Distribution of Protein Precipitation Capacity within Variable Proanthocyanidin Fingerprints
Abstract
1. Introduction
2. Results and Discussion
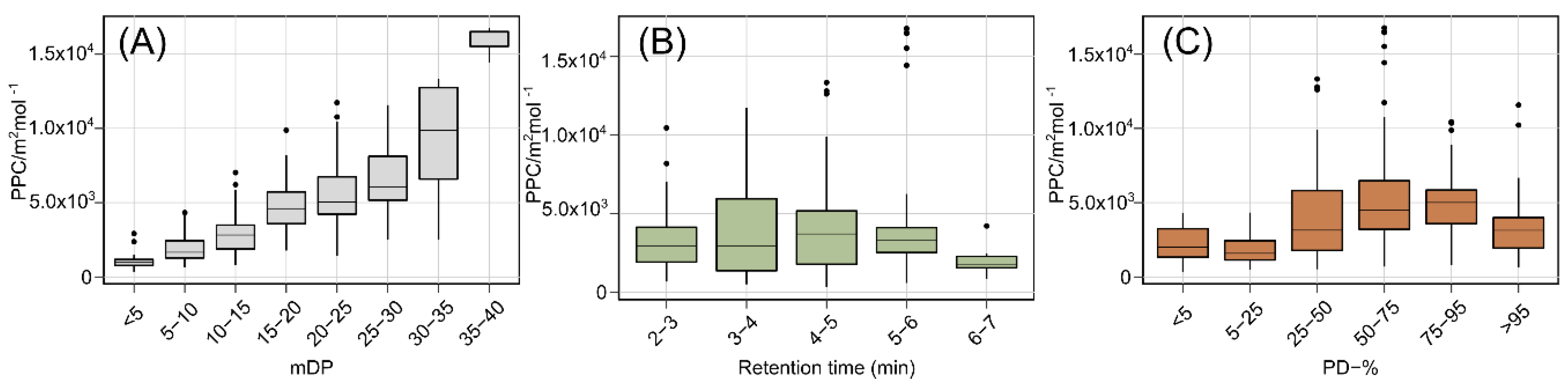
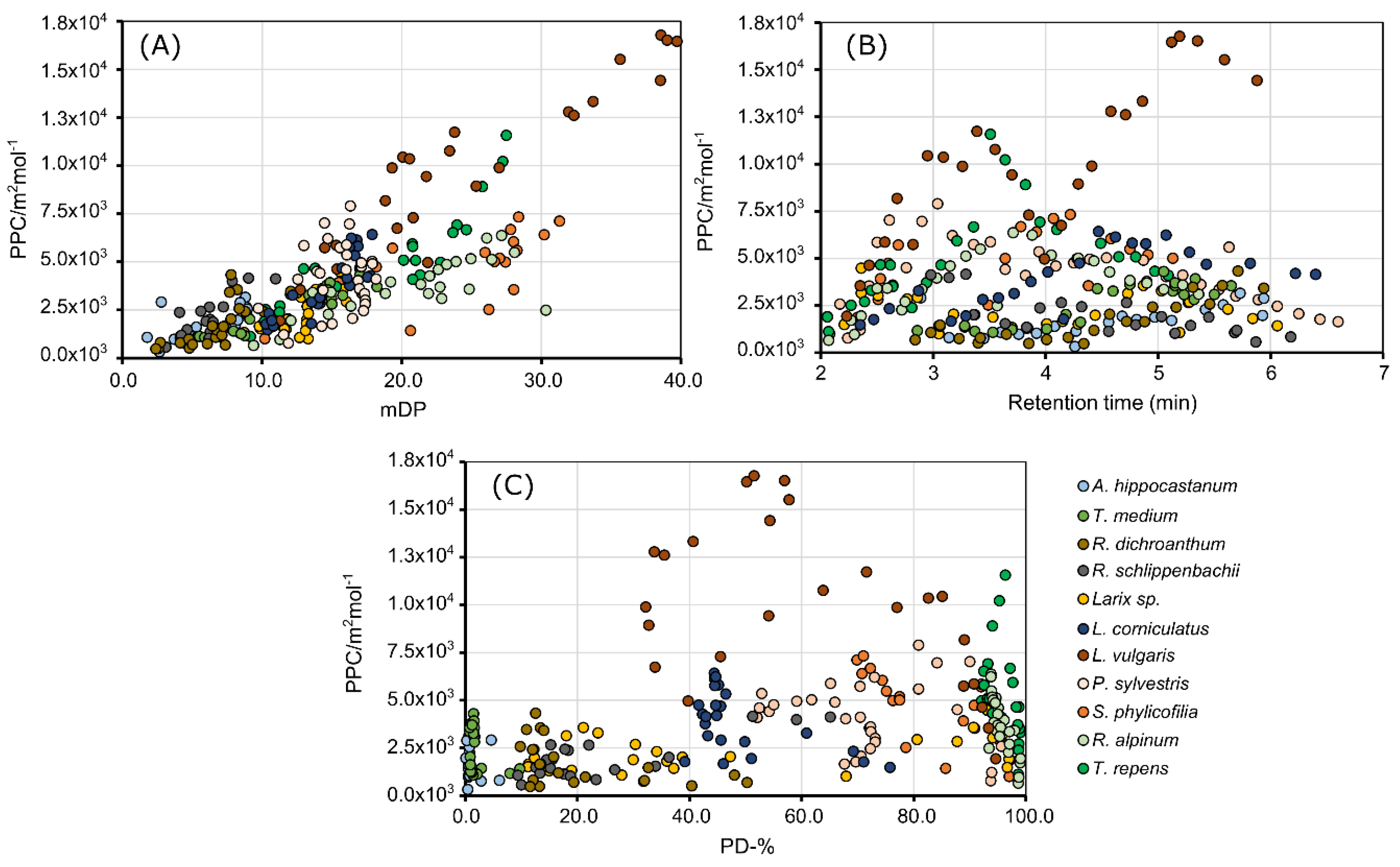
2.1. The Effect of Polymer Size, Prodelphinidin Proportion and Retention Time on the Protein Precipitation Capacity
2.2. Partial Least Square Regression of the Protein Precipitation Capacity of the Proanthocyanidins
2.3. The distribution of the Protein Precipitation Capacity within Proanthocyanidin Fingerprints
2.4. Fraction by Fraction Comparison with High-Resolution Mass Data
3. Materials and Methods
3.1. Chemicals
3.2. Plant Material
3.3. Proanthocyanidin Isolation and Fractionation
3.4. UPLC-DAD-MS/MS Analyses
3.5. UPLC-DAD-HRMS Analyses
3.6. Protein Precipitation Capacity via Well-Plate Assay
3.7. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McDougall, G.J.; Ross, H.A.; Ikeji, M.; Stewart, D. Berry Extracts Exert Different Antiproliferative Effects against Cervical and Colon Cancer Cells Grown in Vitro. J. Agric. Food Chem. 2008, 56, 3016–3023. [Google Scholar] [CrossRef] [PubMed]
- Tatsuno, T.; Jinno, M.; Arima, Y.; Kawabata, T.; Hasegawa, T.; Yahagi, N.; Takano, F.; Ohta, T. Anti-inflammatory and Anti-melanogenic Proanthocyanidin Oligomers from Peanut Skin. Biol. Pharm. Bull. 2012, 35, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Sánchez, D.O.; Jimenez-Ferrer, I.; Sánchez-Sánchez, V.; Zamilpa, A.; González-Cortázar, M.; Tortoriello, J.; Herrera-Ruiz, M. Anti-Inflammatory Activity of a Polymeric Proanthocyanidin from Serjania schiedeana. Molecules 2017, 22, 863. [Google Scholar] [CrossRef] [PubMed]
- Athanasiadou, S.; Kyriazakis, I.; Jackson, F.; Coop, R. Direct anthelmintic effects of condensed tannins towards different gastrointestinal nematodes of sheep: In vitro and in vivo studies. Vet. Parasitol. 2001, 99, 205–219. [Google Scholar] [CrossRef]
- Hoste, H.; Jackson, F.; Athanasiadou, S.; Thamsborg, S.M.; Hoskin, S.O. The effects of tannin-rich plants on parasitic nematodes in ruminants. Trends Parasitol. 2006, 22, 253–261. [Google Scholar] [CrossRef]
- Jayanegara, A.; Goel, G.; Makkar, H.; Becker, K. Divergence between purified hydrolysable and condensed tannin effects on methane emission, rumen fermentation and microbial population in vitro. Anim. Feed. Sci. Technol. 2015, 209, 60–68. [Google Scholar] [CrossRef]
- Aerts, R.J.; Barry, T.N.; McNabb, W.C. Polyphenols and agriculture: Beneficial effects of proanthocyanidins in forages. Agric. Ecosyst. Environ. 1999, 75, 1–12. [Google Scholar] [CrossRef]
