and hemicellulolytic bacterial cultures through solid state fermentation. Sci. World J. 2014, 2014, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.; Teixeira, J.; Oliveira, R.; Van Der Mei, H.C. Response surface optimization of the medium components for the production of biosurfactants by probiotic bacteria. Process. Biochem. 2006, 41, 1–10. [Google Scholar] [CrossRef]
- Ooi, M.F.; Nurzafirah, M.; Foo, H.L.; Loh, T.C.; Rosfarizan, M.; Raha, A.R.; Arbakariya, A. Effects of carbon and nitrogen sources on bacteriocin-inhibitory activity of postbiotic metabolites produced by Lactobacillus plantarum I-UL4. Malays. J. Microbiol. 2015, 11, 176–184. [Google Scholar]
- Lim, Y.H.; Foo, H.L.; Loh, T.C.; Rosfarizan, M.; Raha, A.R.; Zulkifli, I. Optimized medium via statistical approach enhanced threonine production by Pediococcus pentosaceus TL-3 isolated from Malaysian food. Microb. Cell Fact. 2019, 18, 1–19. [Google Scholar] [CrossRef]
- Hwang, C.F.; Chang, J.H.; Houng, J.Y.; Tsai, C.C.; Lin, C.K.; Tsen, H.Y. Optimization of medium composition for improving biomass production of Lactobacillus plantarum Pi06 using the Taguchi array design and the Box-Behnken method. Biotechnol. Bioprocess. Eng. 2012, 17, 827–834. [Google Scholar] [CrossRef]
- Hutkins, R.W. Microbiology and Technology of Fermented Foods; Blackwell Publishing: Ames, IA, USA, 2008; Volume 22. [Google Scholar]
- de Carvalho, I.P.C.; Detmann, E.; Mantovani, H.C.; Paulino, M.F.; Valadares Filho, S.D.C.; Costa, V.A.C.; Gomes, D.I. Growth and antimicrobial activity of lactic acid bacteria from rumen fluid according to energy or nitrogen source. Rev. Bras. Zootec. 2011, 40, 1260–1265. [Google Scholar] [CrossRef]
- Thu, T.V.; Foo, H.L.; Loh, T.C.; Bejo, M.H. Inhibitory activity and organic acid concentrations of metabolite combinations produced by various strains of Lactobacillus plantarum. Afr. J. Biotechnol. 2011, 10, 1359–1363. [Google Scholar]
- Tomas, J.M.S.; Bru, E.; Nader-Macia, M.E. Different combinations of salts affect the growth and bacteriocin production by Lactobacillus salivarius CRL 1328. J. Chem. Technol. Biotechnol. 2010, 85, 91–99. [Google Scholar] [CrossRef]
- Foucaud, C.; Francois, A.; Richard, J. Development of a chemically defined medium for the growth of Leuconostoc mesenteroides. Appl. Environ. Microbiol. 1997, 63, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Zhang, L.W.; Du, M.; Han, X.; Yi, H.X.; Guo, C.F.; Zhang, Y.C.; Luo, X.; Zhang, Y.H.; Shan, Y.J.; et al. Effect of tween series on growth and cis-9, trans-11 conjugated linoleic acid production of Lactobacillus acidophilus F0221 in the presence of bile salts. Int. J. Mol. Sci. 2011, 12, 9138–9154. [Google Scholar] [CrossRef]
- Oh, S.; Rheem, S.; Sim, J.; Kim, S.; Baek, Y. Optimizing conditions for the growth of Lactobacillus casei YIT 9018 in tryptone-yeast extract-glucose medium by using response surface methodology. Appl. Environ. Microbiol. 1995, 61, 3809–3814. [Google Scholar] [CrossRef] [PubMed]
