Abstract
Multivalent interactions frequently occur in biological systems and typically provide higher binding affinity and selectivity in target recognition than when only monovalent interactions are operative. Thus, taking inspiration by nature, bivalent or multivalent nucleic acid aptamers recognizing a specific biological target have been extensively studied in the last decades. Indeed, oligonucleotide-based aptamers are suitable building blocks for the development of highly efficient multivalent systems since they can be easily modified and assembled exploiting proper connecting linkers of different nature. Thus, substantial research efforts have been put in the construction of dimeric/multimeric versions of effective aptamers with various degrees of success in target binding affinity or therapeutic activity enhancement. The present review summarizes recent advances in the design and development of dimeric and multimeric DNA-based aptamers, including those forming G-quadruplex (G4) structures, recognizing different key proteins in relevant pathological processes. Most of the designed constructs have shown improved performance in terms of binding affinity or therapeutic activity as anti-inflammatory, antiviral, anticoagulant, and anticancer agents and their number is certainly bound to grow in the next future.
Keywords:
aptamer; G-quadruplex; design; dimerization; multivalency; molecular recognition; protein target; therapy 1. Introduction
Nucleic acid-based aptamers are short single-stranded DNA or RNA molecules which, upon folding in their peculiar three-dimensional structure, can bind with high affinity and specificity a selected target of biological interest. They are also called “chemical antibodies”, but compared to protein-based molecules, oligonucleotide aptamers generally show lower immunogenicity, higher stability in a wide range of pH and temperature and the possibility to be easier modified or conjugated. Indeed, site-specific chemical modifications can be easily inserted in oligonucleotide aptamers to improve their stability to nuclease digestion or modulate binding affinity to their target [1,2,3,4,5,6]. These intriguing properties make oligonucleotide aptamers very attractive tools in both therapeutic [2,7,8,9,10,11,12,13,14,15] and diagnostic [16,17,18,19,20,21,22] applications [23].
Starting from a large pool of random oligonucleotide sequences, high affinity aptamers for a given target are generally identified through an in vitro selection process named Systematic Evolution of Ligands by Exponential Enrichment (SELEX) [24,25]. The outstanding progress achieved in this field resulted in a variety of selection methods and a large number of aptamers specific for very different kinds of targets—from small molecules, ions, proteins, cells, to even whole organisms, such as viruses or bacteria—have been thus far fished out [26,27,28,29,30,31,32].
Moreover, several aptamers are specifically internalized upon binding to cell membrane receptors and thus can serve as ideal selective delivery systems for different therapeutic targets, from small, conventional drugs to microRNAs or small interfering RNAs (siRNAs) [10,33,34].
Notably, among combinatorially selected aptamers, most of the oligonucleotides endowed with valuable biological activity are able to adopt stem-loop or G-quadruplex (G4) structures. The simplest architecture is represented by the stem-loop or hairpin rearrangement, i.e., an intramolecular conformation based on the coupling of complementary nucleobases in a single-stranded DNA sequence [35].
In contrast, oligonucleotides featured by guanine-rich sequences generally share the ability to fold into peculiar G4 structures [36,37,38,39,40]. The central core of a G4 architecture is the G-tetrad, a structural motif also named G-quartet, which consists of a cyclic planar arrangement of four guanine bases associated through Hoogsteen-type hydrogen bonds [38,40,41,42,43]. Stacking of two or more G-tetrads provides central cavities with a strong negative electrostatic potential, in which cations can be well accommodated, strongly influencing the formation, stability, and topology of the resulting G4 structure [44,45,46,47].
Considering that protein targets involved in specific diseases can (i) have more than one potential binding site recognized by different aptamers, (ii) be dimeric, tetrameric, or in general multimeric, (iii) dimerize or multimerize as a consequence of physiological or pathological events, multivalent aptamer constructs, especially in the simple dimeric forms, are of particular interest [48,49,50,51]. Specifically, a multivalent aptamer is a construct composed of two or more units of the same or different aptamer motifs, containing or not additional structural elements or functional linkers, able to interact simultaneously with more protein binding sites, generally improving its overall efficacy.
Remarkably, SELEX often identifies oligonucleotide aptamers with a repeated sequence, suggesting high affinity recognition ability by dimeric aptamers for a given protein. As an alternative, since aptamers are largely amenable to chemical modifications [1,2,3,4,5,6], the oligonucleotide sequences initially discovered by SELEX can be easily modified to give dimeric or multimeric aptamers without linkers or using proper spacers of different nature (nucleotidic or not), length and flexibility, and exploiting different kinds of connecting interactions (base-pairs recognitions, covalent chemical linkages). Therefore, oligonucleotide aptamers represent a rich arsenal of finely tunable building blocks, which can be profitably joined to generate suitable constructs with improved functions and properties.
Most exploited strategies involve the simple combination of two of more aptamer units concurrently binding two different domains of a target protein with key biological functions in physiological and pathological conditions.
This review is focused on dimeric and multimeric DNA-based aptamers as therapeutic tools targeting key proteins in different relevant diseases, such as inflammation, viral infection, thrombosis and cancer. We here describe the selection, design and properties of multivalent DNA-based aptamers in terms of binding affinity and therapeutic efficacy.
2. Anti-Inflammatory Aptamers
L-, E-, and P-selectins are calcium-dependent cell surface lectins that mediate leukocyte extravasation from the vasculature into surrounding tissues [52]. All three selectins share a similar structure with an amino-terminal calcium-dependent lectin domain, an epidermal growth factor-like domain and complement binding-like domains [53]. In particular, L-selectin (CD62L) is constitutively expressed on the surface of most circulating leukocytes [54], while the expression of E- and P-selectins on endothelial cells and/or platelets can be induced [52].
Having the ability to modulate leukocyte extravasation—a hallmark of inflammation and homeostatic trafficking—L-selectin plays a key role in inflammation and injury [55,56,57]. Therefore, it is considered a fundamental therapeutic target in inflammatory diseases, stimulating the research of suitable aptamers designed to specifically target and inhibit its function [58].
In the mid-1990s, Hicke and coworkers first identified three L-selectin binding DNA aptamers through SELEX procedures. These 39-mer oligodeoxyribonucleotides (ODNs)—named LD201, LD174, and LD196—proved to be specific for L-selectin over P- and E-species, showing similar binding affinity, in the nanomolar range, to the lectin domain of L-selectin (Table 1). These aptamers were also able to inhibit lymphocyte adhesion and trafficking in vitro and in vivo [59].
A slightly modified and shorter version of the LD201 aptamer (i.e., LD201*, Figure 1a and Table 1) was then specifically designed for an efficient affinity-based purification of L-selectin [60].

Figure 1.
Schematic representation of the secondary structure of the L-selectin targeting aptamers. (a) Structure of the monomeric aptamers LD201* and mΔ1, as predicted by the mfold software for nucleic acid folding prediction [64]. The blue box in LD201* sequence represents the nucleotides removed in the truncated mΔ1 analogue (G3-C8 and G33-C34); (b) structure of the mΔ1 dimer in which two aptamer units are linked with poly(dA) linkers of different length. Figures were adapted from Riese et al. [61] with permission.
Subsequently, Riese and colleagues evolved a truncated form of LD201*, i.e., mΔ1, devoid of 8 bases (G3-C8 and G33-C34, Figure 1a) with respect to the starting sequence. The obtained 28-mer oligonucleotide showed IC50 values comparable to LD201* along with improved thermal stability, expressed in terms of melting temperature values (Tm, Table 1) [61]. Therefore, mΔ1 was selected as a starting building block for the construction of dimeric and trimeric constructs [61]. Specifically, two mΔ1 units were linked via flexible poly(dA) spacers of different length (i.e., containing 3, 9, 15, and 20 deoxyadenosines; Figure 1b), obtaining dΔ1-A3, dΔ1-A9, dΔ1-A15, and dΔ1-A20, respectively [61]. All these dimers were tested for their inhibitory activity on L-selectin by means of competitive surface plasmon resonance (SPR) measurements using well-known L-selectin binders as competitive ligands [61]. Compared to the parent mΔ1, dΔ1-A3 dimer exhibited higher IC50 values, suggesting that the length of the exploited linker was too short and thus detrimental for the aptamer activity. On the contrary, improved inhibitory activity was found for the other three dimers, with the best results obtained for dΔ1-A9 showing the lowest IC50 value in the series (0.3 nM, Table 1) [61].
Stimulated by these intriguing findings, the authors also prepared a trimeric oligonucleotide in which 3 mΔ1 units were connected one to each other through a 9-mer poly(dA) spacers, providing tΔ1-A9. Compared to its monomeric counterpart, this trimeric construct exhibited improved inhibitory activity (IC50 = 0.8 nM, Table 1) which was however lower than that of the dΔ1-A9 dimer, probably for steric hindrance effects [61]. Both the dimer and trimer with (dA)9 spacers demonstrated leukocyte blocking capability in vitro and the trivalent aptamer also efficiently inhibited leukocyte rolling in model mice [61].
In a different design, in order to improve the selective and high affinity targeting of L-selectin, Chang and colleagues generated multivalent forms of the LD201 aptamer [62] using circular DNA templates and exploiting the rolling circle amplification (RCA) method. RCA is a simple and powerful isothermal enzymatic reaction in which a DNA polymerase extends the DNA sequence from a primer by replicating a circular DNA template many times to yield a single stranded(ss) DNA product [63].
Chang et al. used a circular template corresponding to the complementary sequence of the L-selectin aptamer and designed aptamer units connected by a 20 nucleotide poly(T) sequence in the RCA product, reflecting the poly(dA) tract in the circle template. The resulting long, linear ssDNA product (termed LS-Multi-Aptamer) incorporated ca. 30 consecutive copies of the L-selectin aptamer [62]. LS-Multi-Aptamer exhibited approximately a 103 higher affinity for L-selectin than the monovalent aptamer, as evaluated by flow cytometry. Additionally, it showed IC50 values of ~0.75 nM on Jurkat cells, showing a sensibly improved activity than the corresponding monomer counterpart (>1 μM). Finally, LS-Multi-Aptamer proved to inhibit at nanomolar concentrations the interaction between L-selectin and endogenous ligands on the cell surface and block cell homing in secondary lymphoid tissues in mice [62].
4. Anticoagulant Aptamers
The most popular protein target for anticoagulant therapies is thrombin, a multifunctional “trypsin-like” serine protease able to bind fibrinogen and thus catalyze its conversion to fibrin clots in the last step of blood coagulation [83,84,85,86,87,88].
Considering its pivotal role in the coagulation cascade, the inhibition of thrombin activity is one of the most efficient antithrombotic strategies [89,90,91,92]. In this context, the development of effective antithrombin aptamers has been the focus of several investigations [92,93,94,95,96].
The most popular antithrombin aptamers able to inhibit thrombin activity are TBA15, TBA29, and TBA27 [92,96]. Besides differing for their overall length, these aptamers show distinct three-dimensional structures and recognize different thrombin binding sites [96].
Indeed, TBA29 and TBA27—also known as HD22-29 and HD22-27—adopt a mixed duplex/G4 architecture able to bind the exosite II of thrombin (heparin-binding site or ABE II) with high affinity (Kd values of 0.5 and 0.7 nM, respectively for TBA29 and TBA27) [97,98]. In particular, TBA27 is a truncated form of TBA29 (5′-AGTCCGTGGTAGGGCAGGTTGGGGTGACT-3′) lacking the first and last residue of the parent aptamer. In contrast, TBA15 or simply TBA, of sequence 5′-GGTTGGTGTGGTTGG-3′, folds into a stable chair-like, antiparallel G4 structure able to inhibit the conversion of soluble fibrinogen into insoluble fibrin strands by binding to thrombin exosite I (fibrinogen-binding site or ABE I) with a Kd of 26 nM [99,100,101,102,103,104,105,106].
Starting from these G-rich oligomers, several homo and heterodimeric constructs were developed as effective antithrombin agents [48,96].
RA-36 is the simplest TBA-based homodimeric aptamer. This 31-mer oligonucleotide comprises two TBA15 units, both in 5′→3′ direction, covalently linked through one thymidine residue at position 16 (Figure 3a). As its monomeric precursor, RA-36 recognizes thrombin exosite I, inhibiting the binding between the protein and fibrinogen [107]. Notably, this dimer was able to exert its activity only in a K+-rich solution, suggesting that the formation of a stable G4 structure is strictly required for effective inhibition of thrombin catalytic action [107,108].
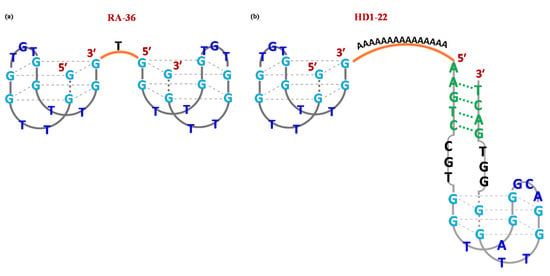
Figure 3.
Schematic representation of the G-quadruplex structure formed in solution by the homodimeric RA-36 (a) and heterodimeric HD1-22 (b) thrombin-targeting aptamers. Figures were redrawn from Amato et al. [109] and Müller et al. [110] respectively.
The intriguing properties of RA-36 stimulated the design of other dimeric TBA15 variants, obtained by joining the 3′-ends of each G4 module and introducing inversion of polarity sites in the overall sequence. The connection between TBA15 motifs was realized using various symmetric linkers—i.e., deoxyadenosine or thymidine residues and/or a glycerol moiety—in place of the thymidine at position 16. Unfortunately, the direct comparison of the anticoagulant properties of the newly developed derivatives with the parent RA-36 was not performed, but the evaluation of prothrombin times revealed improved anticoagulant activity and higher Tm values for most of the designed dimers, compared to unmodified TBA15 (Table 1) [109].
