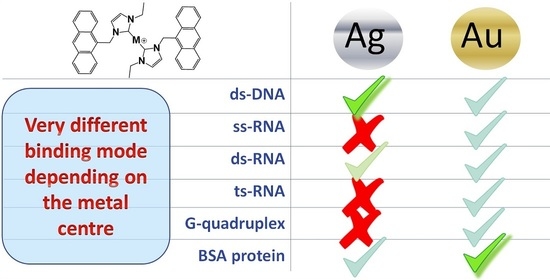
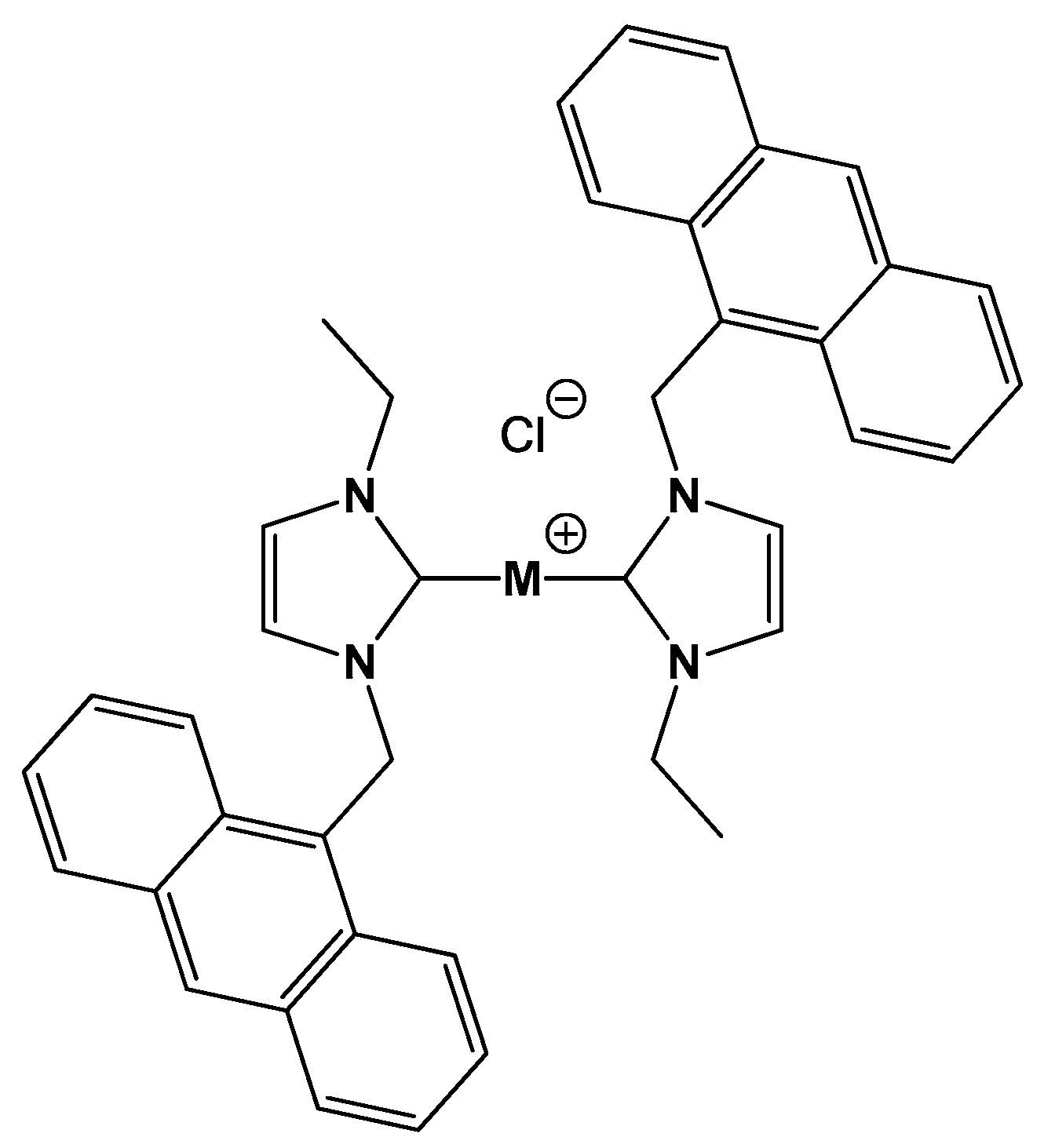
On the Different Mode of Action of Au(I)/Ag(I)-NHC Bis-Anthracenyl Complexes Towards Selected Target Biomolecules
Abstract
1. Introduction
2. Results and Discussion
2.1. Interaction with Natural Double-Stranded DNA
2.1.1. Absorbance and Fluorescence Titrations
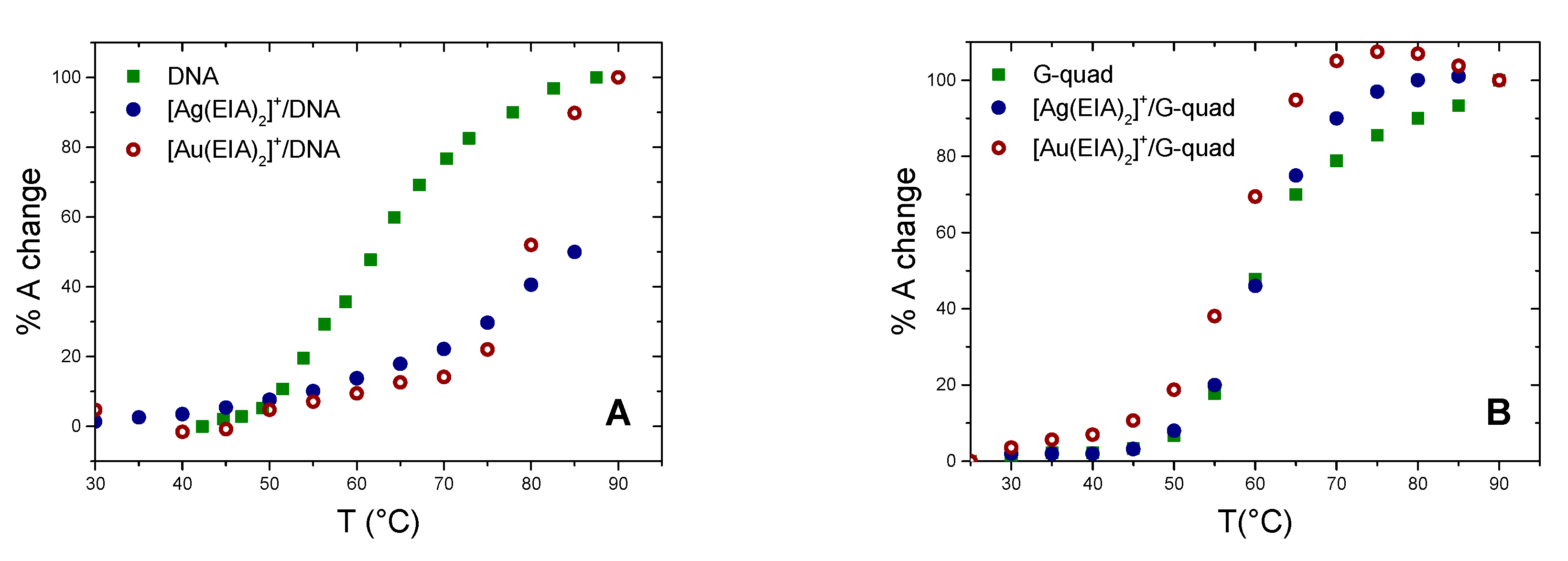
2.1.2. Thermal Denaturation Experiments
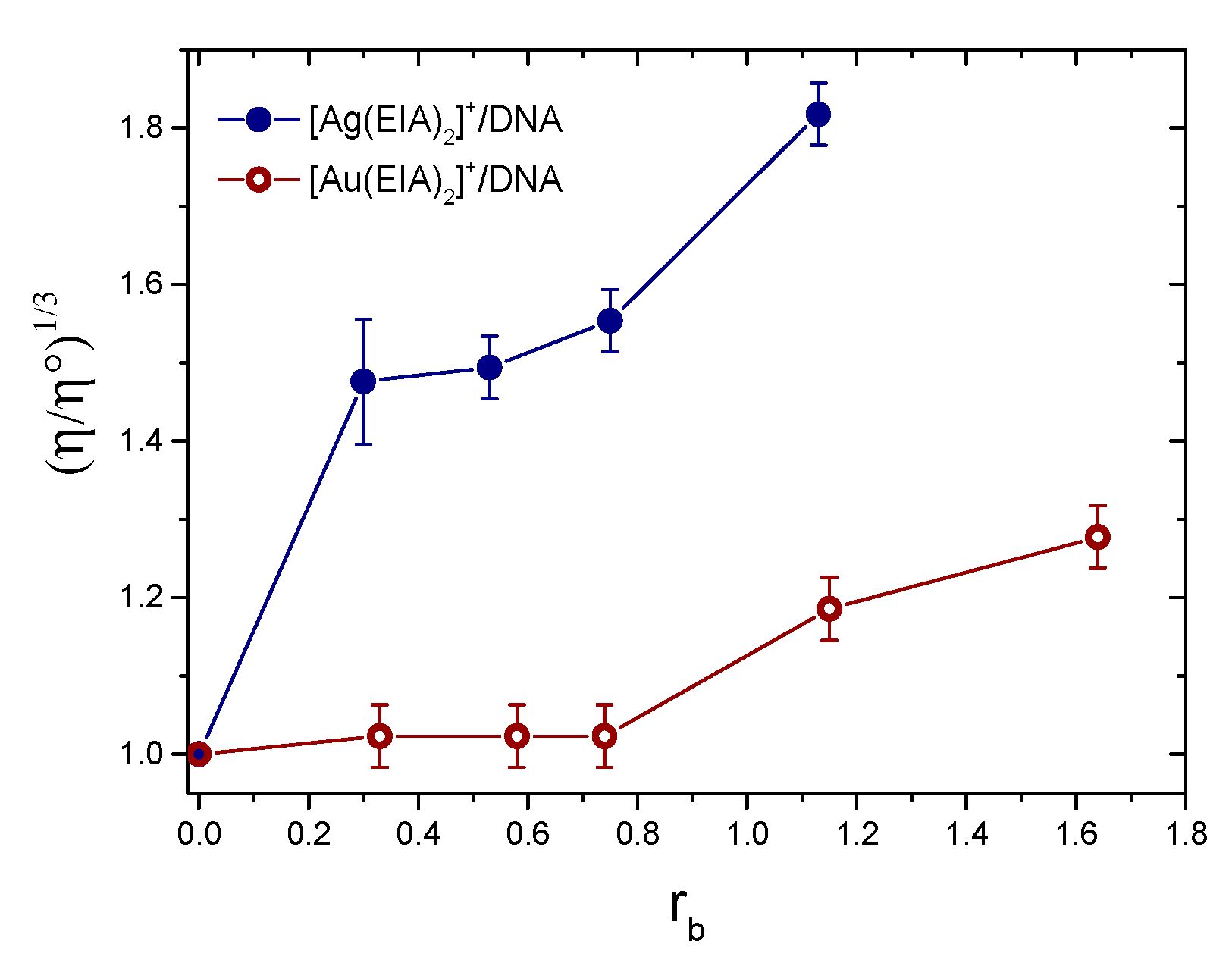
2.1.3. Viscosity
2.2. Interaction with Synthetic RNAs
2.3. Interaction with DNA G-Quadruplexes (G4s)
2.4. Interaction with Bovine Serum Albumin (BSA) Protein
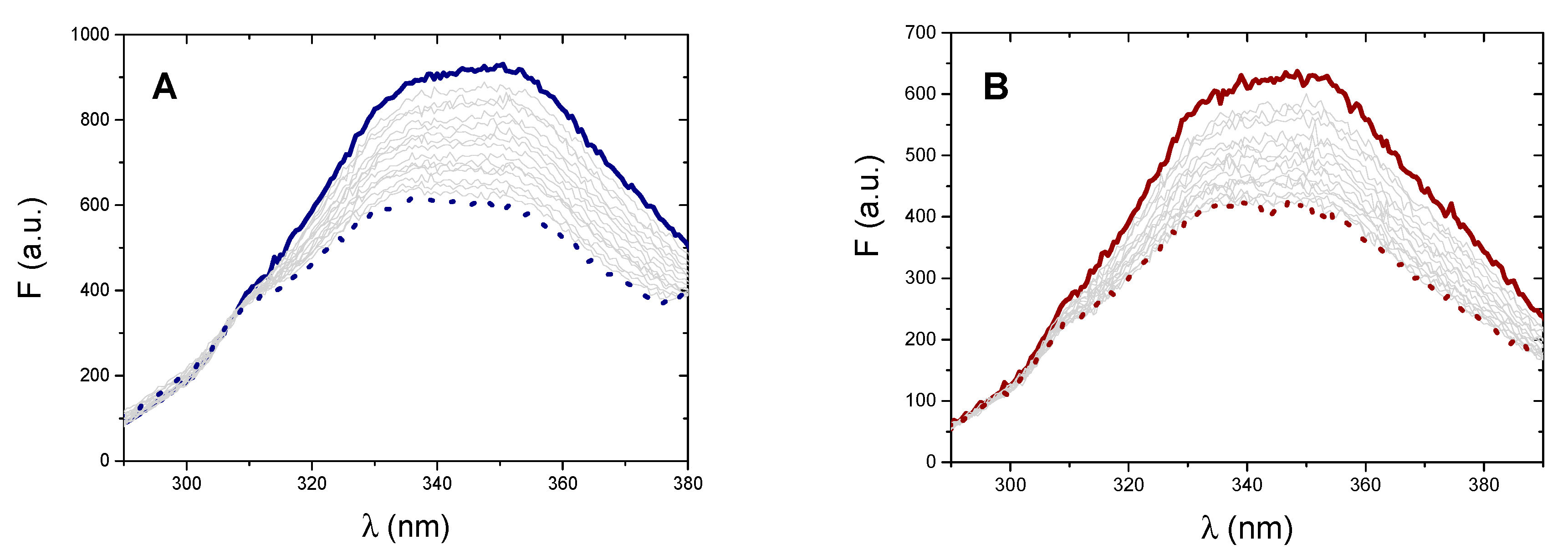
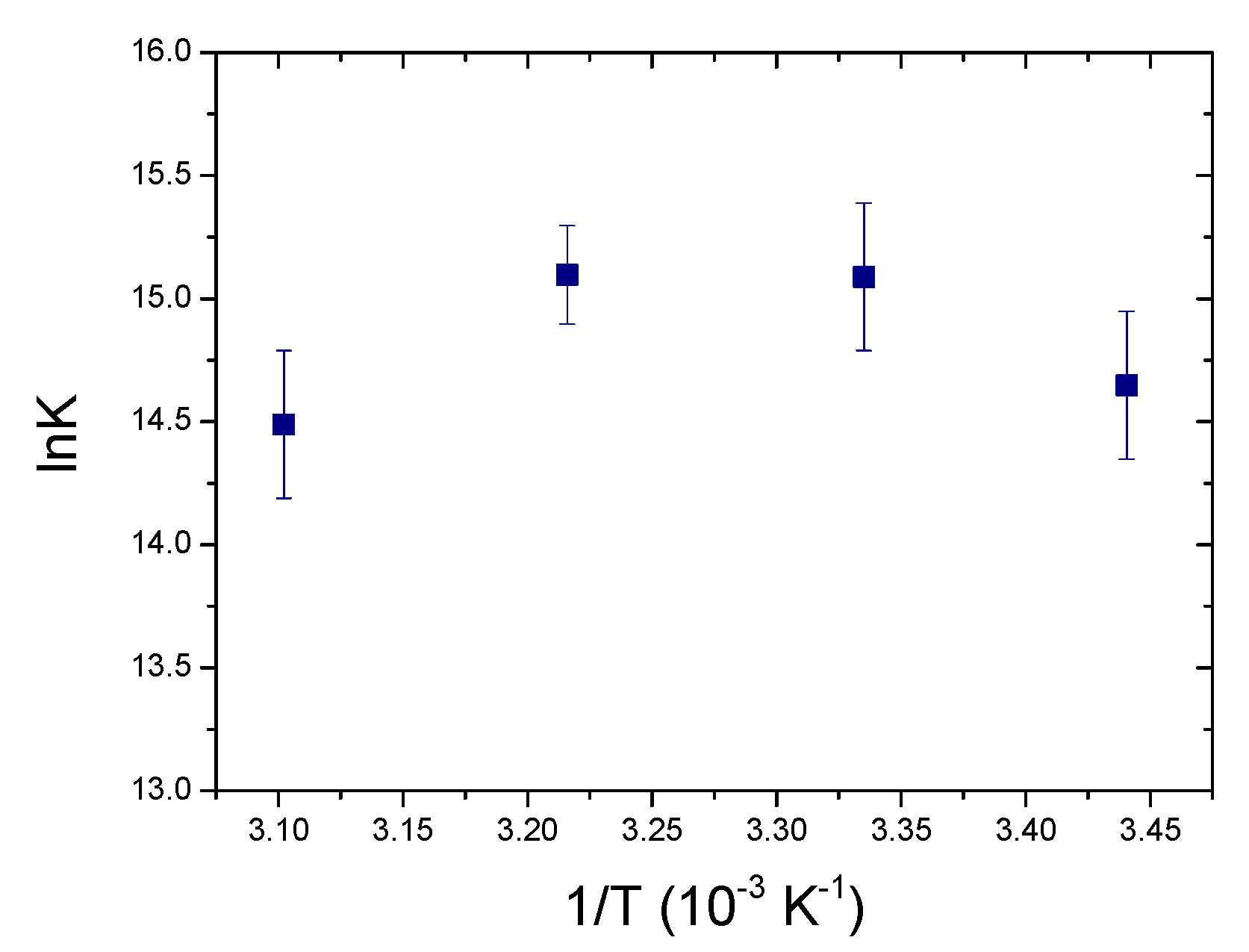
2.4.1. Fluorescence Titrations
2.4.2. Synchronous Fluorescence Spectra
2.4.3. Mass Spectrometry
3. Materials and Methods
3.1. Materials
3.2. Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ott, I. Metal N-Heterocyclic Carbene Complexes in Medicinal Chemistry, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 75, ISBN 9780128191965. [Google Scholar]
- Mora, M.; Gimeno, M.C.; Visbal, R. Recent advances in gold-NHC complexes with biological properties. Chem. Soc. Rev. 2019, 48, 447–462. [Google Scholar] [CrossRef]
- Velazquez, H.D.; Verpoort, F. N-heterocyclic carbene transition metal complexes for catalysis in aqueous media. Chem. Soc. Rev. 2012, 41, 7032–7060. [Google Scholar] [CrossRef]
- Nelson, D.J.; Nolan, S.P.; Nelson, D.J.; Nolan, S.P. Quantifying and understanding the electronic properties of N-heterocyclic carbenes. Chem. Soc. Rev. 2013. [Google Scholar] [CrossRef]
- Niu, W.; Chen, X.; Tan, W.; Veige, A.S. N-Heterocyclic Carbene-Gold(I) Complexes Conjugated to a Leukemia-Specific DNA Aptamer for Targeted Drug Delivery. Angew. Chem. Int. Ed. Engl. 2016, 55, 8889–8893. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Lok, C.N.; Wan, P.K.; Zhang, Z.F.; Fung, S.K.; Che, C.M. Anticancer metal-N-heterocyclic carbene complexes of gold, platinum and palladium. Curr. Opin. Chem. Biol. 2018, 43, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Simpson, P.V.; Schmidt, C.; Ott, I.; Bruhn, H.; Schatzschneider, U. Synthesis, cellular uptake and biological activity against pathogenic microorganisms and cancer cells of rhodium and iridium N-heterocyclic carbene complexes bearing charged substituents. Eur. J. Inorg. Chem. 2013, 2013, 5547–5554. [Google Scholar] [CrossRef]