- Tripuraneni, S. Effect of Nutrient Supplements on Cucumber Fermentation by Lactic Acid Bacteria. Master’s Thesis, University of Arkansas, Little Rock, AR, USA, 2011. [Google Scholar]
- Rosen, K. Production of Baker’s Yeast. In Yeast Biotechnology; Berry, D.R., Russell, I., Steward, G.C., Eds.; Unwin Hyman Ltd.: London, UK, 2012; pp. 471–500. [Google Scholar]
- Simova, E.; Simov, Z.; Beshkova, D.; Frengova, G.; Dimitrov, Z.; Spasov, Z. Amino acid profiles of lactic acid bacteria, isolated from kefir grains and kefir starter made from them. Int. J. Food. Microbiol. 2006, 107, 112–123. [Google Scholar] [CrossRef]
- Lim, Y.S. Isolation of Bacteriocinogenic Lactic Acid Bacteria and Purification of Selected Bacteriocins from Traditional Fermented Foods. Master’s Thesis, Universiti Putra Malaysia, Serdang, Malaysia, 2003. [Google Scholar]
- Kareem, K.Y.; Foo, H.L.; Loh, T.C.; Ooi, M.F.; Asmara, S.A. Inhibitory activity of postbiotic produced by strains of Lactobacillus plantarum using reconstituted media supplemented with inulin. Gut. Pathog. 2014, 6, 1–7. [Google Scholar] [CrossRef]
- Azin, A.; Rosfarizan, M.; Raha, A.R.; Rosli, M.I.; Farideh, N.; Tan, J.S.; Sahar, A. Cyclodextrin glycosyltransferase biosynthesis improvement by recombinant Lactococcus lactis NZ: NSP: CGT: Medium formulation and culture condition optimization. Biotechnol. Biotechnol. Equip. 2015, 29, 555–563. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |
| Std Run | A | B | C | D | E | F | G | H | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Tryptophan Production (mg/L) | Cell Population (logCFU/mL) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 1 | 1 | -1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 0.00 ± 0.00 H | 8.47 ± 0.04 I |
| 2 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | 0.00 ± 0.00 H | 9.26 ± 0.01 B |
| 3 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | 0.42 ± 0.05 G | 8.82 ± 0.03 F |
| 4 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | 0.34 ± 0.02 G | 8.77 ± 0.04 F |
| 5 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | 9.35 ± 0.05 D | 9.10 ± 0.02 D |
| 6 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 0.00 ± 0.00 H | 9.04 ± 0.01 E |
| 7 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | 6.81 ± 0.07 E | 8.70 ± 0.01 G |
| 8 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 16.69 ± 0.51 C | 8.75 ± 0.02 FG |
| 9 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 0.00 ± 0.00 H | 7.33 ± 0.01 M |
| 10 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 0.00 ± 0.00 H | 7.89 ± 0.02 L |
| 11 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 0.00 ± 0.00 H | 9.22 ± 0.02 BC |
| 12 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 0.00 ± 0.00 H | 8.79 ± 0.02 F |
| 13 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | 0.00 ± 0.00 H | 9.02 ± 0.03 E |
