Synthesis of Conformationally Constrained d-Glu-meso-DAP Analogs as Innate Immune Agonists
Abstract
1. Introduction
2. Results and Discussion
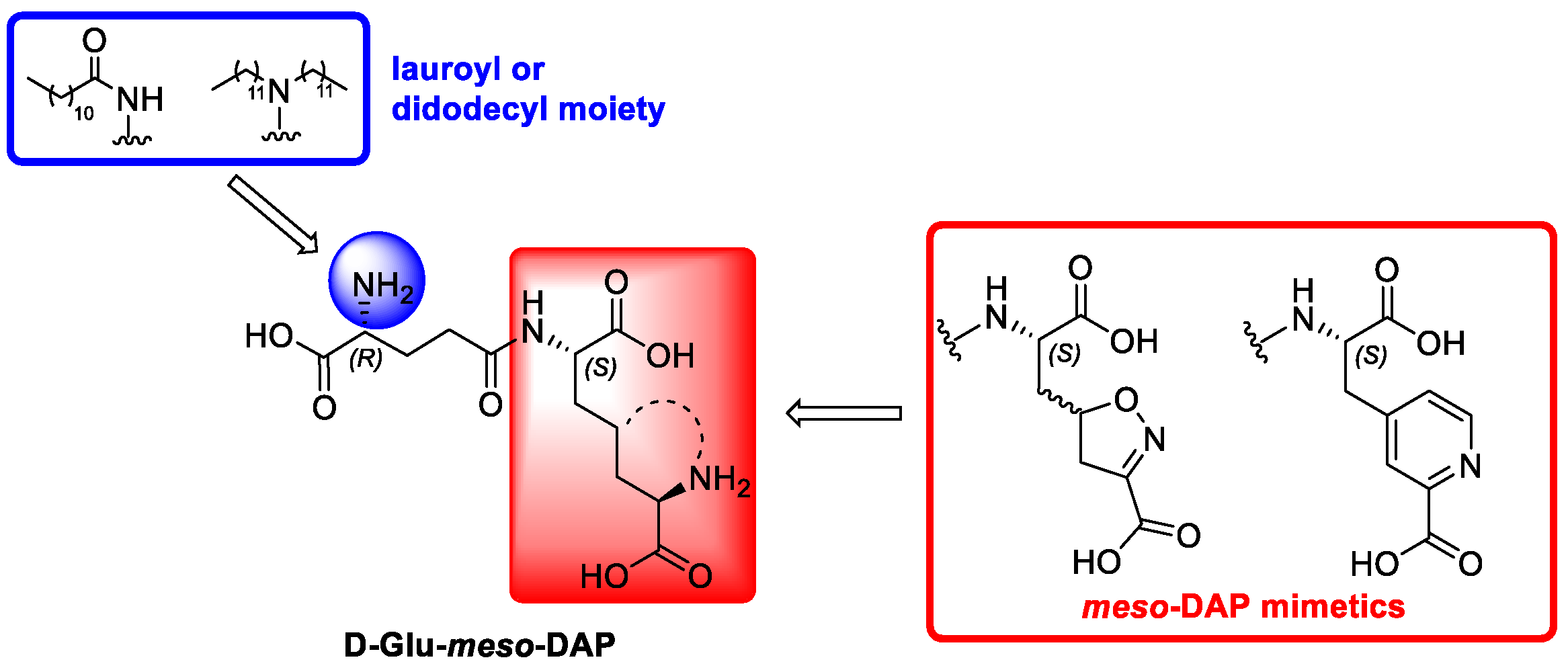
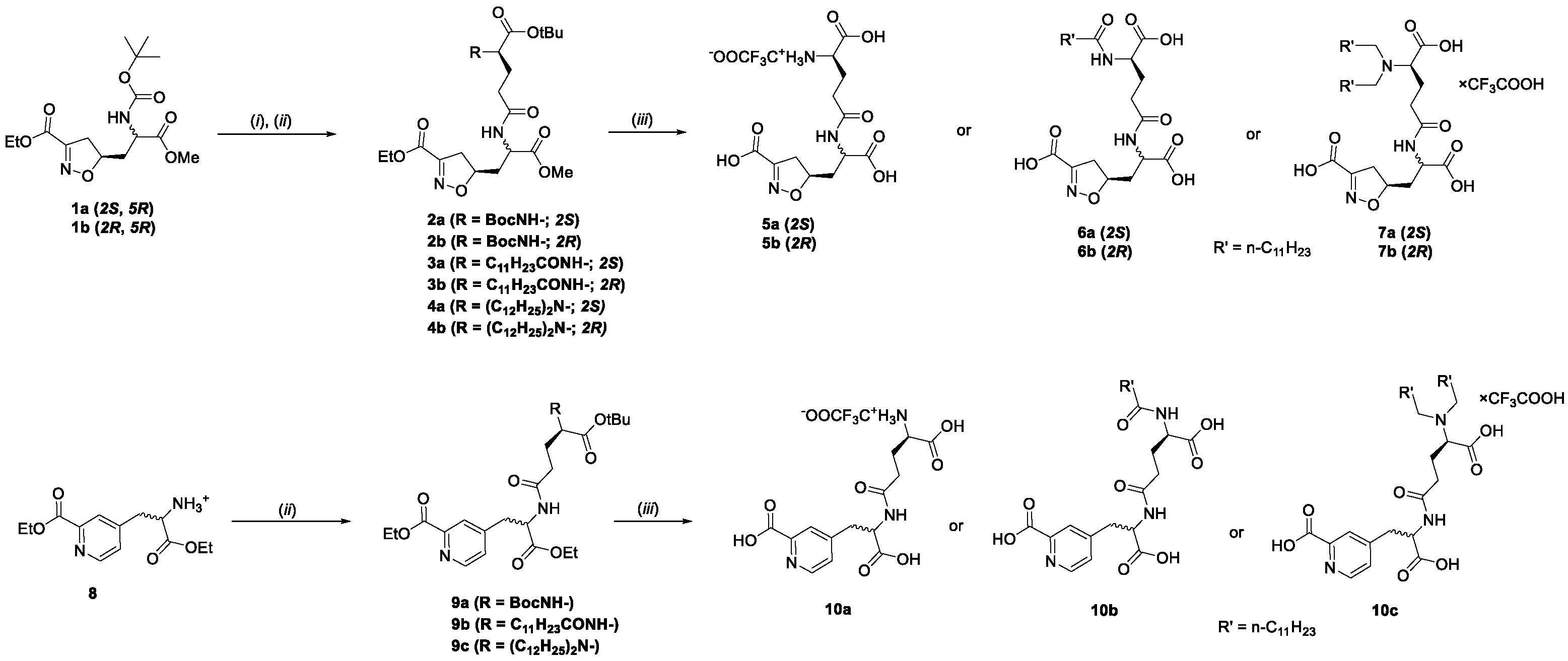
2.1. Design and Chemistry
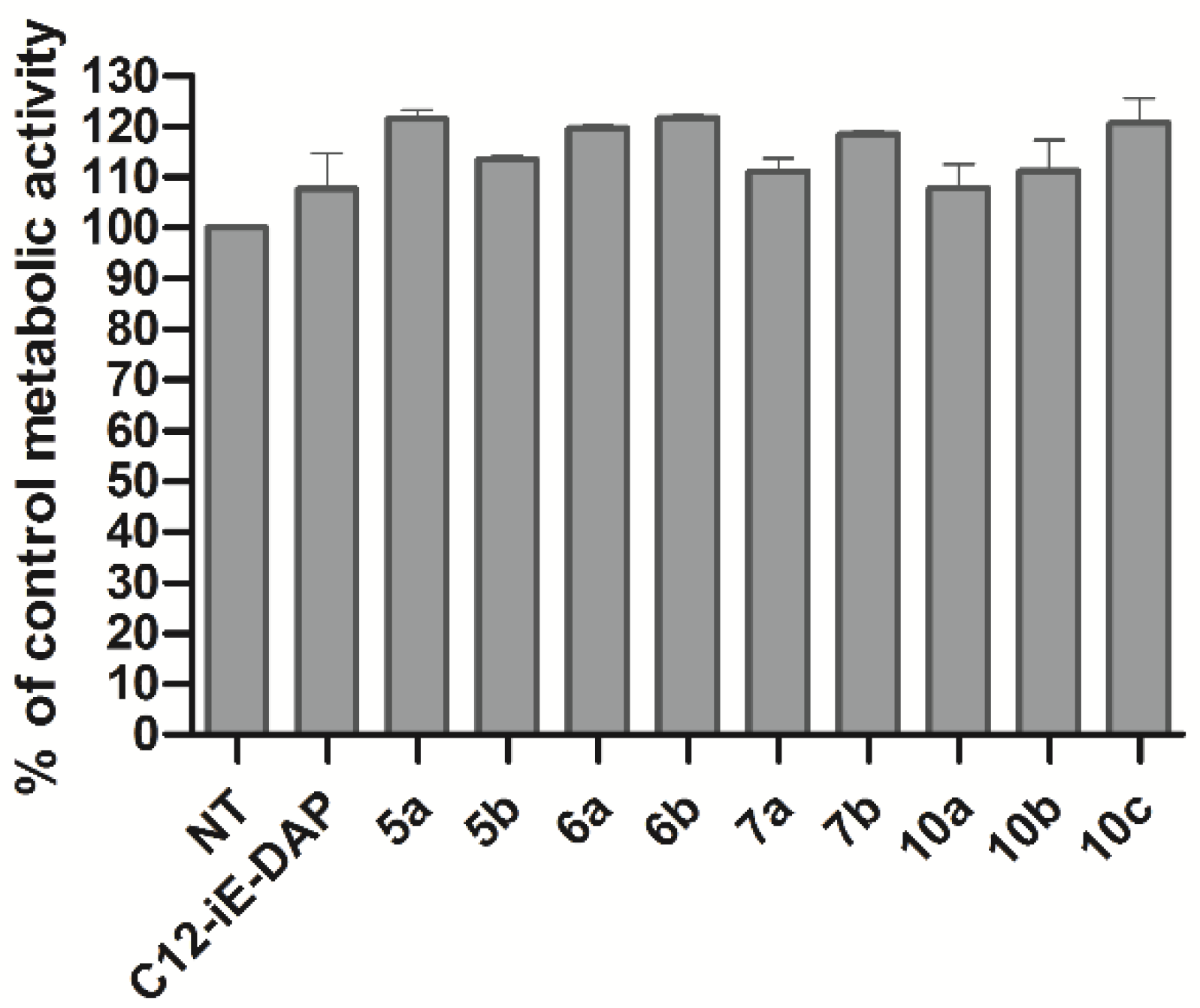
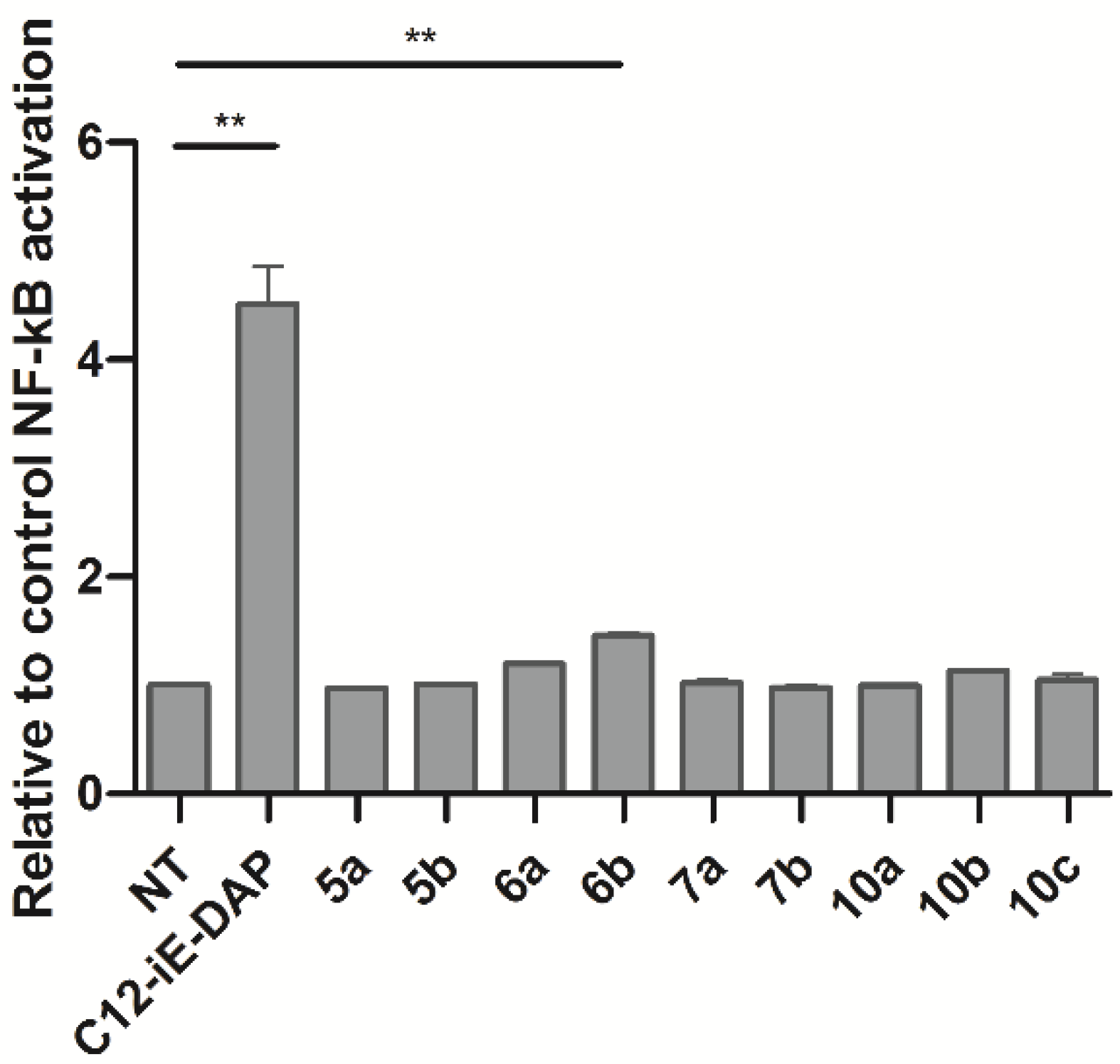
2.2. Biological Characterization
3. Materials and Methods
3.1. General Information
3.2. General Procedures
3.2.1. General Procedure for the Acidolytic Cleavage of Boc Protecting Groups and the Subsequent TBTU-Mediated Coupling
3.2.2. Final Cleavage of Methyl, Ethyl and Tert-Butyl Esters
3.3. Characterization of Compounds
3.4. Cytotoxicity Assay
3.5. Measurement of NF-κB Transcriptional Activity (Quanti-Blue Assay)
3.6. Data Analysis and Statistics
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| C12-iE-DAP | lauroyl-γ-d-glutamyl-meso-diaminopimelic acid |
| iE-DAP | γ-d-glutamyl-meso-diaminopimelic acid |
| meso-DAP | meso-diaminopimelic acid |
| NF-κB | nuclear factor κB |
| NLR | NOD-like receptor |
| NOD | nucleotide-binding oligomerization domain |
| SEAP | secreted embryonic alkaline phosphatase |
| TBTU | 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium tetrafluoroborate |
| TFA | trifluoroacetic acid |
References
- Geddes, K.; Magalhães, J.G.; Girardin, S.E. Unleashing the therapeutic potential of NOD-like receptors. Nat. Rev. Drug Disc. 2009, 8, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Fritz, J.H.; Ferrero, R.L.; Philpott, D.J.; Girardin, S.E. Nod-like proteins in immunity, inflammation and disease. Nat. Immunol. 2006, 7, 1250–1257. [Google Scholar] [CrossRef] [PubMed]