- Desrues, O.; Fryganas, C.; Ropiak, H.M.; Mueller-Harvey, I.; Enemark, H.L.; Thamsborg, S.M. Impact of chemical structure of flavanol monomers and condensed tannins on in vitro anthelmintic activity against bovine nematodes. Parasitology 2016, 143, 444–454. [Google Scholar] [CrossRef]
- Hatew, B.; Stringano, E.; Mueller-Harvey, I.; Hendriks, W.H.; Carbonero, C.H.; Smith, L.M.J.; Pellikaan, W.F. Impact of variation in structure of condensed tannins from sainfoin (Onobrychis viciifolia) onin vitroruminal methane production and fermentation characteristics. J. Anim. Physiol. Anim. Nutr. 2015, 100, 348–360. [Google Scholar] [CrossRef]
- Kilmister, R.L.; Faulkner, P.; Downey, M.O.; Darby, S.J.; Falconer, R.J. The complexity of condensed tannin binding to bovine serum albumin—An isothermal titration calorimetry study. Food Chem. 2016, 190, 173–178. [Google Scholar] [CrossRef]
- Zeller, W.E.; Sullivan, M.L.; Mueller-Harvey, I.; Grabber, J.H.; Ramsay, A.; Drake, C.; Brown, R.H. Protein Precipitation Behavior of Condensed Tannins from Lotus pedunculatus and Trifolium repens with Different Mean Degrees of Polymerization. J. Agric. Food Chem. 2015, 63, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Ropiak, H.M.; Lachmann, P.; Ramsay, A.; Green, R.J.; Mueller-Harvey, I. Identification of Structural Features of Condensed Tannins That Affect Protein Aggregation. PLoS ONE 2017, 12, e0170768. [Google Scholar] [CrossRef] [PubMed]
- Osborne, N.J.T.; McNeill, D.M. Characterisation ofLeucaena condensed tannins by size and protein precipitation capacity. J. Sci. Food Agric. 2001, 81, 1113–1119. [Google Scholar] [CrossRef]
- Harbertson, J.F.; Kilmister, R.L.; Kelm, M.A.; Downey, M.O. Impact of condensed tannin size as individual and mixed polymers on bovine serum albumin precipitation. Food Chem. 2014, 160, 16–21. [Google Scholar] [CrossRef]
- Haslman, E. Practical Polyphenolics: From Structure to Molecular Recognition and Physiological Action; Cambridge University Press: Cambridge, UK, 1998. [Google Scholar]
- Zeller, W.E. Activity, Purification, and Analysis of Condensed Tannins: Current State of Affairs and Future Endeavors. Crop. Sci. 2019, 59, 886–904. [Google Scholar] [CrossRef]
- Leppä, M.M.; Karonen, M.; Tähtinen, P.; Engström, M.T.; Salminen, J.-P. Isolation of chemically well-defined semipreparative liquid chromatography fractions from complex mixtures of proanthocyanidin oligomers and polymers. J. Chromatogr. A 2018, 1576, 67–79. [Google Scholar] [CrossRef]