Alternative dimeric constructs were based on the covalent connection between TBA15 and other antithrombin aptamers providing heterodimers able to recognize different thrombin exosites.
In particular, TBA15 and TBA29 were linked exploiting different spacers, such as a 15-nt long poly(dA) linker providing HD1-22 (Figure 3b) [110,111] or poly(T) spacers of various length [112].
In all cases, TBA15/TBA29 dimeric derivatives showed improved thrombin affinity and/or inhibition activity with respect to each monovalent parent aptamer, especially when a poly(T) spacer of 5 residues was explored (Table 1) [110,111,112].
In a different approach, the same aptamers were joined by ethylene glycol spacers of different length. For instance, Tian and Heyduk prepared a covalent dimer of TBA15 and TBA29 featured by flexible connections based on 5′-(OCH2CH2)6-OPO3-3′ (spacer 18) repeated 5 or 10 times (Table 1) [113]. Five repeated units of the spacer provided an overall linker length of 12 nm, while 10 repetitions allowed reaching a 24 nm-long spacer. This longer version was used both to link the 3′-end of TBA29 with the 5′-end of TBA15 and vice versa. In all cases, the designed bivalent analogues proved to be more efficient in terms of thrombin binding affinity than the starting monomeric aptamers (Table 1) [113].
In turn, Hughes et al. inserted an inverted thymidine (iT) at the 3′-end position of TBA15 and TBA29, thus providing RNV216A and RNV219, respectively [114]. These modified versions were then linked by using either a triethylene glycol (TEG) spacer (RNV220) or four thymidine residues (RNV220-T). Compared to both monovalent aptamers, RNV220 and RNV220-T showed significantly improved antithrombin activity in blood plasma (Table 1) [114].
As a valuable alternative to rational design, Ahmad and coworkers used an in vitro selection strategy to identify the optimal sequence joining TBA15 and TBA29 motifs. The randomized linker was 35-nt long, covering the distance between the different thrombin binding exosites [115]. The resulting 119-mer bivalent aptamer (TBV-08) exhibited noteworthy thrombin binding affinity in the picomolar range (Kd of 8.1 pM, Table 1) which well correlated with improved antithrombin activity. Similarly to previous approaches, the authors also prepared bivalent constructs presenting poly(T) or poly(dA) linkers. Interesting results in terms of Kd values were also found for the dimer containing a poly(T) spacer of 16 residues (Table 1) [115].
Only one study reported the covalent connection of TBA15 and the shorter HD22 aptamer, i.e., TBA27. In detail, from 2 to 10 units of the commercially available hexaethylene glycol (HEG)-based phosphoramidite (indicated as S, Table 1) were inserted to join the different monomeric aptamer units. The best results in terms of thrombin inhibitory activity were found for the analogue including 8 repeated units of the linker phosphoramidite 18 (Table 1), corresponding to a length of ca. 16 nm [116]. On the contrary, introduction of the shortest spacer of the series, i.e., the one constituted by 4 repeated units of phosporamidite 18, dramatically reduced the inhibitory activity of the obtained dimer analogue [116].
In order to simultaneously increase the resistance to nuclease degradation and the thrombin binding properties, Di Giusto and colleagues proposed circular multivalent constructs [117,118]. Indeed, circularization is a well-established strategy to improve the performance of aptamers [119,120] and has been efficiently applied to antithrombin aptamers targeting thrombin exosite I [96,121,122,123].
In this work, four different DNA aptamers were exploited as building blocks to obtain circular multivalent constructs: antithrombin aptamers TBA29 and GS-522 (i.e., a 15-mer with the sequence 5′-GGTTGGTGAGGTTGG-3′ able to bind thrombin exosite I [99] ), the L-selectin aptamer [59] and the aptamer against red blood cell marker. A DNA hairpin loop was included as an ancillary module between the aptamer motifs and all the resulting oligonucleotides were also elongated with flanking regions to allow extended stem-loop structure formation. Intra- or intermolecular DNA ligation approaches were used to provide each multivalent circular species, which finally involved two, three or four identical or different aptamer units [117]. The circular multivalent aptamers showed noteworthy stability in both serum and plasma along with improved anticoagulant activity compared to each parent antithrombin aptamer [117].
Another strategy to generate homo- or heteromultimers of a selected aptamer is based on the use of suitable nanoplatforms on which multiple copies of the monomeric aptamer can be linked [48]. This approach has been extensively investigated for TBA15, which thanks to its short sequence and well-known three-dimensional structure, well conserved also when bound to thrombin [124,125], is often exploited as a model system in proof-of-concept studies [48].
In this context, TBA15 was incorporated onto very different nanoplatforms, including magnetic [126,127,128], gold [129,130,131,132,133], silica-based nanoparticles [134,135,136,137], and graphene [138,139].
As a remarkable example, Hsu et al. proposed the multimerization of both TBA15 and TBA29 on the surface of gold nanoparticles (AuNPs). To reach this aim, both aptamers were equipped with thiol end groups allowing their attachment onto the gold surface through Au-thiol interactions [131]. The resulting multivalent nanoparticles, functionalized with about 15 molecules of each aptamer, exhibited an exceptionally high binding affinity for thrombin with a Kd value of 3.4 fM [131].
5. Anticancer Aptamers
DNA aptamers have demonstrated promising potential in biomedicine, especially in cancer therapy, targeting different protein targets [12,140,141,142,143].
5.1. Receptor Tyrosine Kinase PTK7
Receptor tyrosine kinase PTK7 is a cancer-specific cell surface marker overexpressed in several subtypes of leukemia, including T-cell acute lymphoblastic leukemia (ALL) [144]. Leukemia is one of the most common forms of cancer, especially found in children for which the therapeutic options thus far available are essentially unsatisfactory being associated with severe side effects [145].
Using the RCA method, Zhang and coworkers developed a multivalent system comprising multiple copies (ca. 30–40 units) of the PTK7-binding DNA aptamer known as sgc8 (5′-TCTAACTGCTGCGCCGCCGGGAAAATACTGTACGGTTAGA-3′) [146], separated by poly(dA) linkers with 3 d(GC) repeats. To improve its anticancer efficacy, the multivalent aptamer was then hybridized with doxorubicin (Dox)—a well-known DNA-intercalating chemotherapeutic agent—which should well interact with the d(GC) repetitions [147].
Compared to the monovalent species, the designed multivalent aptamer exhibited a sensibly improved binding affinity (Kd of 6.5 nM vs. 260 nM), more efficient cell internalization and selective cytotoxicity against PTK7-expressing T-leukemia CCRF-CEM cell line [147].
In a strategy similar to that explored by Di Giusto et al., the sgc8 aptamer was elaborated into a circular multivalent aptamer. In particular, each aptamer unit was modified with additional flanking sequences of 13 bases which, being complementary and thus able to form duplex tracts, allowed the formation of a circular bivalent sgc8 construct (cb-Sgc8) [148]. Notably, cb-sgc8 showed excellent in vitro resistance to nuclease degradation and proved to bind CCRF-CEM cells more efficiently than the monovalent aptamer (Kd of 0.30 nM vs. 0.86 nM) [148]. The cb-sgc8 aptamer also exhibited enhanced in vitro cell internalization and higher in vivo accumulation and retention in tumor masses compared to its monomeric component [148].
5.2. Nucleolin
Nucleolin is a multifunctional protein overexpressed on the outer membrane of tumor cells which exerts key roles in cell survival, growth, and proliferation [149,150]. One of the most promising aptamers, first entered in clinical trials for cancer therapy is AS1411 [151], a 26-mer G-rich DNA aptamer (5′-GGTGGTGGTGGTTGTGGTGGTGGTGG-3′), forming highly stable G4 structures, very resistant to nuclease degradation, which selectively targets this protein [152,153,154].
For its intriguing properties, AS1411 has been widely used not only as a drug but also as a tumor-targeting agent. Indeed, a huge number of AS1411-based nanosystems have been successfully developed to transport and selectively deliver anticancer agents to cancer cells [154,155,156,157,158,159,160]. Detailed structural studies on this aptamer revealed its high polymorphism, and its peculiar ability to fold into multiple mono- but also dimeric G4 structures (Figure 4a) [161].
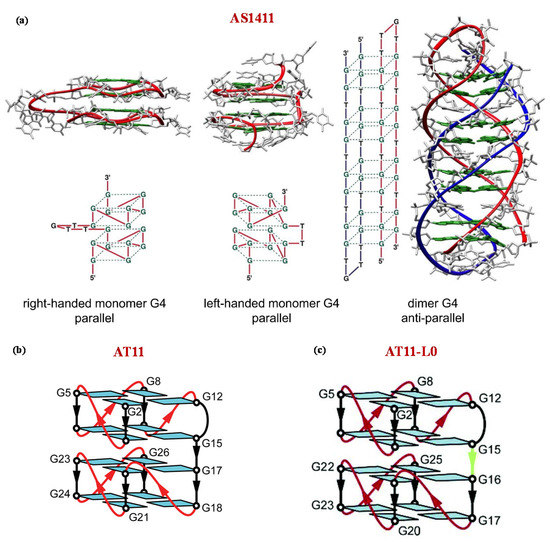
Figure 4.
Schematic representation of the possible G-quadruplex structures of AS1411 (a) and NMR-derived G-quadruplex structures adopted by AT11 (b) and AT11-L0 (c) in K+-solutions as described by Dailey et al. [161], Do et al. [165], and Carvalho et al. [166] respectively. Figures were reproduced from refs [154,165,166] with permission.
To meet the constant requirement of improved analogues with better performance, several modified AS1411 aptamers have been designed [162,163,164]. In particular, to reduce the conformational polymorphism of AS1411, Do and colleagues [165] investigated AS1411-related aptamers with single nucleotide substitution, identifying AT11 (5′-TGGTGGTGGTTGTTGTGGTGGTGGTGGT-3′), able to form a single major G4 architecture with similar antiproliferative activity as AS1411.
Detailed NMR studies proved that AT11 adopts a four-layer G4 structure containing two propeller-type parallel-stranded subunits connected through a central linker (Figure 4b) [165]. Subsequent modifications on AT11 provided the AT11-L0 aptamer (5′-TGGTGGTGGTTGTTGGGTGGTGGGGT-3′), which lacks one thymine residue between the two G4 subunits and forms a parallel-stranded G4 containing four G-quartets (Figure 4c) [165,166].
5.3. Vitronectin
Vitronectin (VN) is a highly glycosylated protein produced essentially in the liver and then secreted into the blood. This protein exists as a monomer, predominantly in serum, or as a multimer [167,168]. VN has a key role in wound healing and is present at elevated levels in breast cancer (BC) cells [169,170].
Thus, VN has been extensively investigated as a therapeutic target in breast cancer, the most common cancer in women representing an extremely heterogeneous malignancy with several subtypes which, growing at different rates, show distinct responses to treatments [171,172,173,174,175].
Combining capillary electrophoresis (CE) and next-generation sequencing (NSG) for aptamer selection [176], Stuart et al. identified a novel DNA aptamer against VN, named VBA-01 (Figure 5a), with a Kd value of 405 nM (Table 1) [177]. Successively, the same authors developed DVBA-01, a dimeric aptamer obtained by extension of each VBA-01 unit with CGGC-dA12-GCCG or CGGC-T12-GCCG tracts in order to obtain a duplex connecting linker (Figure 5b). CG elongation at the ends of the poly(dA) or poly(T) tracts was specifically used since CG tracts are considered preferential binding sites for Dox. The designed dimer proved to be effective as VN-targeting compound, showing very similar binding affinity (Kd = 485 nM, Table 1) compared to the parent monomer [177].

Figure 5.
Schematic representation of the secondary structure of the monomeric and dimeric- vitronectin-targeting aptamers VBA-01 (a) and DVBA-01 (b) as predicted by the mfold software for nucleic acid folding prediction [64]. In DVBA-01 structure, red boxes indicate potential Dox-binding sites. Figures were redrawn from Stuart et al. [177].
Furthermore, using a pH-sensitive approach, DVBA-01 was then complexed with Dox and the obtained complex displayed even higher affinity for the target protein (Kd = 28 nM, Table 1) [177].
5.4. Prostate-Specific Membrane Antigen (PMSA)
Using a strategy very similar to that developed by Stuart and colleagues for the vitronectin aptamers [177], Boyacioglu et al. proposed a novel dimeric DNA aptamer complex bound to Dox through a pH-sensitive method [178]. This construct was specifically designed to target the prostate-specific membrane antigen (PSMA), an exopeptidase highly expressed on the apical plasma membrane of prostate cancer (PC) cells but also found in high percentage in endothelial cells from many different malignancies, including bladder, gastric, and colorectal, as well as hepatocellular, renal, breast, and ovarian cancer [179].
Notably, PSMA is present on the plasma membrane as a dimer [180], and dimeric ligands targeting this protein have shown improved activity compared to their corresponding monomers [181].
