Metabolomics Analysis of the Peels of Different Colored Citrus Fruits (Citrus reticulata cv. ‘Shatangju’) During the Maturation Period Based on UHPLC-QQQ-MS
Abstract
1. Introduction
2. Results
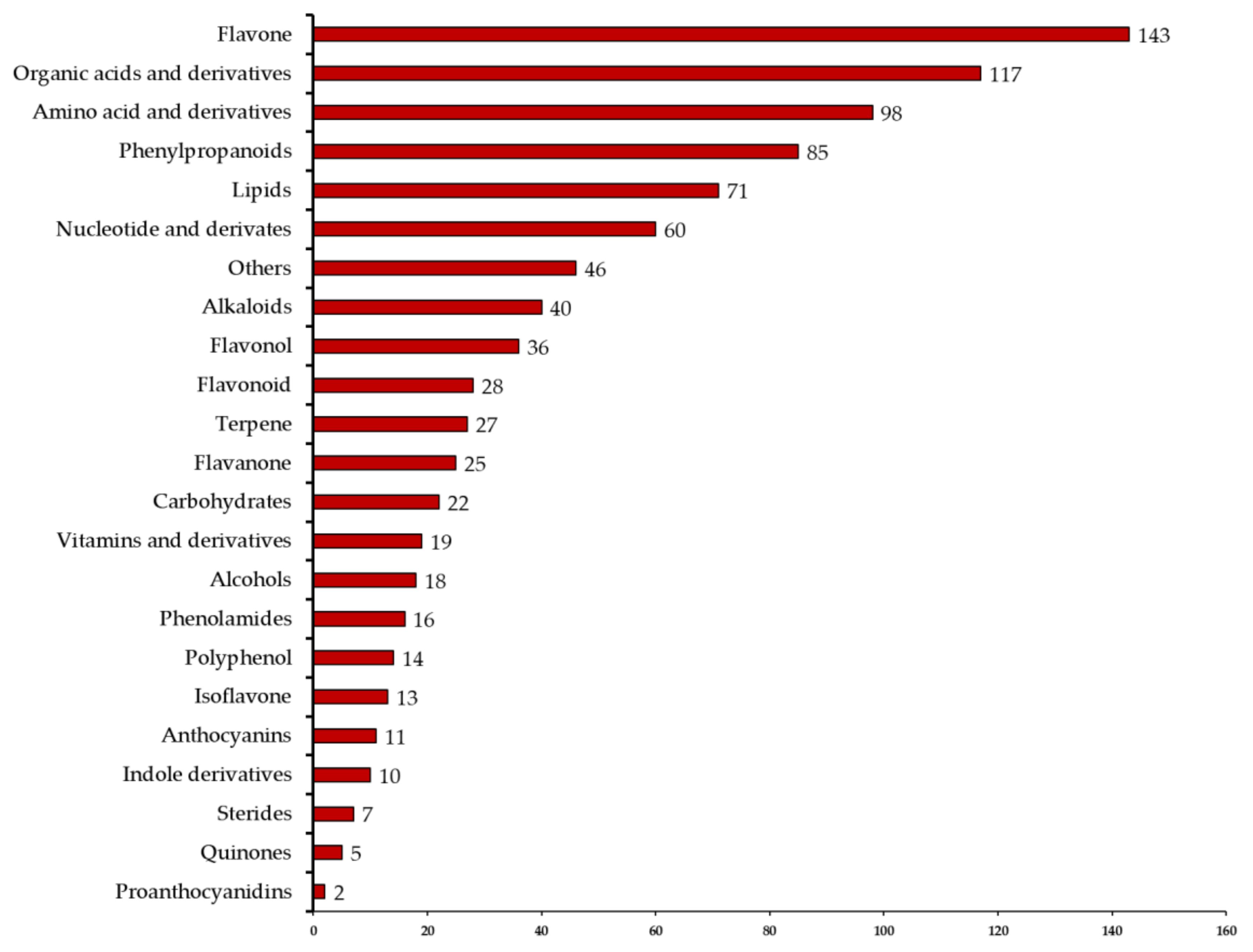
2.1. Qualitative and Quantitative Analysis of Metabolites and Quality Control (QC) Analysis of Samples
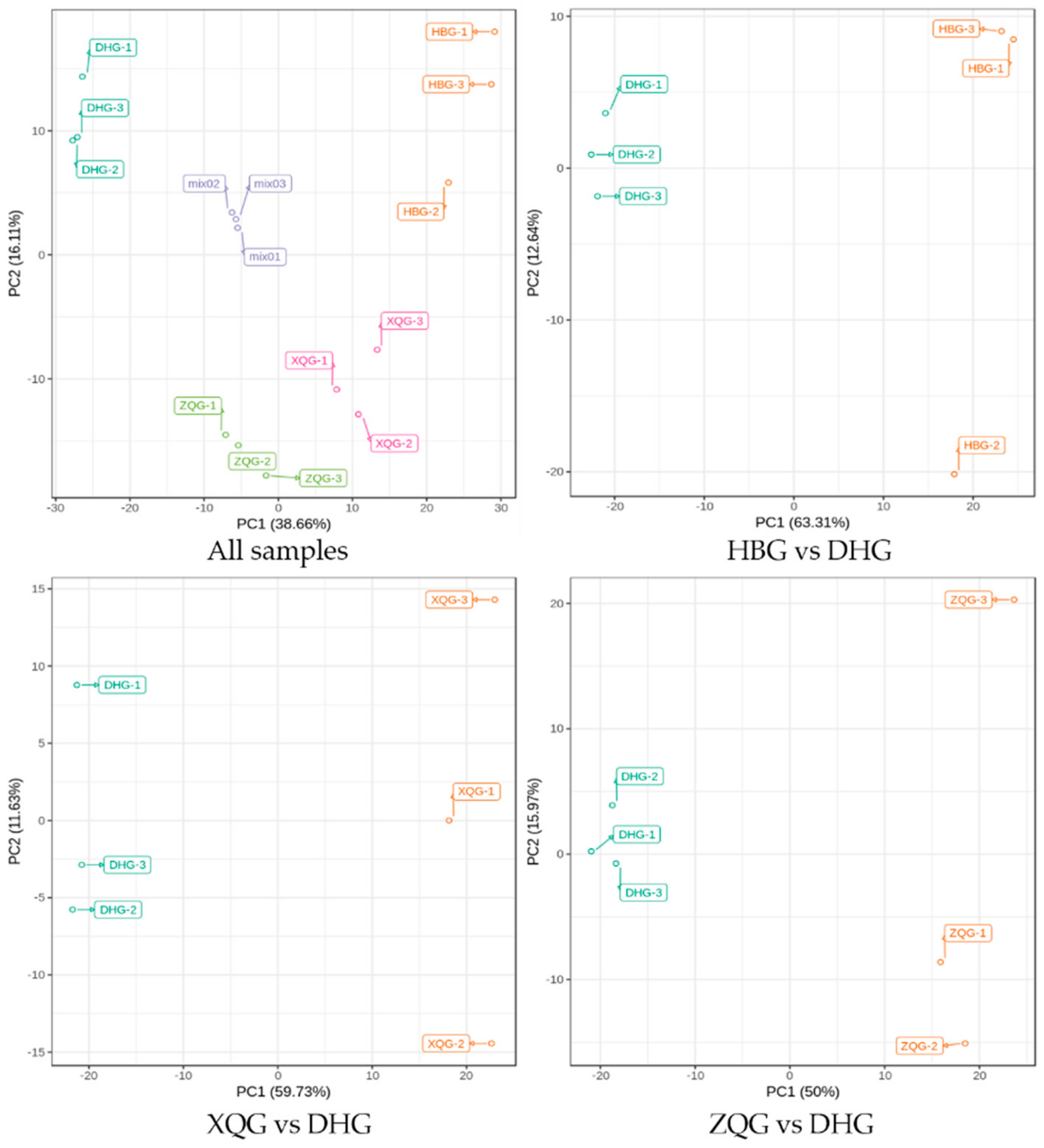
2.2. PCA for the Three Treatment Groups (HBG, XQG, and ZQG) versus the Control Group
2.3. OPLS-DA for the Three Treatment Groups (HBG, XQG, and ZQG) versus the Control (DHG) Group
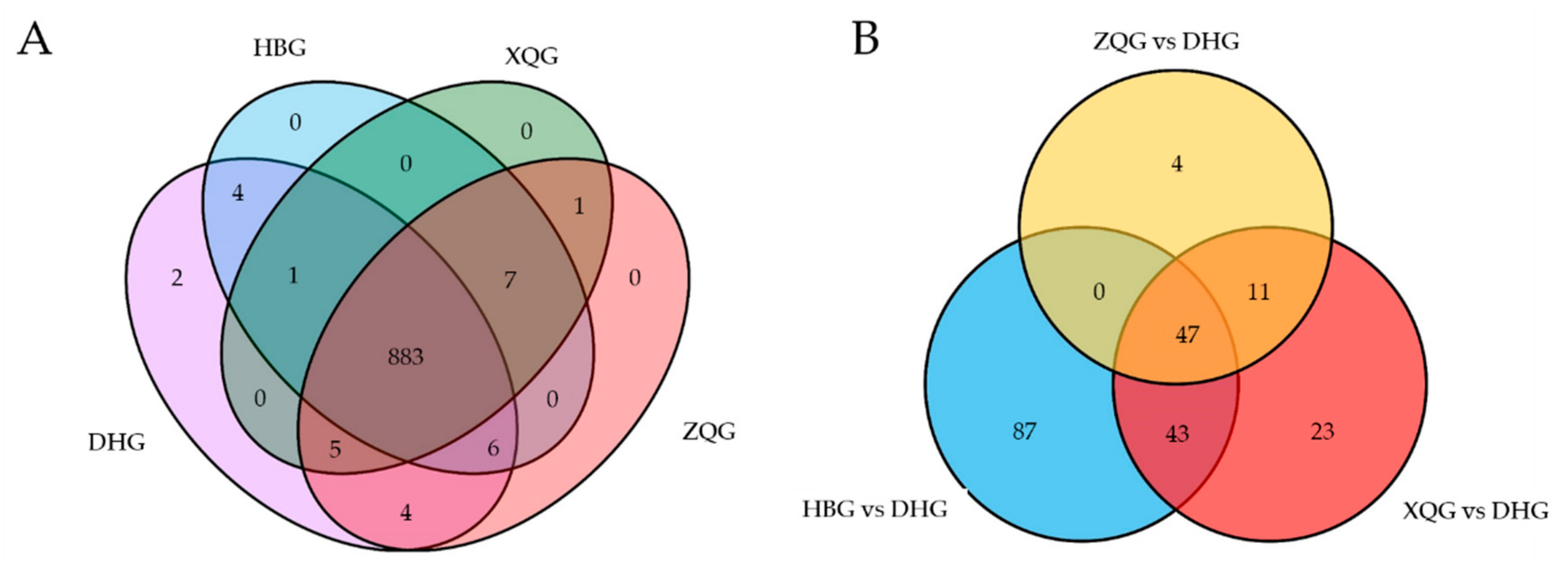
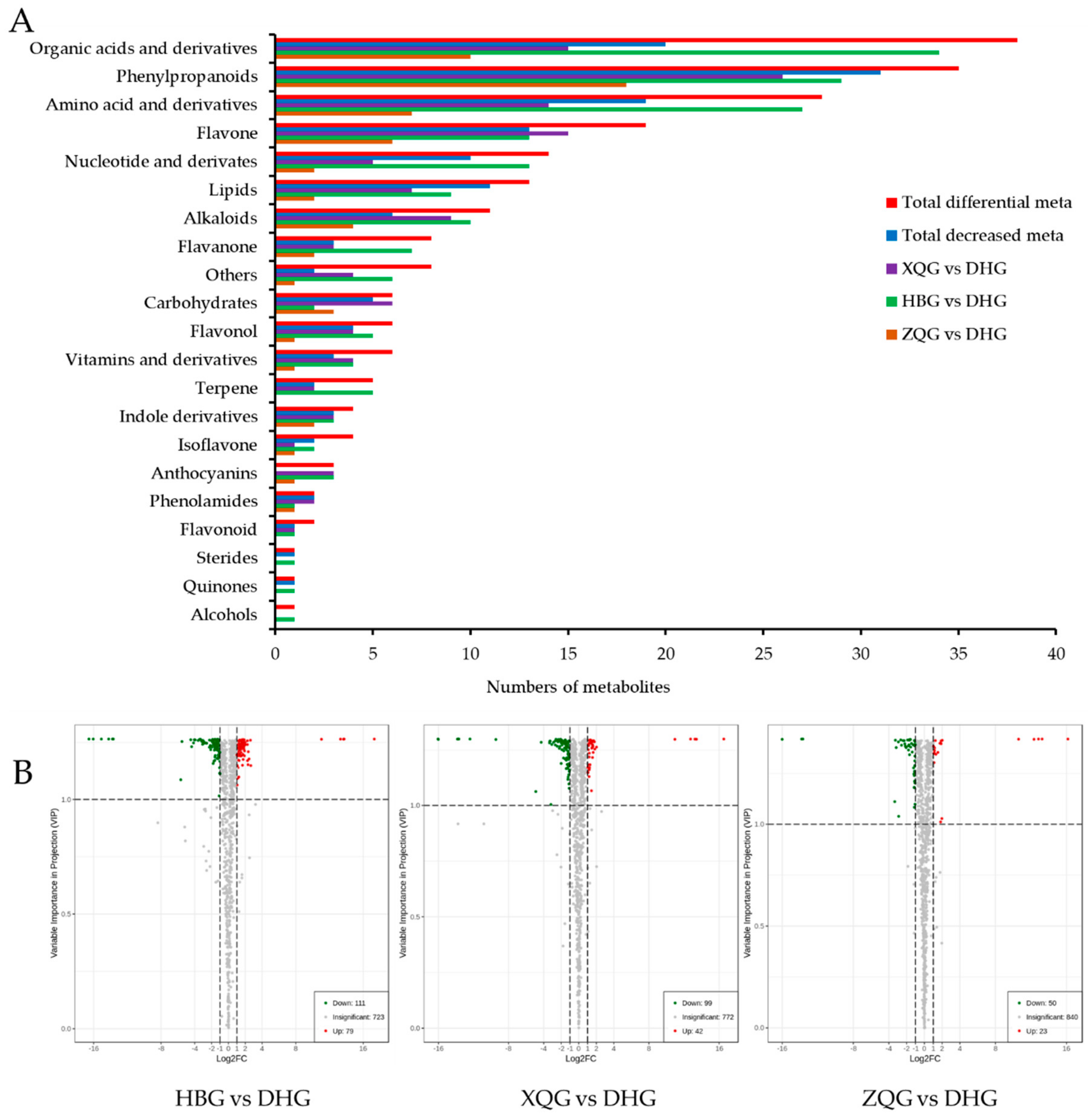
2.4. Univariate Analysis and Screening and Venn and Volcano Plot Generation of Differential Metabolites for the Three Treatment Groups (HBG, XQG, and ZQG) versus the Control Group
2.5. KEGG Annotation and Metabolic Pathway Analysis of Differential Metabolites for the Three Treatment Groups (HBG, XQG, and ZQG) versus the Control Group
2.6. Hierarchical Cluster Analysis of Differential Metabolites for the Three Treatment Groups (HBG, XQG, and ZQG) versus the Control Group
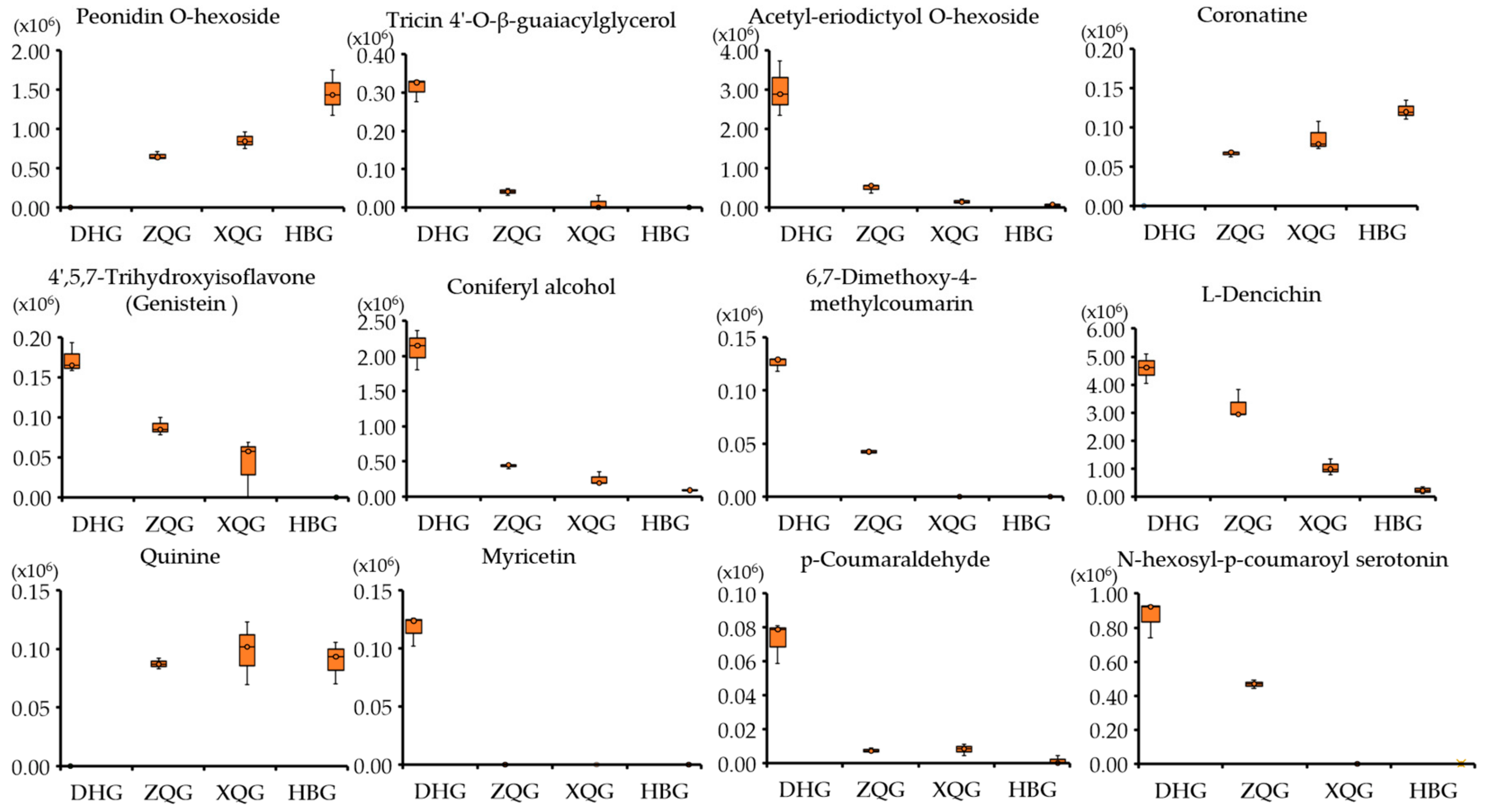
2.7. Boxplots of Differential Metabolites for the Three Treatment Groups (HBG, XQG, and ZQG) versus the Control Group
3. Discussion
3.1. Metabolites Identified in the Four Types of Fruit Peels
3.2. Differential Metabolites between the Three Treatment Groups (HBG, XQG, and ZQG) and the Control Group (DHG)
4. Plant Materials and Treatments
4.1. Samples
4.2. Extraction Process for Metabolite Analysis