- Philpott, D.J.; Sorbara, M.T.; Robertson, S.J.; Croitoru, K.; Girardin, S.E. NOD proteins: Regulators of inflammation in health and disease. Nat. Rev. Immunol. 2014, 14, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Inohara, N.; Ogura, Y.; Chen, F.F.; Muto, A.; Nuñez, G. Human Nod1 confers responsiveness to bacterial lipopolysaccharides. J. Biol. Chem. 2001, 276, 2551–2554. [Google Scholar] [CrossRef]
- Uehara, A.; Sugawara, Y.; Kurata, S.; Fujimoto, Y.; Fukase, K.; Kusumoto, S.; Satta, Y.; Sasano, T.; Sugawara, S.; Takada, H. Chemically synthesized pathogen-associated molecular patterns increase the expression of peptidoglycan recognition proteins via toll-like receptors, NOD1 and NOD2 in human oral epithelial cells. Cell. Microbiol. 2005, 7, 675–686. [Google Scholar] [CrossRef]
- Girardin, S.E.; Boneca, I.G.; Carneiro, L.A.M.; Antignac, A.; Jéhanno, M.; Viala, J.; Tedin, K.; Taha, M.K.; Labigne, A.; Zähringer, U.; et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 2003, 300, 1584–1587. [Google Scholar] [CrossRef]
- Girardin, S.E.; Travassos, L.H.; Hervé, M.; Blanot, D.; Boneca, I.G.; Philpott, D.J.; Sansonetti, P.J.; Mengin-Lecreulx, D. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 2003, 278, 41702–41708. [Google Scholar] [CrossRef]