| 14 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | 4.85 ± 0.04 F | 8.60 ± 0.00 H |
| 15 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 22.94 ± 0.79 B | 9.19 ± 0.01 C |
| 16 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 0.00 ± 0.00 H | 8.75 ± 0.03 FG |
| 17 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | 0.00 ± 0.00 H | 8.75 ± 0.02 FG |
| 18 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 21.27 ± 0.24 B | 7.88 ± 0.02 L |
| 19 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | 10.58 ± 0.25 D | 8.47 ± 0.01 K |
| 20 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 0.00 ± 0.00 H | 8.50 ± 0.01 IJ |
| 21 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 0.00 ± 0.00 H | 9.11 ± 0.01 D |
| 22 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 0.00 ± 0.00 H | 9.34 ± 0.02 AB |
| 23 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 0.35 ± 0.01 G | 9.05 ± 0.01 E |
| 24 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | 0.00 ± 0.00 H | 7.33 ± 0.02 M |
| MRS | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | −1 | −1 | −1 | −1 | 29.41 ± 0.76 A | 9.41 ± 0.01 A |
| Source | Sum of Squares | df | Mean Square | F Value | p-Value Prob > F | |
|---|---|---|---|---|---|---|
| Model | 2428.91 | 21 | 115.66 | 520.43 | <0.01 | significant |
| A-Glucose | 6.13 | 1 | 6.13 | 27.56 | 0.03 | significant |
| B-Sucrose | 433.36 | 1 | 433.36 | 1949.90 | <0.01 | significant |
| C-Fructose | 0.61 | 1 | 0.61 | 2.76 | 0.24 | |
| D-Lactose | 119.36 | 1 | 119.36 | 537.05 | <0.01 | significant |
| E-Molasses | 98.68 | 1 | 98.68 | 444.03 | <0.01 | significant |
| F-Yeast extract | 156.85 | 1 | 156.85 | 705.75 | <0.01 | significant |
| G-Peptone | 70.03 | 1 | 70.03 | 315.12 | <0.01 | significant |
| H-Meat extract | 592.62 | 1 | 592.62 | 2666.50 | <0.01 | significant |
| J-K2HPO4 | 47.76 | 1 | 47.76 | 214.88 | <0.01 | significant |
| L-Urea | 93.75 | 1 | 93.75 | 421.83 | <0.01 | significant |
| M-NH4NO3 | 90.83 | 1 | 90.83 | 408.69 | <0.01 | significant |
| N-(NH4)2SO4 | 54.88 | 1 | 54.88 | 246.92 | <0.01 | significant |
| O-(NH4)2HC6H5O7 | 77.00 | 1 | 77.00 | 346.49 | <0.01 | significant |
| P-NaOAc | 131.64 | 1 | 131.64 | 592.30 | <0.01 | significant |
| Q-MgSO4 | 7.54 | 1 | 7.54 | 33.93 | 0.03 | significant |
| R-MnSO4 | 97.21 | 1 | 97.21 | 437.39 | <0.01 | significant |
| S-Tween 80 | 2.04 | 1 | 2.04 | 9.19 | 0.09 | |
| T-FeSO4 | 188.94 | 1 | 188.94 | 850.15 | <0.01 | significant |
| V-CuSO4 | 92.90 | 1 | 92.90 | 418.02 | <0.01 | significant |
| W-Biotin | 2.09 | 1 | 2.09 | 9.42 | 0.09 | |
| X-dummy | 64.69 | 1 | 64.69 | 291.09 | <0.01 | significant |
| Residual | 0.44 | 2 | 0.22 | |||
| Cor Total | 2429.36 | 23 |
| Source | Sum of Squares | df | Mean Square | F Value | p-Value Prob > F | |
|---|---|---|---|---|---|---|
| Model | 7.15 | 19 | 0.38 | 148.54 | <0.01 | significant |
| A-Glucose | 0.68 | 1 | 0.68 | 269.27 | <0.01 | significant |
| B-Sucrose | 0.16 | 1 | 0.16 | 63.98 | <0.01 | significant |