Firstly, the 48-mer DNA aptamer SZTI01, with sequence 5′-GCGTTTTCGCTTTTGCGTTTTGGGTCATCTGCTTACGATAGCAATGCT-3′, was identified using an affinity matrix system with the extracellular domain of human PSMA [178]. Then, a dimeric aptamer complex (DCA) was designed containing a duplex DNA “bridge” between the aptamer units and including the preferred binding sites for subsequent Dox incorporation (CG-rich sequences). Specifically, the authors elongated each SZTI01 motif inserting GCCG and CGGC sequences respectively to the 5′- and 3′-ends of the A16:T16 DNA duplex-forming sequence (Figure 6), thus providing a 24 base pair DNA duplex with an approximative length of 70 Å, consistent with the size of the PSMA dimer [178].
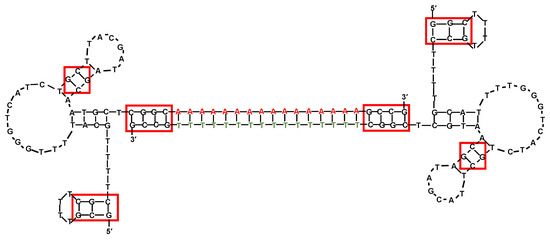
Figure 6.
Schematic representation of the prostate-specific membrane antigen (PSMA)-targeting dimeric aptamer including two SZTI01 motifs linked by a duplex DNA “bridge” containing CG sequences appended to the ends of the dA16 or T16 bases (depicted as red and green, respectively). Red boxes indicate potential Dox-binding sites. Figure was redrawn from Boyacioglu et al. [178].
Dox-loaded dimeric aptamer proved to selectively deliver Dox to C4-2 cells, overexpressing PSMA (PMSA+) with cytotoxic effects similar to free Dox. Notably, no significant reduction of cell viability was observed on PC3 cells, which do not express PSMA, demonstrating high selectivity of the designed complex. Nevertheless, in this work, the binding affinities or IC50 values of these systems were not assessed [178].
5.5. Membrane-Bound Immunoglobulins M (mIgM)
Membrane-bound immunoglobulins M (mIgM), i.e., B-cell receptors, represent a relevant target in non-Hodgkin’s lymphomas (NHLs) [182], a neoplasm of the lymphoid tissues, which originates from B cell precursors, mature B cells, T cell precursors, and mature T cells [183].
Tang and coworkers developed a high-affinity DNA aptamer (TD05, Figure 7a) reactive with Burkitt’s lymphoma cells (Ramos cells) at 4 °C [184].
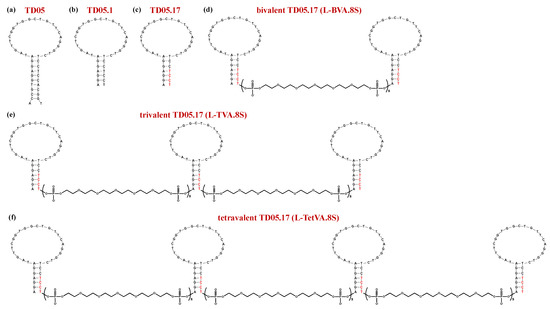
Figure 7.
Schematic representation of the secondary structure of the monomeric and dimeric mIgM-targeting aptamers: TD05 (a), TD05.1 (b), TD05.17 (c), bivalent TD05.17, i.e., L-BVA.8S (d), trivalent TD05.17, i.e., L-TVA.8S (e) and tetravalent TD05.17, i.e., L-TetVA.8S (f). Nucleobases in red color are LNA residues. Figures were redrawn from Mallikaratchy et al. [185].
Unfortunately, this aptamer was not effective in vivo because of its lack of affinity and stability at physiological temperature in human plasma. Thus, Mallikaratchy et al. produced chemically modified versions of the original 45-mer by truncation approaches and introducing locked nucleic acid (LNA) monomers in the oligonucleotide backbone of this aptamer. The best performing analogue proved to be the truncated form TD05.1 (Figure 7b)—lacking 8 nucleotides and including some nucleotide-substitutions—and its LNA derivative TD05.17 (Figure 7c), featured by the insertion of LNA residues in place of the pyrimidine nucleotides in the stem-loop region [185]. Both these derivatives exhibited dramatically enhanced binding affinity with respect to the parent sequence (Table 1) [185].
TD05.1 was then engineered into bivalent, trivalent and tetravalent constructs using HEG-based linkers (indicated as sp18) in order to increase the aptamer binding affinity at physiological temperatures [185]. Specifically, dimers of TD05.1 were prepared with spacers of 6, 8, and 12 units, corresponding to lengths of 12.6, 16.8, and 25.2 nm. All these dimers showed similar binding affinity as the parent monomer, with modest improvement only for the construct including the linker of 8 HEG units. Thus, the authors used this connecting spacer to provide trivalent (TVA.8S) and tetravalent (TetVA.8S) TD05.1-based aptamers. Compared to the monovalent sequence, both trimeric and tetrameric TD05.1 versions exhibited improved binding affinity at 37 °C (Table 1) [185].
Bi-, tri- and tetravalent aptamers were also prepared using TD05.17 (containing LNAs) and 8 HEG units as the starting monomer and the connecting linker, respectively. Trimer (L-TVA.8S) and tetramer (L-TetVA.8S) compounds were also equipped with polyethylene glycol (PEG) appendages at both extremities to improve resistance to nuclease degradation. While the bivalent form (L-BVA.8S, Figure 7d) did not show any relevant increase in binding affinity compared to the monovalent TD05.17 (Table 1), both L-TVA.8S and L-TetVA.8S (Figure 7e,f) showed a ca. 40-fold increased affinity with respect to monovalent TD05.17. In addition, these multivalent LNA-containing constructs showed ca. 2-fold enhanced binding affinity compared to those based on TD05.1 (without LNA bases), suggesting that the conformational constraints introduced by the LNA monomers highly favored the aptamer/protein interactions (Table 1) [185].
Competition and protease studies on Ramos cells also demonstrated that the multimeric aptamer TetVA.8S bound to membrane-associated human mIgM, but not to soluble IgM in plasma, allowing selective targeting of leukemia and lymphoma cells in vivo [185].
In a recent design, using a variant of cell-SELEX—known as LIgand Guided Selection (LIGS) [186,187,188]—a 79-mer aptamer, termed R1, was identified against mIgM. Since its binding affinity proved to be moderate for potential applications (Kd = 315 nM), truncated forms were then evolved [189,190]. These efforts provided two shorter versions: a 42-mer, indicated as R1.2, and a 35-mer, named R1.3, obtained as a further truncation of R1.2. Both aptamers were able to bind mIgM with an improved affinity (Kd values of 35.5 and 134 nM, respectively for R1.2 and R1.3, Table 1) and similar binding specificity compared to the parent R1 aptamer [189,190].
In a subsequent work, three different homodimeric R1.2 variants were designed analyzing the impact of the linker length on the binding affinity. As previously experienced with TD05, HEG-based spacers were used. In particular, 3, 5, or 7 repeated units were inserted to join two R1.2 motifs thus providing DR1.2_3S, DR1.2_5S or DR1.2_7S (Figure 8). In all cases, dimeric constructs exhibited improved binding affinity with respect to the monovalent R1.2 aptamer especially at physiological temperatures, while at 4 °C similar or slightly improved Kd values were found (Table 1) [191].
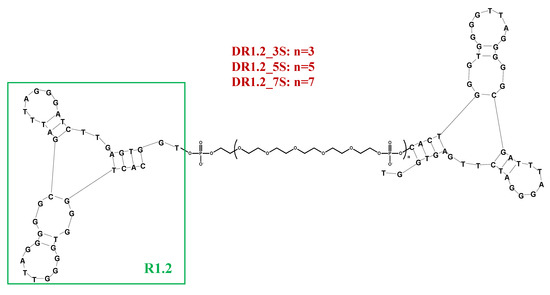
Figure 8.
Schematic representation of the mIgM-targeting dimeric R1.2 aptamer. Figure was redrawn from Batool et al. [191].
5.6. T Cell Receptor Cluster of Differentiation 3 (TCR-CD3)
The T cell receptor cluster of differentiation 3 (TCR-CD3) complex, expressed on human T cells and composed from multiple domains, plays a significant role in immune cell activation [192]. Immunotherapeutic strategies, involving direct targeting of T cells in different pathologies including cancer, have completely revolutionized drug development approaches [193].
Using LIGS approaches, aptamers able to specifically recognize the TCR-CD3 complex were identified. Among these, the 76-mer ZUCH-1 aptamer (Figure 9a) proved to be the optimal one, showing a Kd value of 3.0 nM (Table 1) [194].
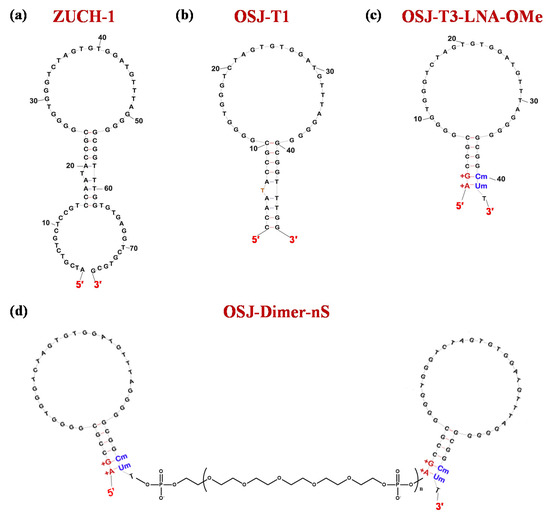
Figure 9.
Schematic representation of the secondary structure of the monomeric and dimeric TCR-CD3-targeting aptamers: ZUCH-1 (a), OSJ-T1 (b), OSJ-T3_LNA-OMe with LNA and 2′-OMe RNA residues (c), bivalent OSJ-T3_LNA-OMe (d). LNA and 2′-OMe RNA nucleobases are marked in red and blue, respectively. Figures were adapted from Freage et al. [195] with permission.
To further enhance the aptamer affinity, in subsequent optimization efforts, Freage et al. designed shorter ZUCH-1 variants and then engineered suitable dimeric derivatives of the most promising truncated versions. In detail, three shorter aptamers, named OSJ-T1, OSJ-T2, and OSJ-T3—composed of 48, 49, and 39 nucleotides, respectively—were designed. These ODNs exhibited similar binding affinity compared to the parent aptamer, with OSJ-T3 (Figure 9b) as the best analogue in the series showing a Kd value of 2.1 nM (Table 1) [195].
Further modifications of OSJ-T3 provided OSJ-T3-LNA-OMe (Figure 9c), with the 1st and 2nd nucleotides from the 5′-end replaced with LNA residues, and the 40th and 41st nucleotides from the 3′-extremity substituted by 2′-OMe RNA monomers. The double modification in the original aptamer sequence proved to be very effective in terms of binding affinity, providing a Kd value of 1.7 nM (Table 1) [195].
Thus, OSJ-T3-LNA-OMe was dimerized by using from 2 (2S) to 8 (8S) repeated units of the HEG-based phosphoramidite 18 spacer (S). Three of the four designed dimers (Figure 9d) exhibited improved binding affinity compared to their monomeric counterpart. The only exception was the longer dimer OSJ-dimer-8S, which showed no improvement in binding ability as a consequence of the dimerization (Table 1) [195].
5.7. Vascular Endothelial Growth Factor (VEGF)
The Vascular Endothelial Growth Factor (VEGF) family is composed by different cytokine proteins involved in both vasculogenesis and angiogenesis processes [196,197,198,199,200,201,202]. VEGF-A, or simply VEGF, is the most important member of this family being able to modulate the proliferation, migration and formation of endothelial cells [203,204]. Its most abundant isoforms are VEGF165 and VEGF121 [205,206,207], which share a common receptor-binding domain (RBD) and play fundamental roles in pathological angiogenesis and vascularization of a large variety of solid tumors [208]. VEGF165 has also a heparin-binding domain (HBD), making this isoform more relevant than VEGF121 from a biological point of view [199,205,206,207].
Due to the high clinical importance of VEGF, a number of anti-VEGF nucleic acid-based aptamers has been identified by SELEX and also suitably engineered to produce homodimeric or multimeric forms [209], also taking into account the homodimeric nature of VEGF165 [200,201].
Through SELEX procedures, Hasegawa et al. identified a 66-mer aptamer, named VEa5 (Figure 10a), able to form a stem-loop structure and showing a Kd value of 130 nM vs. VEGF165 (Table 1) [210,211]. In order to obtain derivatives with increased affinity, VEa5 was subjected to successive optimization through different modifications, especially truncation of the original sequence. The same research group proposed a shorter version, i.e., del5-1 (Figure 10a) [112], while Kaur and colleagues developed the analogue known as SL2-B (Figure 10b) [212]. The latter compound showed a noteworthy enhancement of the binding affinity with a Kd value of 0.5 nM, i.e., 260-fold lower than that found for the parent aptamer (Table 1) [212].
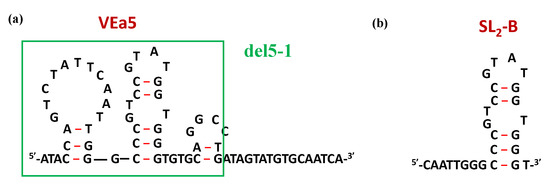
Figure 10.
Schematic representation of the secondary structures of VEa5 (a) and its truncated aptamers del5-1 (green box) and SL2-B (b), as predicted by the mfold software for nucleic acid folding prediction [64]. Figures were redrawn from Hasegawa et al. [211] and Kaur et al. [212], respectively.
Both the original VEa5 and its truncated forms del5-1 and SL2-B aptamers were then used as building blocks in the construction of suitable homodimeric species containing poly(T) linkers of different length or no linker [112,213]. In all cases, the best dimeric versions in terms of binding affinity were those obtained by direct connection of the aptamer units, not containing any spacer in between (Table 1) [112,213].