4.3. HPLC and ESI-Q TRAP-MS/MS Conditions
4.3.1. HPLC Separation Conditions
4.3.2. ESI-Q TRAP-MS/MS
4.4. Qualitative and Quantitative Determination of Metabolites
4.4.1. Qualitative Determination of Metabolites
4.4.2. Quantitative Determination of Metabolites
4.5. Raw Data Preprocessing
4.6. Basis Data Analysis
4.7. Kyoto Encyclopedia of Genes and Genomes (KEGG) Annotation and Metabolic Pathway Analysis of Differential Metabolites
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tripoli, E.; La Guardia, M.; Giammanco, S.; Di Majo, D.; Giammanco, M. Citrus flavonoids: Molecular structure, biological activity and nutritional properties: A review. Food Chem. 2007, 104, 466–479. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Zhang, Q.; Zhang, H.X.; Ma, Q.; Lu, J.Q.; Qiao, Y.J. Characterization of polymethoxylated flavonoids (PMFs) in the peels of ‘Shatangju’ mandarin (Citrus reticulata Blanco) by online high-performance liquid chromatography coupled to photodiode array detection and electrospray tandem mass spectrometry. J. Agric. Food Chem. 2012, 60, 9023–9034. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, Y.; Wang, D.; Jiang, L.; Xu, X. Identification of three kinds of citri reticulatae pericarpium based on deoxyribonucleic acid barcoding and high-performance liquid chromatography-diode array detection-electrospray ionization/mass spectrometry/mass spectrometry combined with chemometric analysis. Pharmacogn. Mag. 2018, 14, 64. [Google Scholar] [PubMed]
- McClean, R.E. Greening or blotchy-mottle disease of citrus. Phytophylactica 1970, 2, 177–194. [Google Scholar]