| C-Fructose | 0.01 | 1 | 0.01 | 4.88 | 0.09 | |
| E-Molasses | 0.27 | 1 | 0.27 | 107.47 | <0.01 | significant |
| F-Yeast extract | 1.83 | 1 | 1.83 | 721.80 | <0.01 | significant |
| G-Peptone | 0.29 | 1 | 0.29 | 112.71 | <0.01 | significant |
| H-Meat extract | 1.05 | 1 | 1.05 | 414.04 | <0.01 | significant |
| J-K2HPO4 | 0.04 | 1 | 0.04 | 15.23 | 0.02 | significant |
| K-KH2PO4 | 0.01 | 1 | 0.01 | 5.82 | 0.07 | |
| N-(NH4)2SO4 | 0.30 | 1 | 0.30 | 119.34 | <0.01 | significant |
| O-(NH4)2HC6H5O7 | 0.46 | 1 | 0.46 | 179.90 | <0.01 | significant |
| P-NaOAc | 0.51 | 1 | 0.51 | 200.63 | <0.01 | significant |
| Q-MgSO4 | 0.32 | 1 | 0.32 | 125.26 | <0.01 | significant |
| R-MnSO4 | 0.25 | 1 | 0.25 | 100.07 | <0.01 | significant |
| S-Tween 80 | 0.03 | 1 | 0.03 | 13.04 | 0.02 | significant |
| U-ZnSO4 | 0.02 | 1 | 0.02 | 9.40 | 0.04 | significant |
| V-CuSO4 | 0.02 | 1 | 0.02 | 8.15 | 0.05 | significant |
| W-Biotin | 0.31 | 1 | 0.31 | 123.62 | <0.01 | significant |
| X-dummy | 0.58 | 1 | 0.58 | 227.63 | <0.01 | significant |
| Residual | 0.01 | 4 | 0.00 | |||
| Cor Total | 7.16 | 23 |
| Media | Cell Population (Log CFU/mL) | Tryptophan Production (mg/L) | Serine Consumption (mg/L) |
|---|---|---|---|
| 1 | 8.95 ± 0.01 B | 26.07 ± 0.86 AB | 9.95 ± 0.43 A |
| 2 | 7.98 ± 0.01 D | 25.16 ± 0.39 B | 11.75 ± 0.86 A |
| 3 | 8.00 ± 0.01 D | 25.00 ± 0.20 B | 10.09 ± 0.73 A |
| 4 | 8.06 ± 0.01 C | 25.89 ± 1.05 AB | 12.43 ± 0.50 A |
| 5 | 8.99 ± 0.02 B | 25.95 ± 0.18 AB | 11.40 ± 0.45 A |
| MRS | 9.43 ± 0.01 A | 26.81 ± 4.86 A | 11.47 ± 1.29 A |
| Run | Cell Population (log CFU/mL) | Tryptophan Production (mg/L) | Serine Consumption (mg/L) |
|---|---|---|---|
| 1 | 9.10 ± 0.02 D | 27.73 ± 0.04 G | 8.72 ± 0.21 D |
| 2 | 9.10 ± 0.01 D | 35.79 ± 0.28 F | 9.05 ± 0.35 DE |
| 3 | 9.14 ± 0.01 CD | 42.64 ± 0.40 E | 9.24 ± 0.39 DE |
| 4 | 9.15 ± 0.01 C | 55.54 ± 0.36 C | 8.93 ± 0.01 DE |
| 5 | 9.24 ± 0.01 B | 69.05 ± 0.55 A | 9.53 ± 0.09 DEF |
| 6 | 9.24 ± 0.01 B | 59.84 ± 0.68 B | 10.15 ± 0.12 F |
| 7 | 9.25 ± 0.01 B | 51.67 ± 0.92 D | 9.65 ± 0.21 EF |
| 8 | 9.18 ± 0.01 C | 43.77 ± 0.23 E | 9.00 ± 0.11 DE |
| 9 | 9.16 ± 0.01 C | 36.74 ± 0.91 F | 7.56 ± 0.16 C |
| 10 | 9.04 ± 0.01 E | 21.98 ± 0.38 H | 4.21 ± 0.10 A |
| 11 | 8.77 ± 0.03 F | 10.02 ± 0.07 I | 5.11 ± 0.45 B |
| MRS | 9.37 ± 0.01 A | 27.69 ± 0.15 G | 7.69 ± 0.37 C |
| Std Run | A | B | C | D | Tryptophan Production (mg/L) | |
|---|---|---|---|---|---|---|
| Experimental | Predicted * | |||||
| 1 | −1 | −1 | −1 | −1 | 56.24 ± 0.04 JKL | 56.86 |
| 2 | 1 | −1 | −1 | −1 | 54.33 ± 0.20 N | 54.44 |
| 3 | −1 | 1 | −1 | −1 | 53.23 ± 0.14 O | 53.54 |
| 4 | 1 | 1 | −1 | −1 | 54.49 ± 0.26 N | 54.72 |
| 5 | −1 | −1 | 1 | −1 | 60.62 ± 0.17 G | 60.38 |
| 6 | 1 | −1 | 1 | −1 | 57.12 ± 0.23 IJK | 57.64 |
| 7 | −1 | 1 | 1 | −1 | 59.14 ± 0.32 H | 59.38 |
| 8 | 1 | 1 | 1 | −1 | 59.69 ± 0.26 GH | 60.24 |
| 9 | −1 | −1 | −1 | 1 | 63.25 ± 0.35 E | 63.20 |