Using a post-SELEX optimization method, i.e., in silico maturation (ISM) approach, Fukaya and coworkers identified the 2G19 aptamer, a 58-mer with VEGF165 binding affinity in the low nanomolar range (Table 1) [214]. In the same work, the authors also designed a 2G19-based bivalent construct with a ca. 26-fold improved binding affinity with respect to its monomeric counterpart (Table 1) [214].
In detail, 2G19 comprises two different stem-loop (sl) regions known as sl2 and sl5. The replacement of sl2 with a sl5 portion provided a bivalent construct with identical stem loop moieties, known as bivalent SL5. The addition of a third sl5 domain at the 3′ end of the bivalent construct provides a trivalent species with highly improved binding affinity in comparison to the starting 2G19 aptamer (Table 1) [214].
In a more recent design, Manochehry and colleagues performed a multi-stream selection strategy, a variant of the classical SELEX procedure in which the concentration of the target protein was varied. Within this approach, they identified novel VEGF165-binding DNA aptamers including H4 with a Kd value of 4 nM [215]. Then, the same research group engineered a H4-based bivalent species by using an unusual, very long poly(T) spacer of 100 residues. The obtained derivative exhibited a ca. 3-fold improved binding affinity compared to the corresponding monovalent sequence (Table 1) [215].
Starting from the 33t aptamer identified by Gold and Janjic [216], Potty et al. evaluated different derivatives obtained by truncation or elongation of the original sequence leading to the +5′G+3′C analogue [217]. This aptamer was then extensively studied by Manochehry and coworkers, who built homodimeric constructs connecting two +5′G+3′C motifs with poly(T) linkers of 20 or 60 units. Both homodimers showed slightly improved binding affinity compared to their monovalent counterpart, with no significant difference associated with the linker length (Table 1) [213].
Besides DNA-based aptamers with stem-loop structures, also high affinity G-rich oligonucleotides were fished out against VEGF165 by SELEX [209].
In this context, Nonaka et al. selected the G-rich aptamer Vap7 able to bind both VEGF121 and VEGF165 isoforms of VEGF-A with high affinity (Kd values of 1.0 and 20 nM, respectively, Table 1) [218]. Successively, the same research group developed a truncated form, named V7t1, containing only the bases presumably involved in the G4 structure formation. Compared to the starting Vap7, V7t1 exhibited higher affinity for VEGF165 (Kd = 1.4 vs. 20 nM; Table 1) associated with comparable affinity for VEGF121 (Kd = 1.1 nM, Table 1) [218]. V7t1 also showed remarkable antiproliferative activity on several cancer cell lines [219]. However, its marked structural polymorphism in K+-containing solutions [220], stimulated the search of improved derivatives, such as the 3R02 aptamer (Table 1), identified by an in silico maturation approach [221]. In order to elucidate the bioactive conformation of V7t1, Moccia and coworkers recently investigated the conformational behavior of this G-rich oligomer in a Na+-rich buffer, mimicking the saline composition of the extracellular environment in which VEGF targeting should occur [222]. In the tested conditions, V7t1 exhibited a different structuring capability dependent on the sample preparation procedure. Indeed, V7t1 samples not subjected to annealing (i.e., directly dissolved in the selected buffer solution without any prior thermal treatment) folded in solution giving mainly dimeric parallel G4 structures, accompanied by low amounts of monomeric G4 forms. In contrast, when subjected to annealing procedures, V7t1 significantly rearranged to give exclusively monomeric G4 structures of mixed topologies [222].
Evidence of the coexistence of both monomeric and dimeric G4 forms has been obtained by native polyacrylamide gel electrophoresis (PAGE), size exclusion chromatography (SE-HPLC) and dynamic light scattering (DLS) experiments, which represent a very useful combination of techniques to verify the formation of multimers or aggregates in solution [164,223,224]. Remarkably, electrophoretic mobility shift assay (EMSA) experiments unambiguously demonstrated that only the dimeric species formed in the not-annealed V7t1 samples efficiently bound VEGF165, whereas the monomeric G4 species did not bind the protein under the same explored conditions [222], thus demonstrating a marked preference of the protein for dimeric G4 structures even when monomolecular forms are concomitantly present [222].
Following these results, the same research group also explored a focused set of three homodimeric V7t1 covalent dimers [225]. In detail, a poly(T) linker of 7 nucleotides, a HEG- or a TEG-based spacer were used, providing the derivatives named bisV7t1T7, bisV7t1HEG2 and bisV7t1TEG2D, respectively. In the latter analogue, i.e., bisV7t1TEG2D, the V7t1 units were connected through the 3′-extremities of each monomeric sequence, thus imposing a 3′-3′ inversion of polarity in the dimeric construct [225].
Compared to the starting V7t1 aptamer, all the covalent dimeric forms showed a slightly lower binding ability to the target protein but similar, if not slightly higher, antiproliferative activity on human breast adenocarcinoma MCF-7 cells. The species with the highest VEGF165 binding affinity was bisV7t1TEG2D, in which the inversion of polarity probably promotes a peculiar spatial arrangement, which is preferred by the protein [225].
Also the V7t1 analogue 3R02 was used to build a homodimeric derivative with a poly(T) spacer of 10 residues, which showed ca. 10-fold higher affinity towards VEGF165 than the corresponding monovalent form (Table 1) [221].
A very interesting VEGF-targeting heterodimeric variants was proposed by Nonaka and coworkers, who connected a DNA-forming steam loop structure with a G4-forming sequence.
In detail, del5-1 and V7t1 were joined by a poly(T) spacer of 10 bases providing a bivalent construct with a noteworthy improvement in binding affinity with respect to each monomeric component (Table 1) [218].
5.8. Hepatocyte Growth Factor Receptor (HGFR)
Hepatocyte growth factor (HGF) regulates cell growth, motility and proliferation upon binding to the cell surface proto-oncogenic c-Met receptor, also called hepatocyte growth factor receptor (HGFR) [226,227,228]. In detail, activation of c-Met through dimerization triggers different signaling cascades involved in angiogenesis, cancer development and metastasis [229,230,231].
As a c-Met binding molecule, the 50-mer DNA aptamer CLN0003_SL1 (or simply SL1) was identified, showing a Kd of 123 nM (Table 1) in SNU-5 cells (c-Met+) [232].

Table 1.
Overview of the here discussed multivalent aptamers developed for different diseases. The specific linker between different aptamer motifs is highlighted in bold. For dimeric and multimeric constructs, the enhancement in binding affinity or activity is also reported. In LD201* aptamer, the underlined sequence is the one conserved from the original LD201 aptamer and the nucleobases in italics are those removed in the design of mΔ1. In the sequence of TD05.17 aptamer, locked nucleic acid (LNA) residues are highlighted in red. In OSJ-T3-LNA-OMe sequence, LNA and 2′-OMe RNA nucleobases are marked in red and blue, respectively. In VEa5 and Vap7 aptamers, the sequence derived from the primer regions are shown in italic lower case letters.
Then, the same research group engineered several dimeric forms of SL1. The most promising analogue was a 100-mer DNA aptamer composed of two different 50-mer units, in which one is SL1 (Table 1). In cell-based experiments, this dimer (named ss-0) proved to induce c-Met activation, also reproducing HGF-induced cellular behaviors. The binding affinity constants in comparison with the starting monomers were not reported in this work [233].
6. Conclusions
To mediate communication between proteins, small molecules and cells, biological systems often take advantage of multivalency, in which many and cooperative low affinity interactions lead to robust and high affinity binding events [234,235].
Since aptamers isolated by SELEX do not always show the desired affinities, refinement of their properties is often necessary. Among the different post-SELEX optimization strategies, such as truncation, and chemical modification, dimerization/multimerization proved to be a very powerful and effective approach to improve aptamer binding affinity and therapeutic efficacy [50,51].
A key role in the design of dimeric or multivalent constructs is played by the connecting linker joining two or more aptamer motifs, whose length, chemical nature, and flexibility strongly influence aptamer-protein interaction.
In this context, it could be noteworthy to underline the analogy with the connecting part of an aptazyme whose design has to mediate flexibility and stability in order to allow the proper transmission of the ligand-aptamer-signal to the catalytic nucleic acid core through a conformational change [236].
In dimeric/multimeric aptamer design, the simplest approach to link two or more aptamer sequences is represented by a simple end-to-end covalent connection, not requiring linkers to control the distance between each individual functional unit. In turn, the presence of linkers can be very effective to obtain optimized multimeric aptamers. The most simple and exploited linkers are based on poly(dA) or poly(T) sequences of variable length, which usually contain 15–20 nucleotides.
As further valuable nucleotide-based linkers, elements capable of forming well-defined secondary structures are also used. Typically, a duplex-like bridge is formed between each aptamer unit allowing their proper positioning and correct activity. Also linkers of non-nucleotidic nature, such as PEG-based ones, are frequently used for their flexibility and can be also added as terminal appendages of the aptamers to improve their enzymatic resistance to nuclease degradation.
Notably, the binding affinity or therapeutic activity of multivalent aptamers are not proportional to the number of connected monomers. Indeed, not always these properties improve as the number of single entities increases, essentially for the steric hindrance or high molecular weight of the obtained construct.
So specific features of the connecting linker have to be often defined by a trial-and-error approach, especially when high-resolution structural data (NMR or crystallographic) for the aptamer/protein complex formation are not available.
In order to improve the cooperative binding of the aptamer units to a selected protein target, there are no simple rules on the length or physico-chemical properties of the connecting linker. Indeed, even using linkers with similar properties, different results have been obtained with different aptamers.
However, the spacer has to be sufficiently long so to allow the correct aptamer structuring and should span the effective distance between the different protein binding sites.
Besides its length, also the linker orientation is a crucial factor to be considered. In fact, the linker is responsible for the spatial presentation of one aptamer unit with respect to the other one. To allow the correct orientation of each aptamer motif vs. the protein binding site, the linker inserted in the dimeric/multimeric aptamer has to be sufficiently flexible to avoid constraints, that could affect the aptamer ability to recognize and thus interact with the protein binding site.
However, bivalent and multivalent aptamers are very promising and versatile tools with potential applications as anticoagulants, anti-inflammatory, antiviral, and anticancer therapeutics. The area of these therapeutic applications and the set of newly designed highly effective multivalent aptamers will certainly be enriched in the foreseeable future.