- Bové, J.M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 2006, 88, 7–37. [Google Scholar]
- Baldwin, E.; Plotto, A.; Manthey, J.; McCollum, G.; Bai, J.; Irey, M.; Cameron, R.; Luzio, G. Effect of Liberibacter infection (Huanglongbing disease) of citrus on orange fruit physiology and fruit/fruit juice quality: chemical and physical analyses. J. Agric. Food Chem. 2009, 58, 1247–1262. [Google Scholar] [CrossRef] [PubMed]
- Polturak, G.; Heinig, U.; Grossman, N.; Battat, M.; Leshkowitz, D.; Malitsky, S.; Rogachev, I.; Aharoni, A. Transcriptome and metabolic profiling provides insights into betalain biosynthesis and evolution in Mirabilis jalapa. Mol. Plant 2018, 11, 189–204. [Google Scholar] [CrossRef]
- Costa, J.H.; Wassano, C.I.; Angolini, C.F.F.; Scherlach, K.; Hertweck, C.; Fill, T.P. Antifungal potential of secondary metabolites involved in the interaction between citrus pathogens. Sci. Rep. UK 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Parhiz, H.; Roohbakhsh, A.; Soltani, F.; Rezaee, R.; Iranshahi, M. Antioxidant and anti-inflammatory properties of the citrus flavonoids hesperidin and hesperetin: an updated review of their molecular mechanisms and experimental models. Phytother. Res. 2015, 29, 323–331. [Google Scholar] [CrossRef]
- Chen, C.; Piero, A.R.L.; Gmitter, F. Pigments in citrus. In Pigments in Fruits and Vegetables; Springer: New York, NY, USA, 2015; pp. 165–187. [Google Scholar]
- Liu, Y.; Liu, Q.; Xiong, J.; Deng, X. Difference of a citrus late-ripening mutant (Citrus sinensis) from its parental line in sugar and acid metabolism at the fruit ripening stage. Sci. China Ser. C Life Sci. 2007, 50, 511–517. [Google Scholar] [CrossRef]