- Brown, R.H.; Mueller-Harvey, I.; Zeller, W.E.; Reinhardt, L.A.; Stringano, E.; Gea, A.; Drake, C.; Ropiak, H.M.; Fryganas, C.; Ramsay, A.; et al. Facile Purification of Milligram to Gram Quantities of Condensed Tannins According to Mean Degree of Polymerization and Flavan-3-ol Subunit Composition. J. Agric. Food Chem. 2017, 65, 8072–8082. [Google Scholar] [CrossRef]
- Saminathan, M.; Tan, H.Y.; Sieo, C.C.; Abdullah, N.; Wong, C.M.V.L.; Abdulmalek, E.; Ho, Y.W. Polymerization degrees, molecular weights and protein-binding affinities of condensed tannin fractions from a leucaena leucocephala hybrid. Molecules 2014, 19, 7990–8010. [Google Scholar] [CrossRef]
- Novobilský, A.; Stringano, E.; Carbonero, C.H.; Smith, L.; Enemark, H.; Mueller-Harvey, I.; Thamsborg, S.M. In vitro effects of extracts and purified tannins of sainfoin (Onobrychis viciifolia) against two cattle nematodes. Veter. Parasitol. 2013, 196, 532–537. [Google Scholar] [CrossRef]
- Trombley, J.D.; Loegel, T.N.; Danielson, N.D.; Hagerman, A.E. Capillary electrophoresis methods for the determination of covalent polyphenol–protein complexes. Anal. Bioanal. Chem. 2011, 401, 1523–1529. [Google Scholar] [CrossRef]
- Appel, H.M. Phenolics in ecological interactions: The importance of oxidation. J. Chem. Ecol. 1993, 19, 1521–1552. [Google Scholar] [CrossRef]
- Oh, H.I.; Hoff, J.E.; Armstrong, G.S.; Haff, L.A. Hydrophobic interaction in tannin-protein complexes. J. Agric. Food Chem. 1980, 28, 394–398. [Google Scholar] [CrossRef]
- Goldstein, J.L.; Swain, T. Changes in tannins in ripening fruits. Phytochemistry 1963, 2, 371–383. [Google Scholar] [CrossRef]
- Hagerman, A.E.; Rice, A.M.E.; Ritchard, N.T. Mechanisms of Protein Precipitation for Two Tannins, Pentagalloyl Glucose and Epicatechin16(4→8) Catechin (Procyanidin). J. Agric. Food Chem. 1998, 46, 2590–2595. [Google Scholar] [CrossRef]
- Zeller, W.E.; Reinhardt, L.A.; Robe, J.T.; Sullivan, M.L.; Panke-Buisse, K. Comparison of Protein Precipitation Ability of Structurally Diverse Procyanidin-Rich Condensed Tannins in Two Buffer Systems. J. Agric. Food Chem. 2020, 68, 2016–2023. [Google Scholar] [CrossRef]
- Charlton, A.J.; Baxter, N.J.; Khan, M.L.; Moir, A.J.G.; Haslam, E.; Davies, A.P.; Williamson, M.P. Polyphenol/Peptide Binding and Precipitation. J. Agric. Food Chem. 2002, 50, 1593–1601. [Google Scholar] [CrossRef]
- Cala, O.; Pinaud, N.; Simon, C.; Fouquet, E.; Laguerre, M.; Dufourc, E.J.; Pianet, I. NMR and molecular modeling of wine tannins binding to saliva proteins: Revisiting astringency from molecular and colloidal prospects. FASEB J. 2010, 24, 4281–4290. [Google Scholar] [CrossRef]