Funding
C.R. is financed by Italian Ministry of Economic Development (MiSE)—Fund for Sustainable Development—Call “HORIZON2020” PON I&C 2014-2020, FOR.TUNA project, code No. F/050347/01_03/X32.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, R.E.; Wu, H.; Niu, Y.; Cai, J. Improving the stability of aptamers by chemical modification. Curr. Med. Chem. 2011, 18, 4126–4138. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liang, C.; Lv, Q.; Li, D.; Xu, X.; Liu, B.; Lu, A.; Zhang, G. Molecular selection, modification and development of therapeutic oligonucleotide aptamers. Int. J. Mol. Sci. 2016, 17, 358. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef] [PubMed]
- Röthlisberger, P.; Hollenstein, M. Aptamer chemistry. Adv. Drug Deliv. Rev. 2018, 134, 3–21. [Google Scholar] [CrossRef]
- Adachi, T.; Nakamura, Y. Aptamers: A review of their chemical properties and modifications for therapeutic application. Molecules 2019, 24, 4229. [Google Scholar] [CrossRef]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Al Bawab, A.; Ismail, S.I. Aptamers chemistry: Chemical modifications and conjugation strategies. Molecules 2020, 25, 3. [Google Scholar] [CrossRef]
- Parashar, A. Aptamers in therapeutics. J. Clin. Diagn. Res. 2016, 10, BE01–BE06. [Google Scholar] [CrossRef]
- Maier, K.E.; Levy, M. From selection hits to clinical leads: Progress in aptamer discovery. Mol. Ther. Methods Clin. Dev. 2016, 5, 16014–16023. [Google Scholar] [CrossRef]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Ismail, S.I.; Alshaer, W. Therapeutic aptamers in discovery, preclinical and clinical stages. Adv. Drug Deliv. Rev. 2018, 134, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Leslie, M.; Kameyama, H.; Volk, D.E.; Tanaka, T. Aptamer therapeutics in cancer: Current and future. Cancers 2018, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.H.; Elsherbiny, M.E.; Emara, M. Updates on aptamer research. Int. J. Mol. Sci. 2019, 20, 2511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef]
- Santosh, B.; Yadava, P.K. Nucleic acid aptamers: Research tools in disease diagnostics and therapeutics. Biomed. Res. Int. 2014, 2014, 50451–50464. [Google Scholar] [CrossRef]
- Ma, H.; Liu, J.; Ali, M.M.; Mahmood, M.A.I.; Labanieh, L.; Lu, M.; Iqbal, S.M.; Zhang, Q.; Zhao, W.; Wan, Y. Nucleic acid aptamers in cancer research, diagnosis and therapy. Chem. Soc. Rev. 2015, 44, 1240–1256. [Google Scholar] [CrossRef]
- Ku, T.H.; Zhang, T.; Luo, H.; Yen, T.M.; Chen, P.W.; Han, Y.; Lo, Y.H. Nucleic acid aptamers: An emerging tool for biotechnology and biomedical sensing. Sensors 2015, 15, 16281–16313. [Google Scholar] [CrossRef]
- Chandola, C.; Kalme, S.; Casteleijn, M.G.; Urtti, A.; Neerathilingam, M. Application of aptamers in diagnostics, drug-delivery and imaging. J. Biosci. 2016, 41, 535–561. [Google Scholar] [CrossRef]
- Musumeci, D.; Platella, C.; Riccardi, C.; Moccia, F.; Montesarchio, D. Fluorescence sensing using DNA aptamers in cancer research and clinical diagnostics. Cancers 2017, 9, 174. [Google Scholar] [CrossRef]
- Dhiman, A.; Kalra, P.; Bansal, V.; Bruno, J.G.; Sharma, T.K. Aptamer-based point-of-care diagnostic platforms. Sens. Actuators B Chem. 2017, 246, 535–553. [Google Scholar] [CrossRef]
- Hori, S.I.; Herrera, A.; Rossi, J.J.; Zhou, J. Current advances in aptamers for cancer diagnosis and therapy. Cancers 2018, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Kulabhusan, P.K.; Hussain, B.; Yüce, M. Current perspectives on aptamers as diagnostic tools and therapeutic agents. Pharmaceutics 2020, 12, 646. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 1992, 355, 850–852. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX-A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef]
- Keefe, A.D.; Cload, S.T. SELEX with modified nucleotides. Curr. Opin. Chem. Biol. 2008, 12, 448–456. [Google Scholar] [CrossRef]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2014, 33, 1141–1161. [Google Scholar] [CrossRef]
- Wu, Y.X.; Kwon, Y.J. Aptamers: The “evolution” of SELEX. Methods 2016, 106, 21–28. [Google Scholar] [CrossRef]
- Antipova, O.M.; Zavyalova, E.G.; Golovin, A.V.; Pavlova, G.V.; Kopylov, A.M.; Reshetnikov, R.V. Advances in the application of modified nucleotides in SELEX technology. Biochemistry 2018, 83, 1161–1172. [Google Scholar] [CrossRef]
- Bayat, P.; Nosrati, R.; Alibolandi, M.; Rafatpanah, H.; Abnous, K.; Khedri, M.; Ramezani, M. SELEX methods on the road to protein targeting with nucleic acid aptamers. Biochimie 2018, 154, 132–155. [Google Scholar] [CrossRef] [PubMed]
- Sola, M.; Menon, A.; Moreno, B.; Meraviglia-Crivelli, D.; Soldevilla, M.; Cartón-García, F.; Pastor, F. Aptamers against live targets: Is in vivo SELEX finally coming to edge? Mol. Ther. Nucleic Acids 2020, 21, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Vandghanooni, S.; Eskandani, M.; Barar, J.; Omidi, Y. Recent advances in aptamer-armed multimodal theranostic nanosystems for imaging and targeted therapy of cancer. Eur. J. Pharm. Sci. 2018, 117, 301–312. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Wen, N.; Xiao, D.; Yan, J.; Xiong, H.; Cai, S.; Liu, Z.; Liu, Y. Aptamer-based targeted drug delivery systems: Current potential and challenges. Curr. Med. Chem. 2020, 27, 2189–2219. [Google Scholar] [CrossRef] [PubMed]
- Broude, N.E. Stem-loop oligonucleotides: A robust tool for molecular biology and biotechnology. Trends Biotechnol. 2002, 20, 249–256. [Google Scholar] [CrossRef]
- Musumeci, D.; Riccardi, C.; Montesarchio, D. G-quadruplex forming oligonucleotides as anti-HIV agents. Molecules 2015, 20, 17511–17532. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 2016, 59, 5987–6011. [Google Scholar] [CrossRef]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1429–1447. [Google Scholar] [CrossRef]
- Kwok, C.K.; Merrick, C.J. G-Quadruplexes: Prediction, characterization, and biological application. Trends Biotechnol. 2017, 35, 997–1013. [Google Scholar] [CrossRef]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G-quadruplex forming aptamers characteristics, applications, and perspectives. Molecules 2019, 24, 3781. [Google Scholar] [CrossRef]
- Gatto, B.; Palumbo, M.; Sissi, C. Nucleic acid aptamers based on the G-quadruplex structure: Therapeutic and diagnostic potential. Curr. Med. Chem. 2009, 16, 1248–1265. [Google Scholar] [CrossRef]
- Collie, G.W.; Parkinson, G.N. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011, 40, 5867–5892. [Google Scholar] [CrossRef]
- Dolinnaya, N.G.; Ogloblina, A.M.; Yakubovskaya, M.G. Structure, properties, and biological relevance of the DNA and RNA G-quadruplexes: Overview 50 years after their discovery. Biochemistry 2016, 81, 1602–1649. [Google Scholar] [CrossRef]
- Tucker, W.O.; Shum, K.T.; Tanner, J.A. G-quadruplex DNA aptamers and their ligands: Structure, function and application. Curr. Pharm. Des. 2012, 18, 2014–2026. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Arachchilage, G.M.; Basu, S. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef]
- Largy, E.; Marchand, A.; Amrane, S.; Gabelica, V.; Mergny, J.L. Quadruplex turncoats: Cation-dependent folding and stability of quadruplex-DNA double switches. J. Am. Chem. Soc. 2016, 138, 2780–2792. [Google Scholar] [CrossRef]
- Largy, E.; Mergny, J.L.; Gabelica, V. Role of alkali metal ions in G-quadruplex nucleic acid structure and stability. Met. Ions Life Sci. 2016, 16, 203–258. [Google Scholar] [CrossRef]
- Musumeci, D.; Montesarchio, D. Polyvalent nucleic acid aptamers and modulation of their activity: A focus on the thrombin binding aptamer. Pharmacol. Ther. 2012, 136, 202–215. [Google Scholar] [CrossRef]
- Vorobyeva, M.; Vorobjev, P.; Venyaminova, A. Multivalent aptamers: Versatile tools for diagnostic and therapeutic applications. Molecules 2016, 21, 1613. [Google Scholar] [CrossRef]
- Gao, S.; Zheng, X.; Jiao, B.; Wang, L. Post-SELEX optimization of aptamers. Anal. Bioanal. Chem. 2016, 408, 4567–4573. [Google Scholar] [CrossRef]
- Hasegawa, H.; Savory, N.; Abe, K.; Ikebukuro, K. Methods for improving aptamer binding affinity. Molecules 2016, 21, 421. [Google Scholar] [CrossRef]
- Bevilacqua, M.P.; Nelson, R.M. Selectins. J. Clin. Investig. 1993, 91, 379–387. [Google Scholar] [CrossRef]
- Rosen, S.D.; Bertozzi, C.R. The selectins and their ligands. Curr. Opin. Cell Biol. 1994, 6, 663–673. [Google Scholar] [CrossRef]
- Rainer, T.H. L-selectin in health and disease. Resuscitation 2002, 52, 127–141. [Google Scholar] [CrossRef]
- Raffler, N.A.; Rivera-Nieves, J.; Ley, K. L-selectin in inflammation, infection and immunity. Drug Discov. Today Ther. Strateg. 2005, 2, 213–220. [Google Scholar] [CrossRef]
- Ivetic, A. A head-to-tail view of L-selectin and its impact on neutrophil behaviour. Cell Tissue Res. 2018, 371, 437–453. [Google Scholar] [CrossRef]
- Ivetic, A.; Green, H.L.H.; Hart, S.J. L-selectin: A major regulator of leukocyte adhesion, migration and signaling. Front. Immunol. 2019, 14, 1068. [Google Scholar] [CrossRef]
- Watson, S.R.; Chang, Y.F.; O’Connell, D.; Weigand, L.; Ringquist, S.; Parma, D.H. Anti-L-selectin aptamers: Binding characteristics, pharmacokinetic parameters, and activity against an intravascular target in vivo. Antisense Nucleic Acid Drug Dev. 2000, 10, 63–75. [Google Scholar] [CrossRef]
- Hicke, B.J.; Watson, S.R.; Koenig, A.; Lynott, C.K.; Bargatze, R.F.; Chang, Y.F.; Ringquist, S.; Moon-McDermott, L.; Jennings, S.; Fitzwater, T.; et al. DNA aptamers block L-selectin function in vivo: Inhibition of human lymphocyte trafficking in SCID mice. J. Clin. Investig. 1996, 98, 2688–2692. [Google Scholar] [CrossRef]
- Romig, T.S.; Bell, C.; Drolet, D.W. Aptamer affinity chromatography: Combinatorial chemistry applied to protein purification. J. Chromatogr. B Biomed. Sci. Appl. 1999, 731, 275–284. [Google Scholar] [CrossRef]
- Riese, S.B.; Buscher, K.; Enders, S.; Kuehne, C.; Tauber, R.; Dernedde, J. Structural requirements of mono- and multivalent L-selectin blocking aptamers for enhanced receptor inhibition in vitro and in vivo. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 901–908. [Google Scholar] [CrossRef]
- Chang, E.K.; Eckert, M.A.; Ali, M.M.; Riazifar, H.; Pone, E.J.; Liu, L.; Zhao, W. Facile supermolecular aptamer inhibitors of L-selectin. PLoS ONE 2015, 10, e0123034. [Google Scholar] [CrossRef]
- Ali, M.M.; Kang, D.-K.; Zhao, W.; Li, F.; Zhang, Z.; Zhang, K.; Le, X.C.; Ankrum, J.A. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Bala, J.; Chinnapaiyan, S.; Dutta, R.K.; Unwalla, H. Aptamers in HIV research diagnosis and therapy. RNA Biol. 2018, 15, 327–337. [Google Scholar] [CrossRef]
- Ruggiero, E.; Richter, S.N. Survey and summary G-quadruplexes and G-quadruplex ligands: Targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef]
- Zou, X.; Wu, J.; Gu, J.; Shen, L.; Mao, L. Application of aptamers in virus detection and antiviral therapy. Front. Microbiol. 2019, 10, 1462. [Google Scholar] [CrossRef]
- De Soultrait, V.R.; Lozach, P.Y.; Altmeyer, R.; Tarrago-Litvak, L.; Litvak, S.; Andréola, M.L. DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J. Mol. Biol. 2002, 324, 195–203. [Google Scholar] [CrossRef]
- Andreola, M.-L. Closely related antiretroviral agents as inhibitors of two HIV-1 enzymes, ribonuclease H and integrase: “Killing two birds with one stone”. Curr. Pharm. Des. 2005, 10. [Google Scholar] [CrossRef]
- Métifiot, M.; Leon, O.; Tarrago-Litvak, L.; Litvak, S.; Andréola, M.L. Targeting HIV-1 integrase with aptamers selected against the purified RNase H domain of HIV-1 RT. Biochimie 2005, 87, 911–919. [Google Scholar] [CrossRef]
- Ojwang, J.O.; Buckheit, R.W.; Pommier, Y.; Mazumder, A.; De Vreese, K.; Este, J.A.; Reymen, D.; Pallansch, L.A.; Lackman-Smith, C.; Wallace, T.L.; et al. T30177, an oligonucleotide stabilized by an intramolecular guanosine octet, is a potent inhibitor of laboratory strains and clinical isolates of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 1995, 39, 2426–2435. [Google Scholar] [CrossRef]