- Miyazaki, T.; Plotto, A.; Goodner, K.; Gmitter Jr, F.G. Distribution of aroma volatile compounds in tangerine hybrids and proposed inheritance. J. Sci. Food Agr. 2011, 91, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z.; Lu, C.; Zhang, Y.; Zhao, S.; Liu, B.; Xu, X.; Liu, Y. Application of plant metabonomics in quality assessment for large-scale production of traditional Chinese medicine. Planta Med. 2013, 79, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tu, H.; Wan, J.; Chen, W.; Liu, X.; Luo, J.; Zhang, H. Spatio-temporal distribution and natural variation of metabolites in citrus fruits. Food Chem. 2016, 199, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Chen, N.; Hu, N.; Deng, W.; Chen, Z.; Li, Z. Comparative metabolomics and transcriptomic profiling reveal the mechanism of fruit quality deterioration and the resistance of citrus fruit against Penicillium digitatum. Postharvest Biol. Tec. 2018, 145, 61–73. [Google Scholar] [CrossRef]
- Wang, Z.; Cui, Y.; Vainstein, A.; Chen, S.; Ma, H. Regulation of fig (Ficus carica L.) fruit color: metabolomic and transcriptomic analyses of the flavonoid biosynthetic pathway. Front. Plant Sci. 2017, 8, 1990. [Google Scholar] [CrossRef]
- Esposito, D.; Ferriello, F.; Dal Molin, A.; Diretto, G.; Sacco, A.; Minio, A.; Barone, A.; Di Monaco, R.; Cavella, S.; Tardella, L.; et al. Unraveling the complexity of transcriptomic, metabolomic and quality environmental response of tomato fruit. BMC Plant Biol. 2017, 17, 66. [Google Scholar]