- Girardin, S.E.; Jéhanno, M.; Mengin-Lecreulx, D.; Sansonetti, P.J.; Alzari, P.M.; Philpott, D.J. Identification of the critical residues involved in peptidoglycan detection by Nod1. J. Biol. Chem. 2005, 280, 38648–38656. [Google Scholar] [CrossRef]
- Chamaillard, M.; Hashimoto, M.; Horie, Y.; Masumoto, J.; Qiu, S.; Saab, L.; Ogura, Y.; Kawasaki, A.; Fukase, K.; Kusumoto, S.; et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 2003, 4, 702–707. [Google Scholar] [CrossRef]
- Inohara, N.; Koseki, T.; del Peso, L.; Hu, Y.; Yee, C.; Chen, S.; Carrio, R.; Merino, J.; Liu, D.; Ni, J.; et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J. Biol. Chem. 1999, 274, 14560–14567. [Google Scholar] [CrossRef]
- Strober, W.; Murray, P.J.; Kitani, A.; Watanabe, T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat. Rev. Immunol. 2006, 6, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Jakopin, Ž. Nucleotide-binding oligomerization domain (NOD) inhibitors: A rational approach toward inhibition of NOD signaling pathway. J. Med. Chem. 2014, 57, 6897–6918. [Google Scholar] [CrossRef] [PubMed]
- Yoo, N.J.; Park, W.S.; Kim, S.Y.; Reed, J.C.; Son, S.G.; Lee, J.Y.; Lee, S.H. Nod1, a CARD protein, enhances pro-interleukin-1beta processing through the interaction with pro-caspase-1. Biochem. Biophys. Res. Commun. 2002, 299, 652–658. [Google Scholar] [CrossRef]
- Inohara, N.; Nuñez, G. NODs: Intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 2003, 3, 371–382. [Google Scholar] [CrossRef]
- Travassos, L.H.; Carneiro, L.A.; Ramjeet, M.; Hussey, S.; Kim, Y.G.; Magalhães, J.G.; Yuan, L.; Soares, F.; Chea, E.; Le Bourhis, L.; et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 2010, 11, 55–62. [Google Scholar] [CrossRef]