- Hu, D.; Yang, C.; Lok, C.N.; Xing, F.; Lee, P.Y.; Fung, Y.M.E.; Jiang, H.; Che, C.M. An Antitumor Bis(N-Heterocyclic Carbene)Platinum(II) Complex That Engages Asparagine Synthetase as an Anticancer Target. Angew. Chem. Int. Ed. 2019, 58, 10914–10918. [Google Scholar] [CrossRef]
- Dabrowiak, J.C. Metals in Medicine; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2017; ISBN 9780470681961. [Google Scholar]
- Casini, A.; Sun, R.W.-Y.; Ott, I. Medicinal Chemistry of Gold Anticancer Metallodrugs. In Metallo-Drugs: Development and Action of Anticancer Agents; De Gruyter, Inc.: Berlin, Germany, 2018; pp. 199–217. ISBN 9783110470734. [Google Scholar]
- Hu, D.; Lok, C.N.; Che, C.M. Anticancer Gold Compounds. In Metallobiology; Garner, C.D., Wedd, A., Kovacs, J., Ciurli, S., Sun, H., Raven, E., Eds.; RSC: Cambridge, UK, 2019; Volume 2019, pp. 120–142. [Google Scholar]
- Medici, S.; Peana, M.; Nurchi, V.M.; Zoroddu, M.A. Medical Uses of Silver: History, Myths, and Scientific Evidence. J. Med. Chem. 2019, 62, 5923–5943. [Google Scholar] [CrossRef]
- Kascatan-Nebioglu, A.; Panzner, M.J.; Tessier, C.A.; Cannon, C.L.; Youngs, W.J. N-Heterocyclic carbene-silver complexes: A new class of antibiotics. Coord. Chem. Rev. 2007, 251, 884–895. [Google Scholar] [CrossRef]
- Johnson, N.A.; Southerland, M.R.; Youngs, W.J. Recent developments in the medicinal applications of silver-nhc complexes and imidazolium salts. Molecules 2017, 22, 1263. [Google Scholar] [CrossRef]
- Gandin, V.; Pellei, M.; Marinelli, M.; Marzano, C.; Dolmella, A.; Giorgetti, M.; Santini, C. Synthesis and in vitro antitumor activity of water soluble sulfonate- and ester-functionalized silver(I) N-heterocyclic carbene complexes. J. Inorg. Biochem. 2013, 129, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, G.F.; Tan, C.P.; Ji, L.N.; Mao, Z.W. Antitumor properties and mechanisms of mitochondria-targeted Ag(i) and Au(i) complexes containing N-heterocyclic carbenes derived from cyclophanes. Metallomics 2014, 6, 1460–1468. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.J.; Sadiq, M.; Baronou, E.; Cooper, P.A.; Dunnill, C.; Georgopoulos, N.T.; Latif, A.; Shepherd, S.; Shnyder, S.D.; Stratford, I.J.; et al. Preclinical anti-cancer activity and multiple mechanisms of action of a cationic silver complex bearing N-heterocyclic carbene ligands. Cancer Lett. 2017, 403, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Citta, A.; Schuh, E.; Mohr, F.; Folda, A.; Massimino, M.L.; Bindoli, A.; Casini, A.; Rigobello, M.P. Fluorescent