A Comparative Study of Some Procedures for Isolation of Fruit DNA of Sufficient Quality for PCR-Based Assays
Abstract
1. Introduction
2. Results
- (i)
- Isolation of DNA by 5 commercial and 1 in-house method.
- (ii)
- Determination of concentration and purity of DNA by UV-VIS spectrophotometry.
- (iii)
- Analysis of quality of each DNA isolate by real-time PCR using primers specific for ITS2 region of plants.
- (iv)
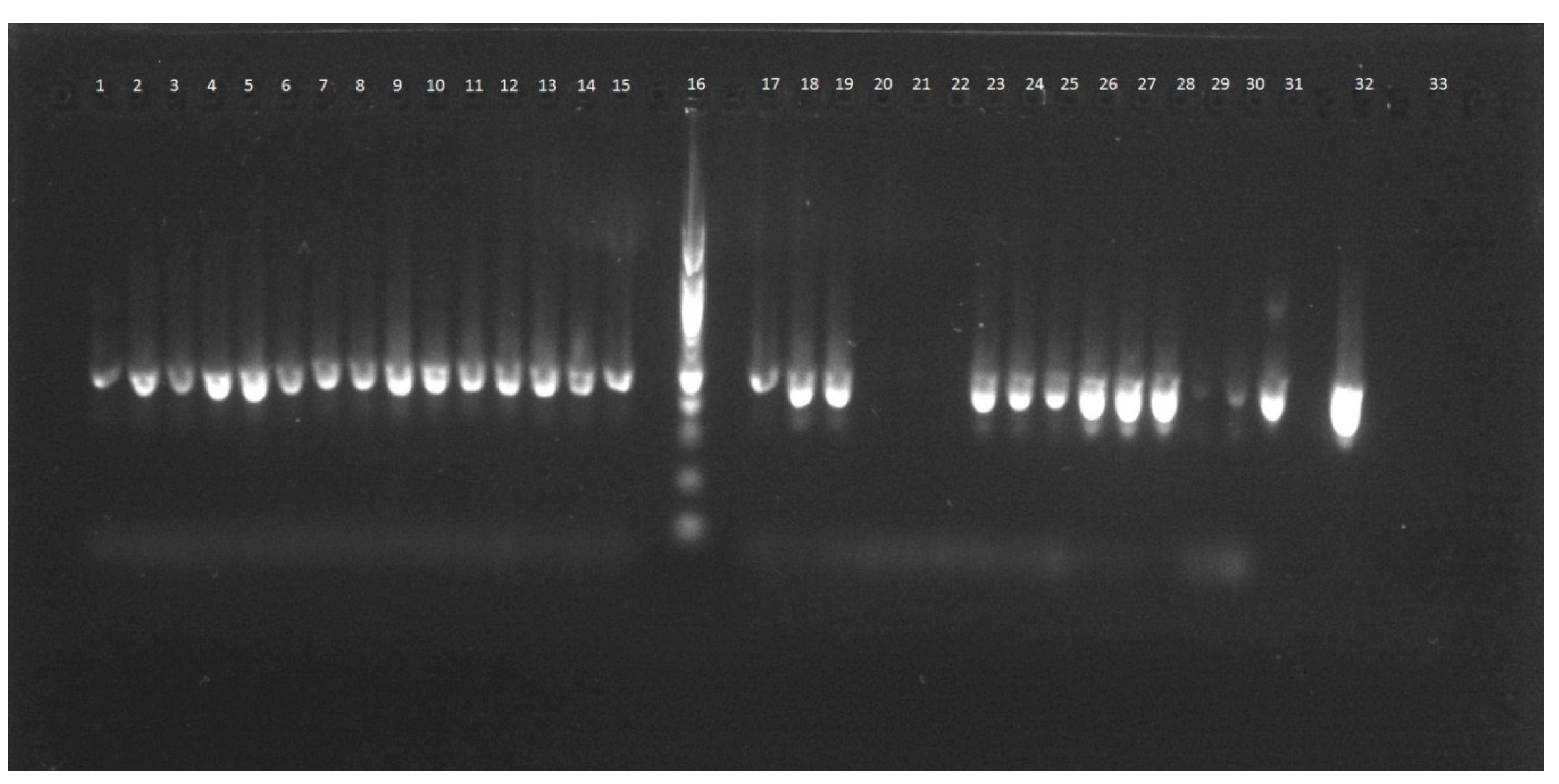
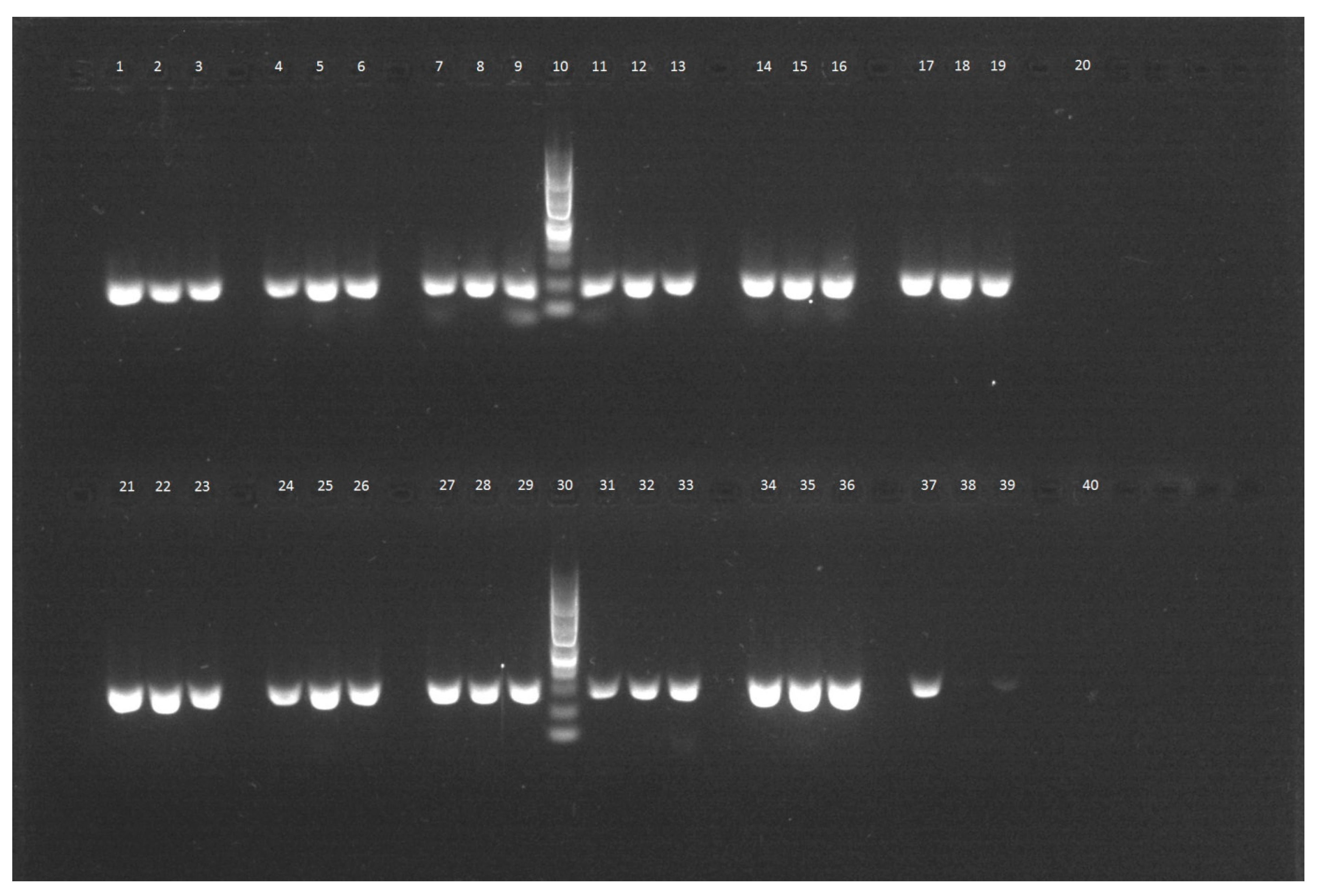
- Analysis of the quality of each DNA isolate by real-time PCR using specific primers for individual fruit species. Every amplicon was analyzed by agarose gel electrophoresis as well.