- Joshi, V.K.; Kumar, A.; Kumar, V. Antimicrobial, antioxidant and phyto-chemicals from fruit and vegetable wastes: A review. Int. J. Food Ferment. Technol. 2012, 2, 123. [Google Scholar]
- Safdar, M.N.; Kausar, T.; Jabbar, S.; Mumtaz, A.; Ahad, K.; Saddozai, A.A. Extraction and quantification of polyphenols from kinnow (Citrus reticulate L.) peel using ultrasound and maceration techniques. J. Food Drug Anal. 2017, 25, 488–500. [Google Scholar] [CrossRef]
- Guccione, C.; Bergonzi, M.C.; Piazzini, V.; Bilia, A.R. A simple and rapid HPLC-PDA MS method for the profiling of citrus peels and traditional Italian liquors. Planta Med. 2016, 82, 1039–1045. [Google Scholar] [CrossRef]
- Wu, T.; Cheng, D.; He, M.; Pan, S.; Yao, X.; Xu, X. Antifungal action and inhibitory mechanism of polymethoxylated flavones from Citrus reticulata Blanco peel against Aspergillus niger. Food Control 2014, 35, 354–359. [Google Scholar] [CrossRef]
- Zulkifli, K.S.; Abdullah, N.; Abdullah, A.; Aziman, N.; Kamarudin, W.S.S.W. Bioactive phenolic compounds and antioxidant activity of selected fruit peels. Int. Conf. Environ. Chem. Biol. 2012, 49, 66–70. [Google Scholar]
- Marzouk, B. Characterization of bioactive compounds in Tunisian bitter orange (Citrus aurantium L.) peel and juice and determination of their antioxidant activities. Biomed. Res. Int. 2013. [Google Scholar] [CrossRef]
- Cheigh, C.I.; Chung, E.Y.; Chung, M.S. Enhanced extraction of flavanones hesperidin and narirutin from Citrus unshiu peel using subcritical water. J. Food Eng. 2012, 110, 472–477. [Google Scholar] [CrossRef]