silver(i) and gold(i)-N-heterocyclic carbene complexes with cytotoxic properties: Mechanistic insights. Metallomics 2013, 5, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Lum, C.T.; Chui, S.S.Y.; Che, C.M. Gold(III) complexes containing N-heterocyclic carbene ligands: Thiol “switch-on” fluorescent probes and anti-cancer agents. Angew. Chem. Int. Ed. 2013, 52, 2930–2933. [Google Scholar] [CrossRef] [PubMed]
- Rubbiani, R.; Salassa, L.; De Almeida, A.; Casini, A.; Ott, I. Cytotoxic gold(I) N-heterocyclic carbene complexes with phosphane ligands as potent enzyme inhibitors. Chem. Med. Chem. 2014, 9, 1205–1210. [Google Scholar] [CrossRef]
- Karaca, Ö.; Meier-Menches, S.M.; Casini, A.; Kühn, F.E. On the binding modes of metal NHC complexes with DNA secondary structures: Implications for therapy and imaging. Chem. Commun. 2017, 53, 8249–8260. [Google Scholar] [CrossRef]
- Huerta-Aguilar, C.; Talamantes Gõmez, J.M.; Thangarasu, P.; Camacho-Arroyo, I.; González-Arenas, A.; Narayanan, J.; Srivastava, R. Synthesis, structural and spectral properties of Au complexes: Luminescence properties and their non-covalent DNA binding studies. Appl. Organomet. Chem. 2013, 27, 578–587. [Google Scholar] [CrossRef]
- Meyer, A.; Oehninger, L.; Geldmacher, Y.; Alborzinia, H.; Wölfl, S.; Sheldrick, W.S.; Ott, I. Gold(I) N-heterocyclic carbene complexes with naphthalimide ligands as combined thioredoxin reductase inhibitors and DNA intercalators. Chem. Med. Chem. 2014, 9, 1794–1800. [Google Scholar] [CrossRef]
- Yan, J.J.; Chow, A.L.F.; Leung, C.H.; Sun, R.W.Y.; Ma, D.L.; Che, C.M. Cyclometalated gold(iii) complexes with N-heterocyclic carbene ligands as topoisomerase i poisons. Chem. Commun. 2010, 46, 3893–3895. [Google Scholar] [CrossRef]
- Bertrand, B.; Stefan, L.; Pirrotta, M.; Monchaud, D.; Bodio, E.; Richard, P.; Le Gendre, P.; Warmerdam, E.; De Jager, M.H.; Groothuis, G.M.M.; et al. Caffeine-based gold(I) N-heterocyclic carbenes as possible anticancer agents: Synthesis and biological properties. Inorg. Chem. 2014, 53, 2296–2303. [Google Scholar] [CrossRef] [PubMed]
- Meier-Menches, S.M.; Aikman, B.; Döllerer, D.; Klooster, W.T.; Coles, S.J.; Santi, N.; Luk, L.; Casini, A.; Bonsignore, R. Comparative biological evaluation and G-quadruplex interaction studies of two new families of organometallic gold(I) complexes featuring N-heterocyclic carbene and alkynyl ligands. J. Inorg. Biochem. 2020, 202, 110844. [Google Scholar] [CrossRef] [PubMed]