- Engström, M.T.; Arvola, J.; Nenonen, S.; Virtanen, V.T.J.; Leppä, M.M.; Tähtinen, P.; Salminen, J.-P. Structural Features of Hydrolyzable Tannins Determine Their Ability to Form Insoluble Complexes with Bovine Serum Albumin. J. Agric. Food Chem. 2019, 67, 6798–6808. [Google Scholar] [CrossRef]
- Dobreva, M.A.; Frazier, R.A.; Mueller-Harvey, I.; Clifton, L.A.; Gea, A.; Green, R.J. Binding of Pentagalloyl Glucose to Two Globular Proteins Occurs via Multiple Surface Sites. Biomacromolecules 2011, 12, 710–715. [Google Scholar] [CrossRef]
- Engström, M.T.; Pälijärvi, M.; Fryganas, C.; Grabber, J.H.; Mueller-Harvey, I.; Salminen, J.-P. Rapid Qualitative and Quantitative Analyses of Proanthocyanidin Oligomers and Polymers by UPLC-MS/MS. J. Agric. Food Chem. 2014, 62, 3390–3399. [Google Scholar] [CrossRef]
- Engström, M.T.; Pälijärvi, M.; Salminen, J.-P. Rapid Fingerprint Analysis of Plant Extracts for Ellagitannins, Gallic Acid, and Quinic Acid Derivatives and Quercetin-, Kaempferol- and Myricetin-Based Flavonol Glycosides by UPLC-QqQ-MS/MS. J. Agric. Food Chem. 2015, 63, 4068–4079. [Google Scholar] [CrossRef]
- Koupai-Abyazani, M.R.; McCallum, J.A.; Bohm, B.A. Identification of the constituent flavanoid units in sainfoin proanthocyanidins by reversed-phase high-performance liquid chromatography. J. Chromatogr. A 1992, 594, 117–123. [Google Scholar] [CrossRef]
- Siebert, K.J. Effects of Protein−Polyphenol Interactions on Beverage Haze, Stabilization, and Analysis. J. Agric. Food Chem. 1999, 47, 353–362. [Google Scholar] [CrossRef]
- Buelga, C.S.; Francia-Aricha, E.; Escribano-Bailón, M. Comparative flavan-3-ol composition of seeds from different grape varieties. Food Chem. 1995, 53, 197–201. [Google Scholar] [CrossRef]
- Salminen, J.-P. Two-Dimensional Tannin Fingerprints by Liquid Chromatography Tandem Mass Spectrometry Offer a New Dimension to Plant Tannin Analyses and Help To Visualize the Tannin Diversity in Plants. J. Agric. Food Chem. 2018, 66, 9162–9171. [Google Scholar] [CrossRef]
- Malisch, C.S.; Lüscher, A.; Baert, N.; Engström, M.T.; Studer, B.; Fryganas, C.; Suter, D.; Mueller-Harvey, I.; Salminen, J.-P. Large Variability of Proanthocyanidin Content and Composition in Sainfoin (Onobrychis viciifolia). J. Agric. Food Chem. 2015, 63, 10234–10242. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2019. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Sanchez, G. Plsdepot: Partial Least Squares (PLS) Data Analysis Methods, Version 0.1.17; 2012. Available online: https://CRAN.R-project.org/package=plsdepot (accessed on 23 September 2020).