| 10 | 1 | −1 | −1 | 1 | 56.32 ± 0.44 JKL | 56.78 |
| 11 | −1 | 1 | −1 | 1 | 59.25 ± 0.25 H | 59.44 |
| 12 | 1 | 1 | −1 | 1 | 55.89 ± 0.85 LM | 56.62 |
| 13 | −1 | −1 | 1 | 1 | 64.47 ± 0.40 D | 64.92 |
| 14 | 1 | −1 | 1 | 1 | 58.01 ± 0.56 I | 58.18 |
| 15 | −1 | 1 | 1 | 1 | 63.13 ± 0.68 EF | 63.48 |
| 16 | 1 | 1 | 1 | 1 | 60.25 ± 0.44 GH | 60.34 |
| 17 | −2 | 0 | 0 | 0 | 62.23 ± 0.29 EF | 61.88 |
| 18 | 2 | 0 | 0 | 0 | 57.19 ± 0.33 IJ | 56.32 |
| 19 | 0 | −2 | 0 | 0 | 56.83 ± 0.12 IJKL | 56.36 |
| 20 | 0 | 2 | 0 | 0 | 55.95 ± 0.13 KLM | 55.20 |
| 21 | 0 | 0 | −2 | 0 | 55.03 ± 0.22 MN | 54.28 |
| 22 | 0 | 0 | 2 | 0 | 61.99 ± 0.42 F | 61.52 |
| 23 | 0 | 0 | 0 | −2 | 59.66 ± 0.27 GH | 59.04 |
| 24 | 0 | 0 | 0 | 2 | 66.07 ± 0.05 C | 65.48 |
| 25 | 0 | 0 | 0 | 0 | 69.33 ± 0.10 AB | 69.42 |
| 26 | 0 | 0 | 0 | 0 | 69.55 ± 0.31 AB | 69.42 |
| 27 | 0 | 0 | 0 | 0 | 70.22 ± 0.07 A | 69.42 |
| 28 | 0 | 0 | 0 | 0 | 68.85 ± 0.39 B | 69.42 |
| 29 | 0 | 0 | 0 | 0 | 69.69 ± 0.55 AB | 69.42 |
| 30 | 0 | 0 | 0 | 0 | 68.88 ± 0.94 B | 69.42 |
| MRS | - | - | - | - | 28.18 ± 0.12 P | - |
| Source | Sequential | Lack of Fit | Adjusted | Predicted | |
|---|---|---|---|---|---|
| p-Value | p-Value | R-Squared | R-Squared | ||
| Linear | 0.1492 | <0.0001 | 0.1059 | 0.0576 | |
| Crossproduct | 0.9724 | <0.0001 | −0.1063 | −0.3333 | |
| Quadratic | <0.0001 | 0.2196 | 0.9837 | 0.9586 | Suggested |
| Cubic | 0.9875 | 0.0265 | 0.9709 | 0.2223 | Aliased |
| Source | Sum of Squares | df | Mean Square | F-Value | p-value Prob > F | |
|---|---|---|---|---|---|---|
| Model | 814.96 | 14 | 58.21 | 126.07 | < 0.01 | significant |
| A | 46.16 | 1 | 46.16 | 99.97 | < 0.01 | significant |
| B | 2.08 | 1 | 2.08 | 4.51 | 0.05 | |
| C | 78.3 | 1 | 78.3 | 169.58 | < 0.01 | significant |
| D | 61.84 | 1 | 61.84 | 133.92 | < 0.01 | significant |
| AB | 12.86 | 1 | 12.86 | 27.84 | < 0.01 | significant |
| AC | 0.11 | 1 | 0.11 | 0.24 | 0.63 | |
| AD | 16.07 | 1 | 16.07 | 34.8 | < 0.01 | significant |
| BC | 5.36 | 1 | 5.36 | 11.62 | <0.01 | significant |
| BD | 0.2 | 1 | 0.2 | 0.42 | 0.53 | |
| CD | 3.17 | 1 | 3.17 | 6.86 | 0.02 | significant |
| A2 | 182.37 | 1 | 182.37 | 394.96 | < 0.01 | significant |
| B2 | 318.65 | 1 | 318.65 | 690.1 | < 0.01 | significant |
| C2 | 227.15 | 1 | 227.15 | 491.94 | < 0.01 | significant |
| D2 | 87.9 | 1 | 87.9 | 190.37 | < 0.01 | significant |
| Residual | 6.93 | 15 | 0.46 | |||
| Lack of Fit | 5.57 | 10 | 0.56 | 2.06 | 0.22 | not significant |
| Pure Error | 1.35 | 5 | 0.27 | |||
| Cor Total | 821.88 | 29 |
| Variables | Symbol Code | Unit | Coded Values | |
|---|---|---|---|---|
| −1 | +1 | |||
| Glucose | A | g/L | 0 | 20 |
| Sucrose | B | g/L | 0 | 17.69 |
| Fructose | C | g/L | 0 | 19.08 |
| Lactose | D | g/L | 0 | 18.86 |
| Molasses | E | g/L | 0 | 25.08 |
| Yeast extract | F | g/L | 0 | 4 |
| Peptone | G | g/L | 0 | 10 |
| Meat extract | H | g/L | 0 | 8 |
| K2HPO4 | J | g/L | 0 | 2 |