- Papi, F.; Bazzicalupi, C.; Ferraroni, M.; Massai, L.; Bertrand, B.; Gratteri, P.; Colangelo, D.; Messori, L. [Au(9-methylcaffein-8-ylidene)2]+/DNA Tel23 System: Solution, Computational, and Biological Studies. Chem. A Eur. J. 2017, 23, 13784–13791. [Google Scholar] [CrossRef] [PubMed]
- Wragg, D.; de Almeida, A.; Bonsignore, R.; Kühn, F.E.; Leoni, S.; Casini, A. On the Mechanism of Gold/NHC Compounds Binding to DNA G-Quadruplexes: Combined Metadynamics and Biophysical Methods. Angew. Chem. Int. Ed. 2018, 57, 14524–14528. [Google Scholar] [CrossRef]
- Massai, L.; Pratesi, A.; Gailer, J.; Marzo, T.; Messori, L. The cisplatin/serum albumin system: A reappraisal. Inorg. Chim. Acta 2019, 495, 118983. [Google Scholar] [CrossRef]
- Pratesi, A.; Cirri, D.; Ciofi, L.; Messori, L. Reactions of Auranofin and Its Pseudohalide Derivatives with Serum Albumin Investigated through ESI-Q-TOF MS. Inorg. Chem. 2018, 57, 10507–10510. [Google Scholar] [CrossRef]
- Fasano, M.; Curry, S.; Terreno, E.; Galliano, M.; Fanali, G.; Narciso, P.; Notari, S.; Ascenzi, P. The extraordinary ligand binding properties of human serum albumin. IUBMB Life 2005, 57, 787–796. [Google Scholar] [CrossRef]
- Fanali, G.; Di Masi, A.; Trezza, V.; Marino, M.; Fasano, M.; Ascenzi, P. Human serum albumin: From bench to bedside. Mol. Asp. Med. 2012, 33, 209–290. [Google Scholar] [CrossRef]
- Tolbatov, I.; Cirri, D.; Marchetti, L.; Marrone, A.; Coletti, C.; Re, N.; La Mendola, D.; Messori, L.; Marzo, T.; Gabbiani, C.; et al. Mechanistic Insights Into the Anticancer Properties of the Auranofin Analog Au(PEt3)I: A Theoretical and Experimental Study. Front. Chem. 2020, 8, 812. [Google Scholar] [CrossRef]
- Pratesi, A.; Cirri, D.; Fregona, D.; Ferraro, G.; Giorgio, A.; Merlino, A.; Messori, L. Structural Characterization of a Gold/Serum Albumin Complex. Inorg. Chem. 2019, 58, 10616–10619. [Google Scholar] [CrossRef]
- Zoppi, C.; Messori, L.; Pratesi, A. ESI MS studies highlight the selective interaction of Auranofin with protein free thiols. Dalton Trans. 2020, 49, 5906–5913. [Google Scholar] [CrossRef] [PubMed]
- McKeage, M.J.; Berners-Price, S.J.; Galettis, P.; Bowen, R.J.; Brouwer, W.; Ding, L.; Zhuang, L.; Baguley, B.C. Role of lipophilicity in determining cellular uptake and antitumour activity of gold phosphine complexes. Cancer Chemother. Pharmacol. 2000, 46, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Sarpong-Kumankomah, S.; Gibson, M.A.; Gailer, J. Organ damage by toxic metals is critically determined by the bloodstream. Coord. Chem. Rev. 2018, 374, 376–386. [Google Scholar] [CrossRef]