- Mazumder, A.; Neamati, N.; Ojwang, J.O.; Sunder, S.; Rando, R.F.; Pommier, Y. Inhibition of the human immunodeficiency virus type 1 integrase by guanosine quartet structures. Biochemistry 1996, 35, 13762–13771. [Google Scholar] [CrossRef]
- Jing, N.; Rando, R.F.; Pommier, Y.; Hogan, M.E. Ion selective folding of loop domains in a potent anti-HIV oligonucleotide. Biochemistry 1997, 36, 12498–12505. [Google Scholar] [CrossRef]
- Jing, N.; Hogan, M.E. Structure-activity of tetrad-forming oligonucleotides as a potent anti-HIV therapeutic drug. J. Biol. Chem. 1998, 273, 34992–34999. [Google Scholar] [CrossRef]
- Do, N.Q.; Lim, K.W.; Teo, M.H.; Heddi, B.; Phan, A.T. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity. Nucleic Acids Res. 2011, 39, 9448–9457. [Google Scholar] [CrossRef]
- Phan, A.T.; Kuryavyi, V.; Ma, J.B.; Faure, A.; Andréola, M.L.; Patel, D.J. An interlocked dimeric parallel-stranded DNA quadruplex: A potent inhibitor of HIV-1 integrase. Proc. Natl. Acad. Sci. USA 2005, 102, 634–639. [Google Scholar] [CrossRef]
- Mukundan, V.T.; Do, N.Q.; Phan, A.T. HIV-1 integrase inhibitor T30177 forms a stacked dimeric G-quadruplex structure containing bulges. Nucleic Acids Res. 2011, 39, 8984–8991. [Google Scholar] [CrossRef]
- Nici, F.; Oliviero, G.; Falanga, A.P.; D’Errico, S.; Marzano, M.; Musumeci, D.; Montesarchio, D.; Noppen, S.; Pannecouque, C.; Piccialli, G.; et al. Anti-HIV activity of new higher order G-quadruplex aptamers obtained from tetra-end-linked oligonucleotides. Org. Biomol. Chem. 2018, 16, 2349–2355. [Google Scholar] [CrossRef]
- Borbone, N.; Amato, J.; Oliviero, G.; D’Atri, V.; Gabelica, V.; De Pauw, E.; Piccialli, G.; Mayol, L. d(CGGTGGT) forms an octameric parallel G-quadruplex via stacking of unusual G(:C):G(:C):G(:C):G(:C) octads. Nucleic Acids Res. 2011, 39, 7848–7857. [Google Scholar] [CrossRef]
- D’Atri, V.; Borbone, N.; Amato, J.; Gabelica, V.; D’Errico, S.; Piccialli, G.; Mayol, L.; Oliviero, G. DNA-based nanostructures: The effect of the base sequence on octamer formation from d(XGGYGGT) tetramolecular G-quadruplexes. Biochimie 2014, 99, 119–128. [Google Scholar] [CrossRef]
- D’Atri, V.; Oliviero, G.; Amato, J.; Borbone, N.; D’Errico, S.; Mayol, L.; Piccialli, V.; Haider, S.; Hoorelbeke, B.; Balzarini, J.; et al. New anti-HIV aptamers based on tetra-end-linked DNA G-quadruplexes: Effect of the base sequence on anti-HIV activity. Chem. Commun. 2012, 48, 9516–9518. [Google Scholar] [CrossRef]
- D’Onofrio, J.; Petraccone, L.; Erra, E.; Martino, L.; Di Fabio, G.; De Napoli, L.; Giancola, C.; Montesarchio, D. 5′-modified G-quadruplex forming oligonucleotides endowed with anti-HIV activity: Synthesis and biophysical properties. Bioconjugate Chem. 2007, 18, 1194–1204. [Google Scholar] [CrossRef]
- Huntington, J.A. Molecular recognition mechanisms of thrombin. J. Thromb. Haemost. 2005, 3, 1861–1872. [Google Scholar] [CrossRef]
- Wolberg, A.S. Thrombin generation and fibrin clot structure. Blood Rev. 2007, 21, 131–142. [Google Scholar] [CrossRef]
- Di Cera, E. Thrombin. Mol. Aspects Med. 2008, 29, 203–254. [Google Scholar] [CrossRef]
- Licari, L.G.; Kovacic, J.P. Thrombin physiology and pathophysiology. J. Vet. Emerg. Crit. Care 2009, 19, 11–22. [Google Scholar] [CrossRef]
- Mazepa, M.; Hoffman, M.; Monroe, D. Superactivated platelets: Thrombus regulators, thrombin generators, and potential clinical targets. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1747–1752. [Google Scholar] [CrossRef]
- Posma, J.J.N.; Posthuma, J.J.; Spronk, H.M.H. Coagulation and non-coagulation effects of thrombin. J. Thromb. Haemost. 2016, 14, 1908–1916. [Google Scholar] [CrossRef]
- Huntington, J.A.; Baglin, T.P. Targeting thrombin rational drug design from natural mechanisms. Trends Pharmacol. Sci. 2003, 24, 589–595. [Google Scholar] [CrossRef]
- Hirsh, J. Current anticoagulant therapy unmet clinical needs. Thromb. Res. 2003, 109, S1–S8. [Google Scholar] [CrossRef]
- Gómez Outes, A.; Suárez Gea, M.L.; Pozo Hernández, C.; Lecumberri, R.; Rocha, E.; Vargas Castrillón, E. New parenteral anticoagulants in development. Ther. Adv. Cardiovasc. Dis. 2011, 5, 33–59. [Google Scholar] [CrossRef]
- Zavyalova, E.G.; Ustinov, N.; Golovin, A.; Pavlova, G.; Kopylov, A. G-quadruplex aptamers to human thrombin versus other direct thrombin inhibitors: The focus on mechanism of action and drug efficiency as anticoagulants. Curr. Med. Chem. 2016, 23, 2230–2244. [Google Scholar] [CrossRef]
- Becker, R.C.; Povsic, T.; Cohen, M.G.; Rusconi, C.P.; Sullenger, B. Nucleic acid aptamers as antithrombotic agents: Opportunities in extracellular therapeutics. Thromb. Haemost. 2010, 103, 586–595. [Google Scholar] [CrossRef]
- Nimjee, S.M.; Povsic, T.J.; Sullenger, B.A.; Becker, R.C. Translation and clinical development of antithrombotic aptamers. Nucleic Acid Ther. 2016, 26, 147–155. [Google Scholar] [CrossRef]
- Ponce, A.T.; Hong, K.L. A mini-review: Clinical development and potential of aptamers for thrombotic events treatment and monitoring. Biomedicines 2019, 7, 55. [Google Scholar] [CrossRef]
- Riccardi, C.; Napolitano, E.; Platella, C.; Musumeci, D.; Montesarchio, D. G-quadruplex-based aptamers targeting human thrombin: Discovery, chemical modifications and antithrombotic effects. Pharmacol. Ther. 2020, 107649. [Google Scholar] [CrossRef]
- Tasset, D.M.; Kubik, M.F.; Steiner, W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J. Mol. Biol. 1997, 272, 688–698. [Google Scholar] [CrossRef]
- Marson, G.; Palumbo, M.; Sissi, C. Folding versus charge: Understanding selective target recognition by the thrombin aptamers. Curr. Pharm. Des. 2012, 18, 2027–2035. [Google Scholar] [CrossRef]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roe, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA 1993, 90, 3745–3749. [Google Scholar] [CrossRef]
- Wang, K.Y.; Bolton, P.H.; McCurdy, S.; Shea, R.G.; Swaminathan, S. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry 1993, 32, 1899–1904. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.Y.; Bolton, P.H.; Krawczyk, S.H.; Bischofberger, N.; Swaminathan, S. The tertiary structure of a DNA aptamer which binds to and inhibits thrombin determines activity. Biochemistry 1993, 32, 11285–11292. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, K.; Padmanabhan, K.P.; Ferrara, J.D.; Sadler, J.E.; Tulinsky, A. The structure of α-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993, 268, 17651–17654. [Google Scholar] [CrossRef] [PubMed]
- Schultze, P.; Macaya, R.F.; Feigon, J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG). J. Mol. Biol. 1994, 235, 1532–1547. [Google Scholar] [CrossRef]
- Padmanabhan, K.; Tulinsky, A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr. Sect. D Biol. Crystallogr. 1996, 52, 272–282. [Google Scholar] [CrossRef]
- Kelly, J.A.; Feigon, J.; Yeates, T.O. Reconciliation of the X-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG). J. Mol. Biol. 1996, 256, 417–422. [Google Scholar] [CrossRef]
- Zavyalova, E.G.; Golovin, A.; Reshetnikov, R.; Mudrik, N.; Panteleyev, D.; Pavlova, G.; Kopylov, A. Novel modular DNA aptamer for human thrombin with high anticoagulant activity. Curr. Med. Chem. 2011, 18, 3343–3350. [Google Scholar] [CrossRef]
- Poniková, S.; Tlučková, K.; Antalík, M.; Víglaský, V.; Hianik, T. The circular dichroism and differential scanning calorimetry study of the properties of DNA aptamer dimers. Biophys. Chem. 2011, 155, 29–35. [Google Scholar] [CrossRef]
- Amato, T.; Virgilio, A.; Pirone, L.; Vellecco, V.; Bucci, M.; Pedone, E.; Esposito, V.; Galeone, A. Investigating the properties of TBA variants with twin thrombin binding domains. Sci. Rep. 2019, 9, 9184. [Google Scholar] [CrossRef]
- Müller, J.; Wulffen, B.; Pötzsch, B.; Mayer, G. Multidomain targeting generates a high-affinity thrombin-inhibiting bivalent aptamer. ChemBioChem 2007, 8, 2223–2226. [Google Scholar] [CrossRef]
- Müller, J.; Freitag, D.; Mayer, G.; Pötzsch, B. Anticoagulant characteristics of HD1-22, a bivalent aptamer that specifically inhibits thrombin and prothrombinase. J. Thromb. Haemost. 2008, 6, 2105–2112. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Taira, K.I.; Sode, K.; Ikebukuro, K. Improvement of aptamer affinity by dimerization. Sensors 2008, 8, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Heyduk, T. Bivalent ligands with long nanometer-scale flexible linkers. Biochemistry 2009, 48, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Hughes, Q.W.; Le, B.T.; Gilmore, G.; Baker, R.I.; Veedu, R.N. Construction of a bivalent thrombin binding aptamer and its antidote with improved properties. Molecules 2017, 22, 1770. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, K.M.; Xiao, Y.; Soh, H.T. Selection is more intelligent than design: Improving the affinity of a bivalent ligand through directed evolution. Nucleic Acids Res. 2012, 40, 11777–11783. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Cao, Z.; Tan, W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc. Natl. Acad. Sci. USA 2008, 105, 5664–5669. [Google Scholar] [CrossRef]
- Di Giusto, D.A.; King, G.C. Construction, stability, and activity of multivalent circular anticoagulant aptamers. J. Biol. Chem. 2004, 279, 46483–46489. [Google Scholar] [CrossRef]
- Di Giusto, D.A.; Knox, S.M.; Lai, Y.; Tyrelle, G.D.; Aung, M.T.; King, G.C. Multitasking by multivalent circular DNA aptamers. ChemBioChem 2006, 7, 535–544. [Google Scholar] [CrossRef]
- Hsiao, K.Y.; Sun, H.S.; Tsai, S.J. Circular RNA—New member of noncoding RNA with novel functions. Exp. Biol. Med. 2017, 242, 1136–1141. [Google Scholar] [CrossRef]
- Li, J.; Mohammed-Elsabagh, M.; Paczkowski, F.; Li, Y. Circular nucleic acids: Discovery, functions and applications. ChemBioChem 2020, 22, 1547–1566. [Google Scholar] [CrossRef]
- Riccardi, C.; Meyer, A.; Vasseur, J.J.; Russo Krauss, I.; Paduano, L.; Oliva, R.; Petraccone, L.; Morvan, F.; Montesarchio, D. Stability is not everything: The case of the cyclization of the thrombin binding aptamer. ChemBioChem 2019, 20, 1789–1794. [Google Scholar] [CrossRef]
- Riccardi, C.; Meyer, A.; Vasseur, J.J.; Russo Krauss, I.; Paduano, L.; Morvan, F.; Montesarchio, D. Fine-tuning the properties of the thrombin binding aptamer through cyclization: Effect of the 5′-3′ connecting linker on the aptamer stability and anticoagulant activity. Bioorg. Chem. 2020, 94, 103379. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Meyer, A.; Vasseur, J.J.; Cavasso, D.; Russo Krauss, I.; Paduano, L.; Morvan, F.; Montesarchio, D. Design, synthesis and characterization of cyclic NU172 analogues: A biophysical and biological insight. Int. J. Mol. Sci. 2020, 21, 3860. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Merlino, A.; Giancola, C.; Randazzo, A.; Mazzarella, L.; Sica, F. Thrombin-aptamer recognition: A revealed ambiguity. Nucleic Acids Res. 2011, 39, 7858–7867. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Merlino, A.; Randazzo, A.; Novellino, E.; Mazzarella, L.; Sica, F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012, 40, 8119–8128. [Google Scholar] [CrossRef]
- Yigit, M.V.; Mazumdar, D.; Lu, Y. MRI detection of thrombin with aptamer functionalized superparamagnetic iron oxide nanoparticles. Bioconjugate Chem. 2008, 19, 412–417. [Google Scholar] [CrossRef]
- Musumeci, D.; Oliviero, G.; Roviello, G.N.; Bucci, E.M.; Piccialli, G. G-quadruplex-forming oligonucleotide conjugated to magnetic nanoparticles: Synthesis, characterization, and enzymatic stability assays. Bioconjugate Chem. 2012, 23, 382–391. [Google Scholar] [CrossRef]
- Yu, J.; Yang, L.; Liang, X.; Dong, T.; Liu, H. Bare magnetic nanoparticles as fluorescence quenchers for detection of thrombin. Analyst 2015, 140, 4114–4120. [Google Scholar] [CrossRef]
- Shiang, Y.C.; Huang, C.C.; Wang, T.H.; Chien, C.W.; Chang, H.T. Aptamer-conjugated nanoparticles efficiently control the activity of thrombin. Adv. Funct. Mater. 2010, 20, 3175–3182. [Google Scholar] [CrossRef]