| KH2PO4 | K | g/L | 0 | 2 |
| Urea | L | g/L | 0 | 3 |
| NH4NO3 | M | g/L | 0 | 5 |
| (NH4)2SO4 | N | g/L | 0 | 5 |
| (NH4)2HC6H5O7 | O | g/L | 0 | 2 |
| NaOAc | P | g/L | 0 | 5 |
| MgSO4 | Q | g/L | 0 | 0.2 |
| MnSO4 | R | g/L | 0 | 0.04 |
| Tween 80 | S | mL/L | 0 | 1 |
| FeSO4 | T | g/L | 0 | 0.01 |
| ZnSO4 | U | g/L | 0 | 0.01 |
| CuSO4 | V | g/L | 0 | 0.01 |
| Biotin | W | g/L | 0 | 0.06 |
| Std Run | A | B | C | D | E | F | G | H | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 |
| 2 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 |
| 3 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 |
| 4 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 |
| 5 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 |
| 6 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 |
| 7 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 |
| 8 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 |
| 9 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 |
| 10 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 |
| 11 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 |
| 12 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 |
| 13 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 |
| 14 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 |
| 15 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 |
| 16 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 |
| 17 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | 1 |
| 18 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 | −1 |
| 19 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1 |
| 20 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 |
| 21 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1 |
| 22 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 |
| 23 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 |
| 24 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 |
| Media Formulation | Medium Composition, g/L |
|---|---|
| Medium 1 | |
| Glucose | 20 |
| Sucrose | 17.69 |
| Lactose | 18.86 |
| Molasses | 25.08 |
| Yeast extract | 4 |
| Peptone | 10 |
| Meat extract | 8 |
| K2HPO4 | 2 |
| Urea | 3 |
| (NH4)2SO4 | 5 |
| (NH4)2HC6H5O7 | 2 |
| NaOAc | 5 |
| MgSO4 | 0.2 |
| MnSO4 | 0.04 |
| FeSO4 | 0.01 |
| CuSO4 | 0.01 |
| Medium 2 | |
| Meat extract | 8 |
| FeSO4 | 0.01 |
| Lactose | 18.86 |
| MnSO4 | 0.04 |
| Urea | 3 |
| CuSO4 | 0.01 |
| K2HPO4 | 2 |
| MgSO4 | 0.2 |
| Medium 3 | |
| Meat extract | 8 |
| FeSO4 | 0.01 |
| Lactose | 18.86 |
| MnSO4 | 0.04 |
| Urea | 3 |
| CuSO4 | 0.01 |
| K2HPO4 | 2 |
| MgSO4 | 0.2 |
| Biotin | 0.06 |
| Tween 80 | 1 |
| Medium 4 | |
| Lactose | 18.86 |
| Meat extract | 8 |
| Urea | 3 |
| FeSO4 | 0.01 |
| Medium 5 | |
| Molasses | 25.08 |
| Meat extract | 8 |
| Urea | 3 |
| FeSO4 | 0.01 |
| No. | Run | Variable level, g/L | |||
|---|---|---|---|---|---|
| Molasses (A) | Meat Extract (B) | Urea (C) | FeSO4 (D) | ||
| Δ | −2.51 | 4 | 0.6 | 0.003 | |