- Matos, M.J.; Labão-Almeida, C.; Sayers, C.; Dada, O.; Tacke, M.; Bernardes, G.J.L. Synthesis and Biological Evaluation of Homogeneous Thiol-Linked NHC*-Au-Albumin and -Trastuzumab Bioconjugates. Chem. A Eur. J. 2018, 24, 12250–12253. [Google Scholar] [CrossRef] [PubMed]
- Han, X.-L.; Tian, F.F.; Ge, Y.S.; Jiang, F.L.; Lai, L.; Li, D.W.; Yu, Q.L.; Wang, J.; Lin, C.; Liu, Y. Spectroscopic, structural and thermodynamic properties of chlorpyrifos bound to serum albumin: A comparative study between BSA and HSA. J. Photochem. Photobiol. B Biol. 2012, 109, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Guarra, F.; Busto, N.; Guerri, A.; Marchetti, L.; Marzo, T.; García, B.; Biver, T.; Gabbiani, C. Cytotoxic Ag(I) and Au(I) NHC-carbenes bind DNA and show TrxR inhibition. J. Inorg. Biochem. 2020, 205, 110998. [Google Scholar] [CrossRef]
- Fabbrini, M.G.; Cirri, D.; Pratesi, A.; Ciofi, L.; Marzo, T.; Guerri, A.; Nistri, S.; Dell’Accio, A.; Gamberi, T.; Severi, M.; et al. A Fluorescent Silver(I) Carbene Complex with Anticancer Properties: Synthesis, Characterization, and Biological Studies. Chem. Med. Chem. 2019, 14, 182–188. [Google Scholar] [CrossRef]
- Tan, W.B.; Bhambhani, A.; Duff, M.R.; Rodger, A.; Kumar, C.V. Spectroscopic Identification of Binding Modes of Anthracene Probes and DNA Sequence Recognition†. Photochem. Photobiol. 2006, 82, 20. [Google Scholar] [CrossRef]
- Modukuru, N.K.; Snow, K.J.; Perrin, B.S.; Bhambhani, A.; Duff, M.; Kumar, C.V. Tuning the DNA binding modes of an anthracene derivative with salt. J. Photochem. Photobiol. A Chem. 2006, 177, 43–54. [Google Scholar] [CrossRef]
- Safiarian, M.S.; Sawoo, S.; Mapp, C.T.; Williams, D.E. Aminomethylanthracene Dyes as High-Ionic-Strength DNA-Photocleaving Agents: Two Rings are Better than One. ChemistrySelect 2018, 4897–4910. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Ali, S.; Badshah, A. Journal of Photochemistry and Photobiology B: Biology Drug—DNA interactions and their study by UV—Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef] [PubMed]
- McGhee, J.D.; Von Hippel, P.H. Theoretical Aspects of DNA-Protein Interactions: Co-operative and Non-co-operative Binding of Large Ligands to a One-dimensional Homogeneous Lattice. J. Mol. Biol. 1974, 86, 469–489. [Google Scholar] [CrossRef]