- Mine, Y.; Yokota, Y.; Wakai, Y.; Fukada, S.; Nishida, M.; Goto, S.; Kuwahara, S. Immunoactive peptides, FK-156 and FK-565. I. Enhancement of host resistance to microbial infection in mice. J. Antibiot. 1983, 36, 1045–1050. [Google Scholar] [CrossRef]
- Mine, Y.; Watanabe, Y.; Tawara, S.; Yokota, Y.; Nishida, M.; Goto, S.; Kuwahara, S. Immunoactive peptides, FK-156 and FK-565. III. Enhancement of host defense mechanisms against infection. J. Antibiot. 1983, 36, 1059–1066. [Google Scholar] [CrossRef]
- da Silva Correia, J.; Miranda, Y.; Austin-Brown, N.; Hsu, J.; Mathison, J.; Xiang, R.; Zhou, H.; Li, Q.; Han, J.; Ulevitch, R.J. Nod1-dependent control of tumor growth. Proc. Natl. Acad. Sci. USA 2006, 103, 1840–1845. [Google Scholar] [CrossRef]
- da Silva Correia, J.; Miranda, Y.; Leonard, N.; Hsu, J.; Ulevitch, R.J. Regulation of Nod1-mediated signaling pathways. Cell Death Differ. 2007, 14, 830–839. [Google Scholar] [CrossRef]
- Nabergoj, S.; Mlinarič-Raščan, I.; Jakopin, Ž. Harnessing the untapped potential of nucleotide-binding oligomerization domain ligands for cancer immunotherapy. Med. Res. Rev. 2019, 39, 1447–1484. [Google Scholar] [CrossRef]
- Agnihotri, G.; Ukani, R.; Malladi, S.S.; Warshakoon, H.J.; Balakrishna, R.; Wang, X.; David, S.A. Structure-activity relationships in nucleotide oligomerization domain 1 (Nod1) agonistic γ-glutamyldiaminopimelic acid derivatives. J. Med. Chem. 2011, 54, 1490–1510. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jakopin, Ž.; Gobec, M.; Kodela, J.; Hazdovac, T.; Mlinarič-Raščan, I.; Sollner Dolenc, M. Synthesis of conformationally constrained γ-D-glutamyl-meso-diaminopimelic acid derivatives as ligands of nucleotide-binding oligomerization domain protein 1 (Nod1). Eur. J. Med. Chem. 2013, 69, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.; Fukase, K. Structures, synthesis, and human Nod1 stimulation of immunostimulatory bacterial peptidoglycan fragments in the environment. J. Nat. Prod. 2011, 74, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Kawasaki, A.; Yang, K.; Fujimoto, Y.; Masumoto, J.; Breukink, E.; Nuñez, G.; Fukase, K.; Inohara, N. A role of lipophilic peptidoglycan-related molecules in induction of Nod1-mediated immune responses. J. Biol. Chem. 2007, 282, 11757–11764. [Google Scholar] [CrossRef] [PubMed]
- Roychowdhury, A.; Wolfert, M.A.; Boons, G.-J. Synthesis and proinflammatory properties of muramyl tripeptides containing lysine and diaminopimelic acid moieties. ChemBioChem 2005, 6, 2088–2097. [Google Scholar] [CrossRef]
- Dzierzbicka, K. Synthesis of 2,6-diaminopimelic acid (DAP) and its analogues. Pol. J. Chem. 2007, 81, 455–473. [Google Scholar] [CrossRef]
- Auger, G.; van Heijenoort, J.; Vederas, J.C.; Blanot, D. Effect of analogues of diaminopimelic acid on the meso-diaminopimelate-adding enzyme from Escherichia coli. FEBS Lett. 1996, 391, 171–174. [Google Scholar] [CrossRef]