- Shiang, Y.C.; Hsu, C.L.; Huang, C.C.; Chang, H.T. Gold nanoparticles presenting hybridized self-assembled aptamers that exhibit enhanced inhibition of thrombin. Angew. Chem. Int. Ed. Eng. 2011, 50, 7660–7665. [Google Scholar] [CrossRef]
- Hsu, C.L.; Chang, H.T.; Chen, C.T.; Wei, S.C.; Shiang, Y.C.; Huang, C.C. Highly efficient control of thrombin activity by multivalent nanoparticles. Chem. A Eur. J. 2011, 17, 10994–11000. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.L.; Wei, S.C.; Jian, J.W.; Chang, H.T.; Chen, W.H.; Huang, C.C. Highly flexible and stable aptamer-caged nanoparticles for control of thrombin activity. RSC Adv. 2012, 2, 1577–1584. [Google Scholar] [CrossRef]
- Huang, S.S.; Wei, S.C.; Chang, H.T.; Lin, H.J.; Huang, C.C. Gold nanoparticles modified with self-assembled hybrid monolayer of triblock aptamers as a photoreversible anticoagulant. J. Control. Release 2016, 221, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Cui, Y.; He, Q.; Yang, Y.; Fei, J.; Li, J. Selective recognition of co-assembled thrombin aptamer and docetaxel on mesoporous silica nanoparticles against tumor cell proliferation. Chemistry 2011, 17, 13170–13174. [Google Scholar] [CrossRef]
- Babu, E.; Mareeswaran, P.M.; Rajagopal, S. Highly sensitive optical biosensor for thrombin based on structure switching aptamer-luminescent silica nanoparticles. J. Fluoresc. 2013, 23, 137–146. [Google Scholar] [CrossRef]
- Yue, Q.; Shen, T.; Wang, L.; Xu, S.; Li, H.; Xue, Q.; Zhang, Y.; Gu, X.; Zhang, S.; Liu, J. A convenient sandwich assay of thrombin in biological media using nanoparticle-enhanced fluorescence polarization. Biosens. Bioelectron. 2014, 56, 231–236. [Google Scholar] [CrossRef]
- Riccardi, C.; Russo Krauss, I.; Musumeci, D.; Morvan, F.; Meyer, A.; Vasseur, J.J.; Paduano, L.; Montesarchio, D. Fluorescent thrombin binding aptamer-tagged nanoparticles for an efficient and reversible control of thrombin activity. ACS Appl. Mater. Interfaces 2017, 9, 35574–35587. [Google Scholar] [CrossRef]
- Lai, P.X.; Mao, J.Y.; Unnikrishnan, B.; Chu, H.W.; Wu, C.W.; Chang, H.T.; Huang, C.C. Self-assembled, bivalent aptamers on graphene oxide as an efficient anticoagulant. Biomater. Sci. 2018, 6, 1882–1891. [Google Scholar] [CrossRef]
- Lin, T.-X.; Lai, P.-X.; Mao, J.-Y.; Chu, H.-W.; Unnikrishnan, B.; Anand, A.; Huang, C.-C. Supramolecular aptamers on graphene oxide for efficient inhibition of thrombin activity. Front. Chem. 2019, 7. [Google Scholar] [CrossRef]
- Ireson, C.R.; Kelland, L.R. Discovery and development of anticancer aptamers. Mol. Cancer Ther. 2006, 5, 2957–2962. [Google Scholar] [CrossRef]
- Lee, J.W.; Kim, H.J.; Heo, K. Therapeutic aptamers: Developmental potential as anticancer drugs. BMB Rep. 2015, 48, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.O.; Kim, G.; Hwang, J.; Han, K.H.; Kwak, M.; Lee, P.C.W. Nucleic acid nanotechnology for cancer treatment. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188377. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Xiang, J. Aptamers, the nucleic acid antibodies, in cancer therapy. Int. J. Mol. Sci. 2020, 21, 2793. [Google Scholar] [CrossRef] [PubMed]
- Barsan, V.; Ramakrishna, S.; Davis, K.L. Immunotherapy for the treatment of acute lymphoblastic leukemia. Curr. Oncol. Rep. 2020, 22, 11. [Google Scholar] [CrossRef]
- Whitehead, T.P.; Metayer, C.; Wiemels, J.L.; Singer, A.W.; Miller, M.D. Childhood leukemia and primary prevention. Curr. Probl. Pediatr. Adolesc. Health Care 2016, 46, 317–352. [Google Scholar] [CrossRef]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef]
- Zhang, Z.; Ali, M.M.; Eckert, M.A.; Kang, D.K.; Chen, Y.Y.; Sender, L.S.; Fruman, D.A.; Zhao, W. A polyvalent aptamer system for targeted drug delivery. Biomaterials 2013, 34, 9728–9735. [Google Scholar] [CrossRef]
- Kuai, H.; Zhao, Z.; Mo, L.; Liu, H.; Hu, X.; Fu, T.; Zhang, X.; Tan, W. Circular bivalent aptamers enable in vivo stability and recognition. J. Am. Chem. Soc. 2017, 139, 9128–9131. [Google Scholar] [CrossRef]
- Fujiki, H.; Watanabe, T.; Suganuma, M. Cell-surface nucleolin acts as a central mediator for carcinogenic, anti-carcinogenic, and disease-related ligands. J. Cancer Res. Clin. Oncol. 2014, 140, 689–699. [Google Scholar] [CrossRef]
- Koutsioumpa, M.; Papadimitriou, E. Cell surface nucleolin as a target for anti-cancer therapies. Recent Pat. Anticancer Drug Discov. 2014, 9, 137–152. [Google Scholar] [CrossRef]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N.J.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Investig. New Drugs 2014, 32, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.J.; Kahlon, J.B.; Thomas, S.D.; Trent, J.O.; Miller, D.M. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J. Biol. Chem. 1999, 274, 26369–26377. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.J.; Laber, D.A.; Miller, D.M.; Thomas, S.D.; Trent, J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009, 86, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.J.; Reyes-reyes, E.M.; Malik, M.T.; Murphy, E.M.; Toole, M.G.O.; Trent, J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1414–1428. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; Zeng, X.; Wu, J.; Zhu, X.; Yu, X.; Zhang, X.; Zhang, J.; Liu, G.; Mei, L. Polydopamine-based surface modification of novel nanoparticle-aptamer bioconjugates for in vivo breast cancer targeting and enhanced therapeutic effects. Theranostics 2016, 6, 470–484. [Google Scholar] [CrossRef]
- Xu, G.; Yu, X.; Zhang, J.; Sheng, Y.; Liu, G.; Tao, W.; Mei, L. Robust aptamer–polydopamine-functionalized M-PLGA–TPGS nanoparticles for targeted delivery of docetaxel and enhanced cervical cancer therapy. Int. J. Nanomed. 2016, 11, 2953–2965. [Google Scholar] [CrossRef]
- Chen, D.; Li, B.; Cai, S.; Wang, P.; Peng, S.; Sheng, Y.; He, Y.; Gu, Y.; Chen, H. Dual targeting luminescent gold nanoclusters for tumor imaging and deep tissue therapy. Biomaterials 2016, 100, 1–16. [Google Scholar] [CrossRef]
- Riccardi, C.; Fàbrega, C.; Grijalvo, S.; Vitiello, G.; D’Errico, G.; Eritja, R.; Montesarchio, D. AS1411-decorated niosomes as effective nanocarriers for Ru(III)-based drugs in anticancer strategies. J. Mater. Chem. B 2018, 6, 5368–5384. [Google Scholar] [CrossRef]
- Yang, S.; Ren, Z.; Chen, M.; Wang, Y.; You, B.; Chen, W.; Qu, C.; Liu, Y.; Zhang, X. Nucleolin targeting AS1411-aptamer-modified graft polymeric micelle with dual pH/redox sensitivity designed to enhance tumor therapy through the codelivery of doxorubicin/TLR4 siRNA and suppression of invasion. Mol. Pharm. 2018, 15, 314–325. [Google Scholar] [CrossRef]
- Chen, W.H.; Yang Sung, S.; Fadeev, M.; Cecconello, A.; Nechushtai, R.; Willner, I. Targeted VEGF-triggered release of an anti-cancer drug from aptamer-functionalized metal–organic framework nanoparticles. Nanoscale 2018, 25. [Google Scholar] [CrossRef]
- Dailey, M.M.; Clarke Miller, M.; Bates, P.J.; Lane, A.N.; Trent, J.O. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010, 38, 4877–4888. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Lee, Y.B.; Lee, J.H.; Lee, D.H.; Cho, E.J.; Yu, S.J.; Kim, Y.J.; Kim, J.I.; Im, J.H.; Lee, J.H.; et al. Modified AS1411 aptamer suppresses hepatocellular carcinoma by up-regulating galectin-14. PLoS ONE 2016, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Sun, L.; Li, K.; Yang, X.; Cai, B.; Zhang, Y.; Zhu, Y.; Ma, Y.; Guan, Z.; Wu, Y.; et al. The bioactivity of D-/L-isonucleoside- and 2′-deoxyinosine-incorporated aptamer AS1411s including DNA replication/microRNA expression. Mol. Ther. Nucleic Acids 2017, 9, 218–229. [Google Scholar] [CrossRef]
- Riccardi, C.; Musumeci, D.; Russo Krauss, I.; Piccolo, M.; Irace, C.; Paduano, L.; Montesarchio, D. Exploring the conformational behaviour and aggregation properties of lipid-conjugated AS1411 aptamers. Int. J. Biol. Macromol. 2018, 118, 1384–1399. [Google Scholar] [CrossRef] [PubMed]
- Do, N.Q.; Chung, W.J.; Truong, T.H.A.; Heddi, B.; Phan, A.T. G-quadruplex structure of an anti-proliferative DNA sequence. Nucleic Acids Res. 2017, 45, 7487–7493. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Lopes-Nunes, J.; Lopes, A.C.; Cabral Campello, M.P.; Paulo, A.; Queiroz, J.A.; Cruz, C. Aptamer-guided acridine derivatives for cervical cancer. Org. Biomol. Chem. 2019, 17, 2992–3002. [Google Scholar] [CrossRef]
- Seiffert, D.; Crain, K.; Wagner, N.V.; Loskutoff, D.J. Vitronectin gene expression in vivo. Evidence for extrahepatic synthesis and acute phase regulation. J. Biol. Chem. 1994, 269, 19836–19842. [Google Scholar] [CrossRef]
- Zhuang, P.; Chen, A.I.; Peterson, C.B. Native and multimeric vitronectin exhibit similar affinity for heparin: Differences in heparin binding properties induced upon denaturation are due to self-association into a multivalent form. J. Biol. Chem. 1997, 272, 6858–6867. [Google Scholar] [CrossRef]
- Aaboe, M.; Offersen, B.V.; Christensen, A.; Andreasen, P.A. Vitronectin in human breast carcinomas. Biochim. Biophys. Acta Mol. Basis Dis. 2003, 1638, 72–82. [Google Scholar] [CrossRef]
- Kadowaki, M.; Sangai, T.; Nagashima, T.; Sakakibara, M.; Yoshitomi, H.; Takano, S.; Sogawa, K.; Umemura, H.; Fushimi, K.; Nakatani, Y.; et al. Identification of vitronectin as a novel serum marker for early breast cancer detection using a new proteomic approach. J. Cancer Res. Clin. Oncol. 2011, 137, 1105–1115. [Google Scholar] [CrossRef]
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar]
- Akram, M.; Iqbal, M.; Daniyal, M.; Khan, A.U. Awareness and current knowledge of breast cancer. Biol. Res. 2017, 50, 33. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Piccolo, M.; Misso, G.; Ferraro, M.G.; Riccardi, C.; Capuozzo, A.; Zarone, M.R.; Maione, F.; Trifuoggi, M.; Stiuso, P.; D’Errico, G.; et al. Exploring cellular uptake, accumulation and mechanism of action of a cationic Ru-based nanosystem in human preclinical models of breast cancer. Sci. Rep. 2019, 9, 7006. [Google Scholar] [CrossRef] [PubMed]
- Riley, K.R.; Gagliano, J.; Xiao, J.; Libby, K.; Saito, S.; Yu, G.; Cubicciotti, R.; Macosko, J.; Colyer, C.L.; Guthold, M.; et al. Combining capillary electrophoresis and next-generation sequencing for aptamer selection. Anal. Bioanal. Chem. 2015, 407, 1527–1532. [Google Scholar] [CrossRef]
- Stuart, C.H.; Riley, K.R.; Boyacioglu, O.; Herpai, D.M.; Debinski, W.; Qasem, S.; Marini, F.C.; Colyer, C.L.; Gmeiner, W.H. Selection of a novel aptamer against vitronectin using capillary electrophoresis and next generation sequencing. Mol. Ther. Nucleic Acids 2016, 5, e386. [Google Scholar] [CrossRef]
- Boyacioglu, O.; Stuart, C.H.; Kulik, G.; Gmeiner, W.H. Dimeric DNA aptamer complexes for high-capacity-targeted drug delivery using ph-sensitive covalent linkages. Mol. Ther. Nucleic Acids 2013, 2, e107. [Google Scholar] [CrossRef]
- Silver, D.A.; Pellicer, I.; Fair, W.R.; Heston, W.D.W.; Cordon-Cardo, C. Prostate-specific membrane antigen expression in normal and malignant human tissues. Clin. Cancer Res. 1997, 3, 81–85. [Google Scholar]
- Schülke, N.; Varlamova, O.A.; Donovan, G.P.; Ma, D.; Gardner, J.P.; Morrissey, D.M.; Arrigale, R.R.; Zhan, C.; Chodera, A.J.; Surowitz, K.G.; et al. The homodimer of prostate-specific membrane antigen is a functional target for cancer therapy. Proc. Natl. Acad. Sci. USA 2003, 100, 12590–12595. [Google Scholar] [CrossRef]
- Aggarwal, S.; Singh, P.; Topaloglu, O.; Isaacs, J.T.; Demneade, S.R. A dimeric peptide that binds selectively to prostate-specific membrane antigen and inhibits its enzymatic activity. Cancer Res. 2006, 66, 9171–9177. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.A.; Glaser, S.L.; Dorfman, R.F.; Bracci, P.M.; Eberle, E.; Holly, E.A. Expert review of non-Hodgkin’s lymphomas in a population-based cancer registry: Reliability of diagnosis and subtype classifications. Cancer Epidemiol. Biomark. Prev. 2004, 13, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Armitage, J.O.; Gascoyne, R.D.; Lunning, M.A.; Cavalli, F. Non-Hodgkin lymphoma. Lancet 2017, 390, 298–310. [Google Scholar] [CrossRef]