- Loizzo, M.R.; Tundis, R.; Bonesi, M.; Menichini, F.; De Luca, D.; Colica, C.; Menichini, F. Evaluation of Citrus aurantifolia peel and leaves extracts for their chemical composition, antioxidant and anti-cholinesterase activities. J. Sci. Food Agr. 2012, 92, 2960–2967. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, Q.; Liu, Z.; Yang, K.; Chen, Z.; Cheng, Q.; Wu, L. The versatile effects of dihydromyricetin in health. Evid. Based Compl. Alt. 2017, 2017, 27–29. [Google Scholar] [CrossRef]
- Wei, L.; Sun, X.; Qi, X.; Zhang, Y.; Li, Y.; Xu, Y. Dihydromyricetin Ameliorates Cardiac Ischemia/ Reperfusion Injury through Sirt3 Activation. Biomed. Res. Int. 2019. [Google Scholar] [CrossRef]
- Xie, S.; Zhao, T.; Zhang, Z.; Meng, J. Reduction of Dihydrokaempferol by Vitis vinfera Dihydroflavonol 4-Reductase to Produce Orange Pelargonidin-Type Anthocyanins. J. Agr. Food Chem. 2018, 66, 3524–3532. [Google Scholar] [CrossRef]
- Martinelli, F.; Ibanez, A.M.; Reagan, R.L.; Davino, S.; Dandekar, A.M. Stress responses in citrus peel: comparative analysis of host responses to Huanglongbing disease and puffing disorder. Sci. Hortic. Amst. 2015, 192, 409–420. [Google Scholar] [CrossRef]
- Ballester, A.R.; Lafuente, M.T.; de Vos, R.C.; Bovy, A.G.; González Candelas, L. Citrus phenylpropanoids and defence against pathogens. Part I: metabolic profiling in elicited fruits. Food Chem. 2013, 136, 178–185. [Google Scholar] [CrossRef]