- Chaires, J.B. A thermodynamic signature for drug-DNA binding mode. Arch. Biochem. Biophys. 2006, 453, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Venturini, M.; Yarmoluk, S. Influence of cyanine dye structure on self-aggregation and interaction with nucleic acids: A kinetic approach to TO and BO binding. Arch. Biochem. Biophys. 2007, 465, 90–100. [Google Scholar] [CrossRef]
- Biagini, S.; Bianchi, A.; Biver, T.; Boggioni, A.; Nikolayenko, I.V.; Secco, F.; Venturini, M. DNA binding of a proflavine derivative bearing a platinum hanging residue. J. Inorg. Biochem. 2011, 105, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Mohsen-Nia, M.; Amiri, H.; Jazi, B. Dielectric constants of water, methanol, ethanol, butanol and acetone: Measurement and computational study. J. Solut. Chem. 2010, 39, 701–708. [Google Scholar] [CrossRef]
- García, B.; Leal, J.M.; Ruiz, R.; Biver, T.; Secco, F.; Venturini, M. Change of the binding mode of the DNA/proflavine system induced by ethanol. J. Phys. Chem. B 2010, 114, 8555–8564. [Google Scholar] [CrossRef]
- Cohen, G.; Eisenberg, H. Viscosity and sedimentation study of sonicated DNA–proflavine complexes. Biopolymers 1969, 8, 45–55. [Google Scholar] [CrossRef]
- Luedtke, N.W.; Hwang, J.S.; Nava, E.; Gut, D.; Kol, M. The DNA and RNA specificity of eilatin Ru(II) complexes as compared to eilatin and ethidium bromide. Nucleic Acids Res. 2014. [Google Scholar] [CrossRef]
- Vuradi, R.K.; Dandu, K.; Yata, P.K.; Radi, V.M.; Mallepally, R.R.; Chintakuntla, N.; Ch, R.; Thakur, S.S.; Rao, C.M.; Satyanarayana, S. Studies on the DNA binding and anticancer activity of Ru (II) polypyridyl complexes by using (2-(4-(diethoxymethyl)-1H-imidazo [4,5-f][1,10] phenanthroline)) intercalative ligand. New J. Chem. 2018, 42, 846–859. [Google Scholar] [CrossRef]
- Vamsikrishna, N.; Kumar, M.P.; Ramesh, G.; Ganji, N.; Daravath, S. DNA interactions and biocidal activity of metal complexes of benzothiazole Schiff bases: Synthesis, characterization and validation. J. Chem. Sci. 2017, 129, 609–622. [Google Scholar] [CrossRef]