Sample Availability: Samples of the compounds are not available from the authors. |
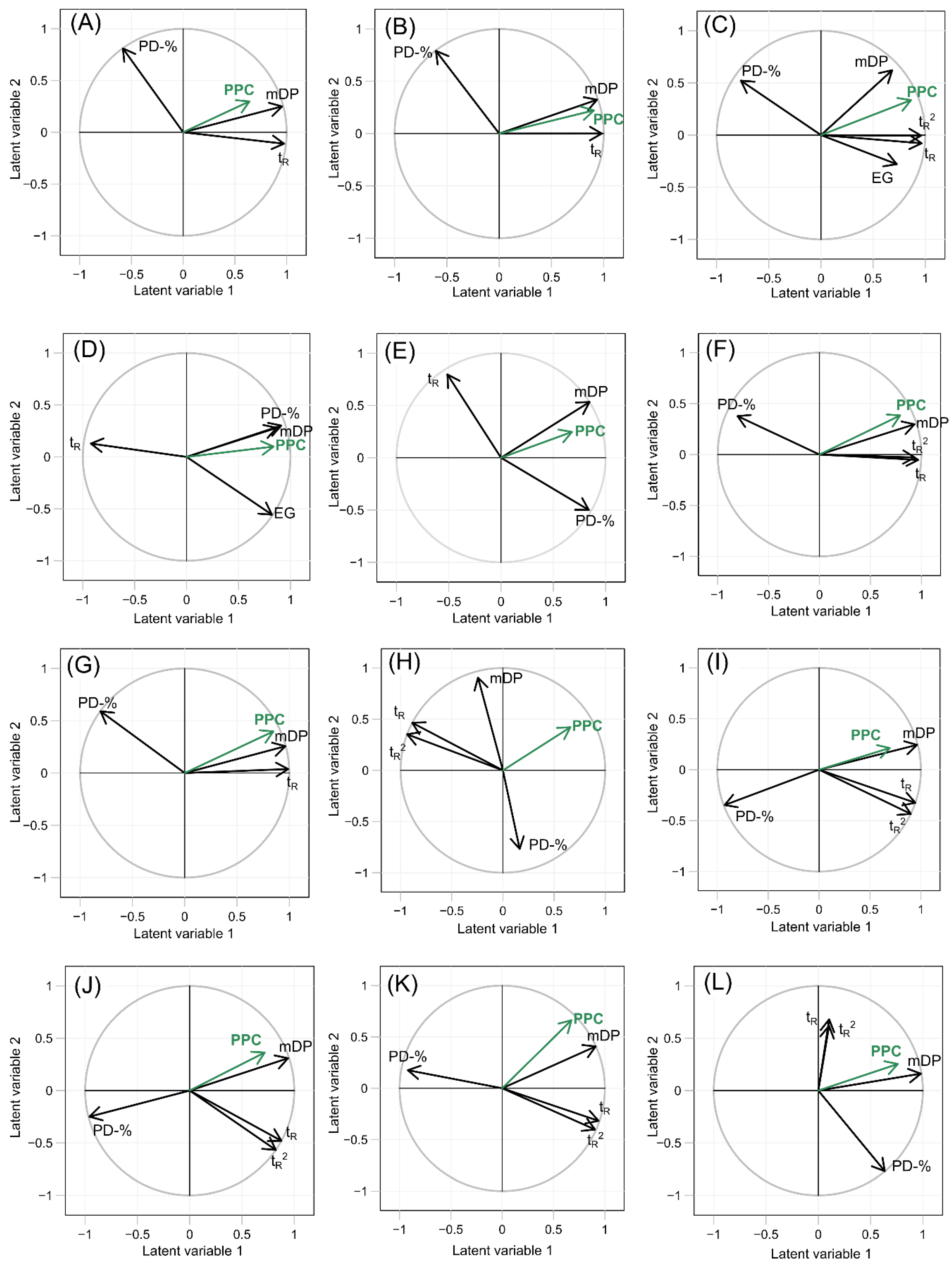


| Plant Species | Variable | Regression Coefficients | R2X | R2Y | Q2 | Plant Species | Variable | Regression Coefficients | R2X | R2Y | Q2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A. hippo-castanum | tR | 0.314 | 0.961 | 0.499 | 0.378 | L. vulgaris | tR | 0.385 | 0.980 | 0.875 | 0.854 |
| mDP | 0.506 | 0.975 | mDP | 0.754 | 0.990 | ||||||
| PD-% | 0.255 | 0.995 | PD-% | 0.322 | 0.997 | ||||||
| T. medium | tR | 0.399 | 0.961 | 0.867 | 0.857 | P. sylvestris | tR2 | −0.532 | 0.999 | 0.611 | 0.483 |
| mDP | 0.577 | 0.979 | tR | −0.408 | 0.999 | ||||||
| PD-% | 0.044 | 0.990 | mDP | 0.607 | 0.879 | ||||||
| R. dichro-anthum | tR2 | 0.365 | 0.920 | 0.856 | 0.829 | PD-% | −0.323 | 0.615 | |||
| tR | 0.295 | 0.936 | S. phylicifolia | tR2 | −0.021 | 0.999 | 0.526 | 0.378 | |||
| mDP | 0.483 | 0.851 | tR | 0.028 | 0.999 | ||||||
| PD-% | 0.091 | 0.861 | mDP | 0.379 | 0.984 | ||||||
| EG | −0.043 | 0.605 | PD-% | −0.347 | 0.981 | ||||||
| R. schlippen-bachii | tR | −0.243 | 0.866 | 0.711 | 0.594 | R. alpinum | tR2 | −0.147 | 0.999 | 0.646 | 0.496 |
| mDP | 0.291 | 0.830 | tR | −0.046 | 0.999 | ||||||
| PD-% | 0.320 | 0.921 | mDP | 0.457 | 0.979 | ||||||
| EG | 0.085 | 0.983 | PD-% | −0.469 | 0.980 | ||||||
| Larix sp. | TR | −0.048 | 0.896 | 0.521 | 0.395 | T. repens | tR2 | −0.331 | 0.977 | 0.892 | 0.858 |