| 1 | Origin | 25.08 | 8 | 3.0 | 0.010 |
| 2 | Origin + Δ | 22.57 | 12 | 3.6 | 0.013 |
| 3 | Origin + 2Δ | 20.06 | 16 | 4.2 | 0.016 |
| 4 | Origin + 3Δ | 17.55 | 20 | 4.8 | 0.019 |
| 5 | Origin + 4Δ | 15.04 | 24 | 5.4 | 0.022 |
| 6 | Origin + 5Δ | 12.53 | 28 | 6.0 | 0.025 |
| 7 | Origin + 6Δ | 10.02 | 32 | 6.6 | 0.028 |
| 8 | Origin + 7Δ | 7.51 | 36 | 7.2 | 0.031 |
| 9 | Origin + 8Δ | 5.00 | 40 | 7.8 | 0.034 |
| 10 | Origin + 9Δ | 2.49 | 44 | 8.4 | 0.037 |
| 11 | Origin + 10Δ | 0 | 48 | 9.0 | 0.040 |
| Variables | Coded Symbol | Coded Values | ||||
|---|---|---|---|---|---|---|
| −α | −1 | 0 | +1 | +α | ||
| Molasses | A | 10.02 | 12.53 | 15.04 | 17.55 | 20.06 |
| Meat extract | B | 16 | 20 | 24 | 28 | 32 |
| Urea | C | 4.2 | 4.8 | 5.4 | 6 | 6.6 |
| FeSO4 | D | 0.016 | 0.019 | 0.022 | 0.025 | 0.028 |
| Std Run | A | B | C | D |
|---|---|---|---|---|
| 1 | −1 | −1 | −1 | −1 |
| 2 | 1 | −1 | −1 | −1 |
| 3 | −1 | 1 | −1 | −1 |
| 4 | 1 | 1 | −1 | −1 |
| 5 | −1 | −1 | 1 | −1 |
| 6 | 1 | −1 | 1 | −1 |
| 7 | −1 | 1 | 1 | −1 |
| 8 | 1 | 1 | 1 | −1 |
| 9 | −1 | −1 | −1 | 1 |
| 10 | 1 | −1 | −1 | 1 |
| 11 | −1 | 1 | −1 | 1 |
| 12 | 1 | 1 | −1 | 1 |
| 13 | −1 | −1 | 1 | 1 |
| 14 | 1 | −1 | 1 | 1 |
| 15 | −1 | 1 | 1 | 1 |
| 16 | 1 | 1 | 1 | 1 |
| 17 | −2 | 0 | 0 | 0 |
| 18 | 2 | 0 | 0 | 0 |
| 19 | 0 | −2 | 0 | 0 |
| 20 | 0 | 2 | 0 | 0 |
| 21 | 0 | 0 | −2 | 0 |
| 22 | 0 | 0 | 2 | 0 |
| 23 | 0 | 0 | 0 | −2 |
| 24 | 0 | 0 | 0 | 2 |
| 25 | 0 | 0 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 |
| 28 | 0 | 0 | 0 | 0 |
| 29 | 0 | 0 | 0 | 0 |
| 30 | 0 | 0 | 0 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, Y.H.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdul Rahim, R. Rapid Evaluation and Optimization of Medium Components Governing Tryptophan Production by Pediococcus acidilactici TP-6 Isolated from Malaysian Food via Statistical Approaches. Molecules 2020, 25, 779. https://doi.org/10.3390/molecules25040779
Lim YH, Foo HL, Loh TC, Mohamad R, Abdul Rahim R. Rapid Evaluation and Optimization of Medium Components Governing Tryptophan Production by Pediococcus acidilactici TP-6 Isolated from Malaysian Food via Statistical Approaches. Molecules. 2020; 25(4):779. https://doi.org/10.3390/molecules25040779
Chicago/Turabian StyleLim, Ye Heng, Hooi Ling Foo, Teck Chwen Loh, Rosfarizan Mohamad, and Raha Abdul Rahim. 2020. "Rapid Evaluation and Optimization of Medium Components Governing Tryptophan Production by Pediococcus acidilactici TP-6 Isolated from Malaysian Food via Statistical Approaches" Molecules 25, no. 4: 779. https://doi.org/10.3390/molecules25040779
APA StyleLim, Y. H., Foo, H. L., Loh, T. C., Mohamad, R., & Abdul Rahim, R. (2020). Rapid Evaluation and Optimization of Medium Components Governing Tryptophan Production by Pediococcus acidilactici TP-6 Isolated from Malaysian Food via Statistical Approaches. Molecules, 25(4), 779. https://doi.org/10.3390/molecules25040779