- Lehrer, S.S. Solute Perturbation of Protein Fluorescence. the Quenching of the Tryptophyl Fluorescence of Model Compounds and of Lysozyme by Iodide Ion. Biochemistry 1971, 10, 3254–3263. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.H.; Maruyama, T.; Okada, T.; Yamasaki, K.; Otagiri, M. Study of interaction of carprofen and its enantiomers with human serum abumin-I. Biochem. Pharmcol. 1993, 46, 1721–1731. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of Protein Association Reactions: Forces Contributing to Stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef] [PubMed]
- Kandagal, P.B.; Ashoka, S.; Seetharamappa, J.; Shaikh, S.M.T.; Jadegoud, Y.; Ijare, O.B. Study of the interaction of an anticancer drug with human and bovine serum albumin: Spectroscopic approach. J. Pharm. Biomed. Anal. 2006, 41, 393–399. [Google Scholar] [CrossRef]
- Suryawanshi, V.D.; Walekar, L.S.; Gore, A.H.; Anbhule, P.V.; Kolekar, G.B. Spectroscopic analysis on the binding interaction of biologically active pyrimidine derivative with bovine serum albumin. J. Pharm. Anal. 2016, 6, 56–63. [Google Scholar] [CrossRef]
- Radisavljević, S.; Petrović, B. Gold(III) Complexes: An Overview on Their Kinetics, Interactions With DNA/BSA, Cytotoxic Activity, and Computational Calculations. Front. Chem. 2020, 8, 379. [Google Scholar] [CrossRef]
- Tolbatov, I.; Coletti, C.; Marrone, A.; Re, N. Reactivity of gold(I) monocarbene complexes with protein targets: A theoretical study. Int. J. Mol. Sci. 2019, 20, 820. [Google Scholar] [CrossRef]
- Felsenfeld, G.; Hirschman, S.Z. A neighbor-interaction analysis of the hypochromism and spectra of DNA. J. Mol. Biol. 1965, 13, 407–427. [Google Scholar] [CrossRef]
- Biver, T.; Secco, F.; Venturini, M. Relaxation kinetics of the interaction between RNA and metal-intercalators: The Poly(A)·Poly(U)/platinum-proflavine system. Arch. Biochem. Biophys. 2005, 437, 215–223. [Google Scholar] [CrossRef]
- Hoyuelos, F.J.; García, B.; Leal, J.M.; Busto, N.; Biver, T.; Secco, F.; Venturini, M. RNA triplex-to-duplex and duplex-to-triplex conversion induced by coralyne. Phys. Chem. Chem. Phys. 2014, 16, 6012–6018. [Google Scholar] [CrossRef] [PubMed]
- Rubio, A.R.; Busto, N.; Leal, J.M.; García, B. Doxorubicin binds to duplex RNA with higher affinity than ctDNA and favours the isothermal denaturation of triplex RNA. RSC Adv. 2016, 6, 101142–101152. [Google Scholar] [CrossRef]
- Campbell, N.H.; Parkinson, G.N. Crystallographic studies of quadruplex nucleic acids. Methods 2007, 43, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Mach, H.; Middaugh, C.R.; Lewis, R.V. Statistical determination of the average values of the extinction coefficients of tryptophan and tyrosine in native proteins. Anal. Biochem. 1992, 200, 74–80. [Google Scholar] [CrossRef]