| mDP | 0.650 | 1.000 | tR | −0.157 | 0.980 | ||||||
| PD-% | 0.124 | 0.954 | mDP | 1.192 | 0.995 | ||||||
| L. cornicula-tus | tR2 | 0.041 | 0.874 | 0.773 | 0.644 | PD-% | −0.038 | 0.875 | |||
| tR | 0.145 | 0.942 | Comple-te data set | tR2 | 0.036 | 0.393 | 0.642 | 0.614 | |||
| mDP | 0.926 | 0.957 | tR | 0.088 | 0.473 | ||||||
| PD-% | 0.311 | 0.789 | mDP | 0.799 | 0.986 | ||||||
| PD-% | −0.056 | 0.996 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leppä, M.M.; Laitila, J.E.; Salminen, J.-P. Distribution of Protein Precipitation Capacity within Variable Proanthocyanidin Fingerprints. Molecules 2020, 25, 5002. https://doi.org/10.3390/molecules25215002
Leppä MM, Laitila JE, Salminen J-P. Distribution of Protein Precipitation Capacity within Variable Proanthocyanidin Fingerprints. Molecules. 2020; 25(21):5002. https://doi.org/10.3390/molecules25215002
Chicago/Turabian StyleLeppä, Milla Marleena, Juuso Erik Laitila, and Juha-Pekka Salminen. 2020. "Distribution of Protein Precipitation Capacity within Variable Proanthocyanidin Fingerprints" Molecules 25, no. 21: 5002. https://doi.org/10.3390/molecules25215002
APA StyleLeppä, M. M., Laitila, J. E., & Salminen, J.-P. (2020). Distribution of Protein Precipitation Capacity within Variable Proanthocyanidin Fingerprints. Molecules, 25(21), 5002. https://doi.org/10.3390/molecules25215002