- Abbott, S.; Lane-Bell, P.; Sidhu, K.P.S.; Vederas, J.C. Synthesis and testing of heterocyclic analogues of diaminopimelic acid (DAP) as inhibitors of DAP dehydrogenase and DAP epimerase. J. Am. Chem. Soc. 1994, 116, 6513–6520. [Google Scholar] [CrossRef]
- Conti, P.; De Amici, M.; Grazioso, G.; Roda, G.; Barberis Negra, F.F.; Nielsen, B.; Stensbøl, T.B.; Madsen, U.; Bräuner-Osborne, H.; Frydenvang, K.; et al. Design, synthesis, and pharmacological characterization of novel, potent NMDA receptor antagonists. J. Med. Chem. 2004, 47, 6740–6748. [Google Scholar] [CrossRef]
- Hilpert, H. Synthese von 3-(2-carboxy-4-pyridyl)- und 3-(6-carboxy-3-pyridyl)-DL-alanin. Helv. Chim. Acta 1987, 70, 1307–1311. [Google Scholar] [CrossRef]
- Diaper, C.M.; Sutherland, A.; Pillai, B.; James, M.N.; Semchuk, P.; Blanchard, J.S.; Vederas, J.C. The stereoselective synthesis of aziridine analogues of diaminopimelic acid (DAP) and their interaction with DAP epimerase. Org. Biomol. Chem. 2005, 3, 4402–4411. [Google Scholar] [CrossRef] [PubMed]
- Uehara, A.; Fujimoto, Y.; Kawasaki, A.; Kusumoto, S.; Fukase, K.; Takada, H. Meso-diaminopimelic acid and meso-lanthionine, amino acids specific to bacterial peptidoglycans, activate human epithelial cells through NOD1. J. Immunol. 2006, 177, 1796–1804. [Google Scholar] [CrossRef] [PubMed]
- Soni, A.S.; Lin, C.S.; Murphy, M.E.P.; Tanner, M.E. Peptides containing meso-oxa-diaminopimelic acid as substrates for the cell-shape-determining proteases Csd6 and Pgp2. ChemBioChem 2019, 20, 1591–1598. [Google Scholar] [CrossRef] [PubMed]