- Zoppi, C.; Nocentini, A.; Supuran, C.T.; Pratesi, A.; Messori, L. Native mass spectrometry of human carbonic anhydrase I and its inhibitor complexes. JBIC J. Biol. Inorg. Chem. 2020, 1, 3. [Google Scholar] [CrossRef]
- Coffer, M.T.; Shaw, C.F.; Eidsness, M.K.; Watkins, J.W.; Elder, R.C. Reactions of Auranofin and Et3PAuCl with Bovine Serum Albumin. Inorg. Chem. 1986, 25, 333–339. [Google Scholar] [CrossRef]
- Dixon, J.M.; Egusa, S. Conformational Change-Induced Fluorescence of Bovine Serum Albumin-Gold Complexes. J. Am. Chem. Soc. 2018, 140, 2265–2271. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds [Au(EIA)2]Cl (bis(1-(anthracen-9-ylmethyl)-3-ethylimidazol-2-ylidene) gold chloride) and [Ag(EIA)2]Cl (bis(1-(anthracen-9-ylmethyl)-3-ethylimidazol-2-ylidene) silver chloride) are available from the authors. |



| T (°C) | KABS (103 M−1) | KFLUO (103 M−1) |
|---|---|---|
| 15.0 | 10 ± 1 | 27 ± 2 |
| 25.0 | 7.4 ± 0.2 | 20 ± 3 |
| 35.0 | 5.1 ± 0.3 | 10 ± 2 |
| 48.0 | 3.7 ± 0.4 | 5.4 ± 0.3 |
| ΔH (kJ/mol) | −23.5 ± 0.3 | −37.9 ± 0.9 |
| ΔS (J/molK) | −5 ± 3 | −46 ± 3 |
| −TΔS (kJ/mol) | 2 ± 1 | 14 ± 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Binacchi, F.; Guarra, F.; Cirri, D.; Marzo, T.; Pratesi, A.; Messori, L.; Gabbiani, C.; Biver, T. On the Different Mode of Action of Au(I)/Ag(I)-NHC Bis-Anthracenyl Complexes Towards Selected Target Biomolecules. Molecules 2020, 25, 5446. https://doi.org/10.3390/molecules25225446
Binacchi F, Guarra F, Cirri D, Marzo T, Pratesi A, Messori L, Gabbiani C, Biver T. On the Different Mode of Action of Au(I)/Ag(I)-NHC Bis-Anthracenyl Complexes Towards Selected Target Biomolecules. Molecules. 2020; 25(22):5446. https://doi.org/10.3390/molecules25225446
Chicago/Turabian StyleBinacchi, Francesca, Federica Guarra, Damiano Cirri, Tiziano Marzo, Alessandro Pratesi, Luigi Messori, Chiara Gabbiani, and Tarita Biver. 2020. "On the Different Mode of Action of Au(I)/Ag(I)-NHC Bis-Anthracenyl Complexes Towards Selected Target Biomolecules" Molecules 25, no. 22: 5446. https://doi.org/10.3390/molecules25225446
APA StyleBinacchi, F., Guarra, F., Cirri, D., Marzo, T., Pratesi, A., Messori, L., Gabbiani, C., & Biver, T. (2020). On the Different Mode of Action of Au(I)/Ag(I)-NHC Bis-Anthracenyl Complexes Towards Selected Target Biomolecules. Molecules, 25(22), 5446. https://doi.org/10.3390/molecules25225446