- German-Ponciano, L.J.; Rosas-Sánchez, G.U.; Rivadeneyra-Domínguez, E.; Rodríguez-Landa, J.F. Advances in the preclinical study of some flavonoids as potential antidepressant agents. Scientifica 2018. [Google Scholar] [CrossRef]
- Munakata, R.; Inoue, T.; Koeduka, T.; Sasaki, K.; Tsurumaru, Y.; Sugiyama, A.; Uto, Y.; Hori, H.; Azuma, J.; Yazaki, K. Characterization of coumarin-specific prenyltransferase activities in Citrus limon peel. Biosci. Biotech. Bioch. 2012, 76, 1389–1393. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Shan, Y.; Wu, Y.; Liu, G.; Chen, B.; Yao, S. Simultaneous determination of flavanones, hydroxycinnamic acids and alkaloids in citrus fruits by HPLC-DAD-ESI/MS. Food Chem. 2011, 127, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Massenti, R.; Lo Bianco, R.; Sandhu, A.K.; Gu, L.; Sims, C. Huanglongbing modifies quality components and flavonoid content of ‘Valencia’ oranges. J. Sci. Food Agr. 2016, 96, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Fraga, C.G.; Clowers, B.H.; Moore, R.J.; Zink, E.M. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Anal. Chem. 2010, 82, 4165–4173. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060. [Google Scholar] [CrossRef]
- Eriksson, L.; Johansson, E.; Kettaneh-Wold, N.; Trygg, J.; Wikström, C.; Wold, S. Multi-and Megavariate Data Analysis; Part I and II; Umetrics AB: Umeå, Sweden, 2006. [Google Scholar]
- Chen, Y.; Zhang, R.; Song, Y.; He, J.; Sun, J.; Bai, J.; An, Z.; Dong, L.; Zhan, Q.; Abliz, Z. RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: finding potential biomarkers for breast cancer. Analyst 2009, 134, 2003–2011. [Google Scholar] [CrossRef]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Saccenti, E.; Hoefsloot, H.C.