- Apostolos, J.A.; Nelson, J.M.; Pires, M.M. Facile synthesis and metabolic incorporation of m-DAP bioisosteres into cell walls of live bacteria. ACS Chem. Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bixler, R.L.; Niemann, C. Synthesis of β-(4-pyridyl)-DL-alanine and of β-(4-pyridyl-1-oxide)-DL-, D.-, and L-alanine. J. Org. Chem. 1958, 23, 575–584. [Google Scholar] [CrossRef]
- Vijayrajratnam, S.; Pushkaran, A.C.; Balakrishnan, A.; Vasudevan, A.K.; Biswas, R.; Mohan, C.G. Bacterial peptidoglycan with amidated meso-diaminopimelic acid evades NOD1 recognition: An insight into NOD1 structure-recognition. Biochem. J. 2016, 473, 4573–4592. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guzelj, S.; Jakopin, Ž. Synthesis of Conformationally Constrained d-Glu-meso-DAP Analogs as Innate Immune Agonists. Molecules 2020, 25, 5228. https://doi.org/10.3390/molecules25225228
Guzelj S, Jakopin Ž. Synthesis of Conformationally Constrained d-Glu-meso-DAP Analogs as Innate Immune Agonists. Molecules. 2020; 25(22):5228. https://doi.org/10.3390/molecules25225228
Chicago/Turabian StyleGuzelj, Samo, and Žiga Jakopin. 2020. "Synthesis of Conformationally Constrained d-Glu-meso-DAP Analogs as Innate Immune Agonists" Molecules 25, no. 22: 5228. https://doi.org/10.3390/molecules25225228
APA StyleGuzelj, S., & Jakopin, Ž. (2020). Synthesis of Conformationally Constrained d-Glu-meso-DAP Analogs as Innate Immune Agonists. Molecules, 25(22), 5228. https://doi.org/10.3390/molecules25225228