- Tang, Z.; Shangguan, D.; Wang, K.; Shi, H.; Sefah, K.; Mallikratchy, P.; Chen, H.W.; Li, Y.; Tan, W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007, 79, 4900–4907. [Google Scholar] [CrossRef]
- Mallikaratchy, P.R.; Ruggiero, A.; Gardner, J.R.; Kuryavyi, V.; Maguire, W.F.; Heaney, M.L.; McDevitt, M.R.; Patel, D.J.; Scheinberg, D.A. A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic Acids Res. 2011, 39, 2458–2469. [Google Scholar] [CrossRef]
- Mallikaratchy, P. Evolution of complex target SELEX to identify aptamers against mammalian cell-surface antigens. Molecules 2017, 22, 215. [Google Scholar] [CrossRef]
- Zumrut, H.E.; Ara, M.N.; Fraile, M.; Maio, G.; Mallikaratchy, P. Ligand-guided selection of target-specific aptamers: A screening technology for identifying specific aptamers against cell-surface proteins. Nucleic Acid Ther. 2016, 26, 190–198. [Google Scholar] [CrossRef]
- Zumrut, H.E.; Mallikaratchy, P.R. Ligand Guided Selection () of artificial nucleic acid ligands against cell surface targets. ACS Appl. Bio Mater. 2020, 3, 2545–2552. [Google Scholar] [CrossRef]
- Zümrüt, H.E.; Batool, S.; Van, N.; George, S.; Bhandari, S.; Mallikaratchy, P. Structural optimization of an aptamer generated from Ligand-Guided Selection (LIGS) resulted in high affinity variant toward mIgM expressed on Burkitt’s lymphoma cell lines. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1825–1832. [Google Scholar] [CrossRef]
- Moccia, F.; Platella, C.; Musumeci, D.; Batool, S.; Zumrut, H.; Bradshaw, J.; Mallikaratchy, P.; Montesarchio, D. The role of G-quadruplex structures of LIGS-generated aptamers R1.2 and R1.3 in IgM specific recognition. Int. J. Biol. Macromol. 2019, 133, 839–849. [Google Scholar] [CrossRef]
- Batool, S.; Argyropoulos, K.V.; Azad, R.; Okeoma, P.; Zumrut, H.; Bhandari, S.; Dekhang, R.; Mallikaratchy, P.R. Dimerization of an aptamer generated from ligand-guided selection (LIGS) yields a high affinity scaffold against B-cells. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Kuhns, M.S.; Badgandi, H.B. Piecing together the family portrait of TCR-CD3 complexes. Immunol. Rev. 2012, 250, 120–143. [Google Scholar] [CrossRef] [PubMed]
- Inthagard, J.; Edwards, J.; Roseweir, A.K. Immunotherapy: Enhancing the efficacy of this promising therapeutic in multiple cancers. Clin. Sci. 2019, 133, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Zumrut, H.E.; Batool, S.; Argyropoulos, K.V.; Williams, N.; Azad, R.; Mallikaratchy, P.R. Integrating ligand-receptor interactions and in vitro evolution for streamlined discovery of artificial nucleic acid ligands. Mol. Ther. Nucleic Acids 2019, 17, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Freage, L.; Jamal, D.; Williams, N.B.; Mallikaratchy, P.R. A homodimeric aptamer variant generated from ligand-guided selection activates T-cell receptor cluster of differentiation three complex. Mol. Ther. Nucleic Acids 2020, 22, 167–178. [Google Scholar] [CrossRef]
- Senger, D.R.; Van De Water, L.; Brown, L.F.; Nagy, J.A.; Yeo, K.T.; Yeo, T.K.; Berse, B.; Jackman, R.W.; Dvorak, A.M.; Dvorak, H.F. Vascular permeability factor (VPF, VEGF) in tumor biology. Cancer Metastasis Rev. 1993, 12, 303–324. [Google Scholar] [CrossRef]
- Dvorak, H.F. Vascular permeability factor/vascular endothelial growth factor: A critical cytokine in tumor angiogenesis and a potential target for diagnosis and therapy. J. Clin. Oncol. 2002, 20, 4368–4380. [Google Scholar] [CrossRef]
- Guo, P.; Fang, Q.; Tao, H.Q.; Schafer, C.A.; Fenton, B.M.; Ding, I.; Hu, B.; Cheng, S.Y. Overexpression of vascular endothelial growth factor by MCF-7 breast cancer cells promotes estrogen-independent tumor growth in vivo. Cancer Res. 2003, 63, 4684–4691. [Google Scholar]
- Ferrara, N.; Gerber, H.P.; LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef]
- Holmes, D.I.R.; Zachary, I. The vascular endothelial growth factor (VEGF) family: Angiogenic factors in health and disease. Genome Biol. 2005, 6, 209. [Google Scholar] [CrossRef][Green Version]
- Shibuya, M. Vascular Endothelial Growth Factor (VEGF) Signaling in Tumour Vascularization: Potential and Challenges. Genes Cancer 2011, 2, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Siveen, K.S.; Prabhu, K.; Krishnankutty, R.; Kuttikrishnan, S.; Tsakou, M.; Alali, F.Q.; Dermime, S.; Mohammad, R.M.; Uddin, S. Vascular endothelial growth factor (VEGF) signaling in tumour vascularization: Potential and challenges. Curr. Vasc. Pharmacol. 2017, 15, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Ema, M. Roles of VEGF-A signalling in development, regeneration, and tumours. J. Biochem. 2014, 156, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in signaling and disease: Beyond discovery and development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.L.; Rak, J.W.; Klement, G.; Kerbel, R.S. Vascular endothelial growth factor isoform expression as a determinant of blood vessel patterning in human melanoma xenografts. Cancer Res. 2002, 62, 1838–1846. [Google Scholar]
- Catena, R.; Muniz-Medina, V.; Moralejo, B.; Javierre, B.; Best, C.J.M.; Emmert-Buck, M.R.; Green, J.E.; Baker, C.C.; Calvo, A. Increased expression of VEGF121/VEGF165-189 ratio results in a significant enhancement of human prostate tumor angiogenesis. Int. J. Cancer 2007, 120, 2096–2109. [Google Scholar] [CrossRef]
- Harper, S.J.; Bates, D.O. VEGF-A splicing: The key to anti-angiogenic therapeutics? Nat. Rev. Cancer 2008, 8, 880–887. [Google Scholar] [CrossRef]
- Meadows, K.L.; Hurwitz, H.I. Anti-VEGF therapies in the clinic. Cold Spring Harb. Perspect. Med. 2012, 2, a006577. [Google Scholar] [CrossRef]
- Riccardi, C.; Napolitano, E.; Platella, C.; Musumeci, D.; Melone, M.A.B.; Montesarchio, D. Anti-VEGF DNA-based aptamers in cancer therapeutics and diagnostics. Med. Res. Rev. 2020. [Google Scholar] [CrossRef]
- Ikebukuro, K.; Hasegawa, H.; Sode, K. Selection and characterization of DNA aptamers against VEGF165 with aptamer blotting method and its application. Nucleic Acids Symp. Ser. 2007, 51, 399–400. [Google Scholar] [CrossRef]
- Hasegawa, H.; Sode, K.; Ikebukuro, K. Selection of DNA aptamers against VEGF165 using a protein competitor and the aptamer blotting method. Biotechnol. Lett. 2008, 30, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Yung, L.Y.L. Probing high affinity sequences of DNA aptamer against VEGF165. PLoS ONE 2012, 7, e31196. [Google Scholar] [CrossRef] [PubMed]
- Manochehry, S.; Mcconnell, E.M.; Li, Y. Unraveling determinants of affinity enhancement in dimeric aptamers for a dimeric protein. Sci. Rep. 2019, 9, 17824. [Google Scholar] [CrossRef] [PubMed]
- Fukaya, T.; Abe, K.; Savory, N.; Tsukakoshi, K.; Yoshida, W.; Ferri, S.; Sode, K.; Ikebukuro, K. Improvement of the VEGF binding ability of DNA aptamers through in silico maturation and multimerization strategy. J. Biotechnol. 2015, 212, 99–105. [Google Scholar] [CrossRef]
- Manochehry, S.; Gu, J.; McConnell, E.; Salena, B.; Li, Y. In vitro selection of new DNA aptamers for human vascular endothelial growth factor 165. ChemBioChem 2020, 21, 2029–2036. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Janjic, N. High-Affinity Oligonucleotide Ligands to Vascular Endothelial Growth Factor (VEGF). U.S. Patent 7,153,948, 26 December 2006. [Google Scholar]
- Potty, A.S.R.; Kourentzi, K.; Fang, H.; Jackson, G.W.; Zhang, X.; Legge, G.B.; Willson, R.C. Biophysical characterization of DNA aptamer interactions with vascular endothelial growth factor. Biopolymers 2009, 91, 145–156. [Google Scholar] [CrossRef]
- Nonaka, Y.; Sode, K.; Ikebukuro, K. Screening and improvement of an anti-VEGF DNA aptamer. Molecules 2010, 15, 215–225. [Google Scholar] [CrossRef]
- Edwards, S.L.; Poongavanam, V.; Kanwar, J.R.; Roy, K.; Hillman, K.M.; Prasad, N.; Leth-Larsen, R.; Petersen, M.; Marušič, M.; Plavec, J.; et al. Targeting VEGF with LNA-stabilized G-rich oligonucleotide for efficient breast cancer inhibition. Chem. Commun. 2015, 51, 9499–9502. [Google Scholar] [CrossRef]
- Marušič, M.; Veedu, R.N.; Wengel, J.; Plavec, J. G-rich VEGF aptamer with locked and unlocked nucleic acid modifications exhibits a unique G-quadruplex fold. Nucleic Acids Res. 2013, 41, 9524–9536. [Google Scholar] [CrossRef]
- Nonaka, Y.; Yoshida, W.; Abe, K.; Ferri, S.; Schulze, H.; Bachmann, T.T.; Ikebukuro, K. Affinity improvement of a VEGF aptamer by in silico maturation for a sensitive VEGF-detection system. Anal. Chem. 2013, 85, 1132–1137. [Google Scholar] [CrossRef]
- Moccia, F.; Riccardi, C.; Musumeci, D.; Leone, S.; Oliva, R.; Petraccone, L.; Montesarchio, D. Insights into the G-rich VEGF-binding aptamer V7t1: When two G-quadruplexes are better than one! Nucleic Acids Res. 2019, 47, 8318–8331. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, D.; Roviello, G.N.; Rigione, G.; Capasso, D.; Di Gaetano, S.; Riccardi, C.; Roviello, V.; Montesarchio, D. Benzodifuran derivatives as potential antiproliferative agents: Possible correlation between their bioactivity and aggregation properties. ChemPlusChem 2017, 82, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Vicidomini, C.; Cioffi, F.; Broersen, K.; Roviello, V.; Riccardi, C.; Montesarchio, D.; Capasso, D.; Gaetano, S.D.; Roviello, G.N. Benzodifurans for biomedical applications: BZ4, a selective anti-proliferative and anti-amyloid lead compound. Future Med. Chem. 2019, 11, 285–302. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Musumeci, D.; Platella, C.; Gaglione, R.; Arciello, A.; Montesarchio, D. Tuning the polymorphism of the anti-VEGF G-rich V7t1 aptamer by covalent dimeric constructs. Int. J. Mol. Sci. 2020, 21, 1963. [Google Scholar] [CrossRef] [PubMed]
- Bottaro, D.P.; Rubin, J.S.; Faletto, D.L.; Chan, A.M.L.; Kmiecik, T.E.; Vande Woude, G.F.; Aaronson, S.A. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 1991, 251, 802–804. [Google Scholar] [CrossRef]
- Gentile, A.; Trusolino, L.; Comoglio, P.M. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008, 27, 85–94. [Google Scholar] [CrossRef]
- Huang, W.; Manglik, A.; Venkatakrishnan, A.J.; Laeremans, T.; Feinberg, E.N.; Sanborn, A.L.; Kato, H.E.; Livingston, K.E.; Thorsen, T.S.; Kling, R.C.; et al. Structural insights into μ-opioid receptor activation. Nature 2015, 90, 972–981. [Google Scholar] [CrossRef]
- Birchmeier, C.; Birchmeier, W.; Gherardi, E.; Vande Woude, G.F. Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol. 2003, 4, 915–925. [Google Scholar] [CrossRef]
- Trusolino, L.; Bertotti, A.; Comoglio, P.M. MET signalling: Principles and functions in development, organ regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 834–848. [Google Scholar] [CrossRef]
- Graveel, C.R.; Tolbert, D.; Vande Woude, G.F. MET: A critical player in tumorigenesis and therapeutic target. Cold Spring Harb. Perspect. Biol. 2013, 5, a009209. [Google Scholar] [CrossRef]
- Ueki, R.; Sando, S. A DNA aptamer to c-Met inhibits cancer cell migration. Chem. Commun. 2014, 50, 13131–13134. [Google Scholar] [CrossRef] [PubMed]
- Ueki, R.; Ueki, A.; Kanda, N.; Sando, S. Oligonucleotide-Based Mimetics of Hepatocyte Growth Factor. Angew. Chem. Int. Ed. 2016, 55, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Mammen, M.; Choi, S.K.; Whitesides, G.M. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. Eng. 1998, 37, 2754–2794. [Google Scholar] [CrossRef]
- Fasting, C.; Schalley, C.A.; Weber, M.; Seitz, O.; Hecht, S.; Koksch, B.; Dernedde, J.; Graf, C.; Knapp, E.W.; Haag, R. Multivalency as a chemical organization and action principle. Angew. Chem. Int. Ed. Eng. 2012, 51, 10472–10498. [Google Scholar] [CrossRef]
- Yokobayashi, Y. Aptamer-based and aptazyme-based riboswitches in mammalian cells. Curr. Opin. Chem. Biol. 2019, 52, 72–78. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).