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M.M. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 2014, 10, 361–374. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015, 44, D457–D462. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Huang, Y.; Wu, W.; Zhu, C.; Zhang, R.; Chen, J.; Zeng, J. Metabolomics Analysis of the Peels of Different Colored Citrus Fruits (Citrus reticulata cv. ‘Shatangju’) During the Maturation Period Based on UHPLC-QQQ-MS. Molecules 2020, 25, 396. https://doi.org/10.3390/molecules25020396
Wang F, Huang Y, Wu W, Zhu C, Zhang R, Chen J, Zeng J. Metabolomics Analysis of the Peels of Different Colored Citrus Fruits (Citrus reticulata cv. ‘Shatangju’) During the Maturation Period Based on UHPLC-QQQ-MS. Molecules. 2020; 25(2):396. https://doi.org/10.3390/molecules25020396
Chicago/Turabian StyleWang, Feiyan, Yongjing Huang, Wen Wu, Congyi Zhu, Ruimin Zhang, Jiezhong Chen, and Jiwu Zeng. 2020. "Metabolomics Analysis of the Peels of Different Colored Citrus Fruits (Citrus reticulata cv. ‘Shatangju’) During the Maturation Period Based on UHPLC-QQQ-MS" Molecules 25, no. 2: 396. https://doi.org/10.3390/molecules25020396
APA StyleWang, F., Huang, Y., Wu, W., Zhu, C., Zhang, R., Chen, J., & Zeng, J. (2020). Metabolomics Analysis of the Peels of Different Colored Citrus Fruits (Citrus reticulata cv. ‘Shatangju’) During the Maturation Period Based on UHPLC-QQQ-MS. Molecules, 25(2), 396. https://doi.org/10.3390/molecules25020396