2.1. Characterization of DNA Isolates by UV-VIS Spectrophotometry
2.2. Real-Time PCR
2.2.1. Red Fruits
2.2.2. Stone Fruits
2.2.3. Tropical Fruit
2.2.4. Pome Fruit
3. Discussion
4. Materials and Methods
4.1. Fruit Species
4.2. Commercial Kits for DNA Isolation
4.3. DNA Isolation
4.4. Real-Time PCR
4.5. Agarose Gel Electrophoresis of PCR Products
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Koswig, S. Determination of Foreign Fruit Types and Fruit Varieties. Analyses, Evaluation and Practical Problems. Fruit Process. J. Fruit Process. Juice Prod. Eur. Overseas Ind. 2006, 16, 401–412. [Google Scholar]
- Zhang, J.; Yu, Q.; Cheng, H.; Ge, Y.; Liu, H.; Ye, X.; Chen, Y. Metabolomic Approach for the Authentication of Berry Fruit Juice by Liquid Chromatography Quadrupole Time-of-Flight Mass Spectrometry Coupled to Chemometrics. J. Agric. Food Chem. 2018, 66, 8199–8208. [Google Scholar] [CrossRef]
- Pardo-Mates, N.; Vera, A.; Barbosa, S.; Hidalgo-Serrano, M.; Núñez, O.; Saurina, J.; Hernández-Cassou, S.; Puignou, L. Characterization, classification and authentication of fruit-based extracts by means of HPLC-UV chromatographic fingerprints, polyphenolic profiles and chemometric methods. Food Chem. 2017, 221, 29–38. [Google Scholar] [CrossRef] [PubMed]
- López-Calleja, I.M.; De La Cruz, S.; Pegels, N.; González, I.; Martín, R.; García, T. Sensitive and specific detection of almond (Prunus dulcis) in commercial food products by real-time PCR. LWT Food Sci. Technol. 2014, 56, 31–39. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors-occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Turci, M.; Sardaro, M.L.S.; Visioli, G.; MAESTRI, E.; Marmiroli, M.; Marmiroli, N. Evaluation of DNA extraction procedures for traceability of various tomato products. Food Control 2010, 21, 143–149. [Google Scholar] [CrossRef]
- Varma, A.; Padh, H.; Shrivastava, N. Plant genomic DNA isolation: An art or a science. Biotechnol. J. 2007, 2, 386–392. [Google Scholar] [CrossRef]
- Paulos, S.; Mateo, M.; De Lucio, A.; Hernández-de Mingo, M.; Bailo, B.; Saugar, J.M.; Cardona, G.A.; Fuentes, I.; Mateo, M.; Carmena, D. Evaluation of five commercial methods for the extraction and purification of DNA from human faecal samples for downstream molecular detection of the enteric protozoan parasites Cryptosporidium spp., Giardia duodenalis, and Entamoeba spp. J. Microbiol. Methods. 2016, 127, 68–73. [Google Scholar] [CrossRef]
- Li, P.; Su, X.; Wang, J.; Yang, S.; Wu, G.; Lin, M.; Zhao, K.; Bezuidenhout, C.; Tang, X. Comparison of genomic DNA extraction commercial kits based on clayey and paddy soils. J. Pure Appl. Microbiol. 2014, 7, 69–75. [Google Scholar]
- Jara, C.; Mateo, E.; Guillamón, J.M.; Toria, M.J.; Mas, A. Analysis of several methods for the extraction of high quality DNA from acetic acid bacteria in wine and vinegar for characterization by PCR-based methods. Int. J. Food Microbiol. 2008, 128, 336–341. [Google Scholar] [CrossRef]
- Vodret, B.; Milia, M.; Orani, M.G.; Serratrice, G.; Mancuso, M.R. Detection of Genetically Modified Organisms in Food: Comparison Among Three Different DNA Extraction Methods. Vet. Res. Commun. 2007, 31, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Saghai-Maroof, M.A.; Soliman, M.A.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer-length polymorphism in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Nail. Acad. Sci. USA 1984, 81, 8014–8019. [Google Scholar] [CrossRef] [PubMed]
- Glyn, M.C.P.; Egertová, M.; Gazdova, B.; Kovarik, A.; Bezdek, M.; Leitch, A.R.; Chen, Y. The influence of 5-azacytidine on the condensation of the short arm of rye chromosome 1R in Triticum aestivum L. root tip meristematic nuclei. Chromosoma 1997, 106, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Primer-BLAST: A tool for finding specific primers. NCBI National Center for Biotechnology Information [online]. Bethesda MD, 20894 USA. Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast/ (accessed on 14 April 2020).
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 Region as a Novel DNA Barcode for Identifying Medicinal Plant Species. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ma, R.; Xu, J.; Yan, J.; Guo, L.; Song, J.; Feng, R.; Yu, M. Genome-wide identification and classification of MYB superfamily genes in peach. PLoS ONE 2018, 13, e0199192. [Google Scholar] [CrossRef]
- Sargent, D.J.; Rys, A.; Nier, S.; Simpson, D.W.; Tobutt, K.R. The development and mapping of functional markers in Fragaria and their transferability and potential for mapping in other genera. Theor. Appl. Genet. 2007, 114, 373–384. [Google Scholar] [CrossRef]
- Wang, F.; Xia, L.; Lv, S.; Xu, C.; Niu, Y.; Liu, W.; Zeng, L.; Zhou, J.; Hu, B. Development of a Mitochondrial SCAR Marker Related to Susceptibility of Banana (Musa AAA Cavendish) to Fusarium oxysporum f. sp. Cubense Race 4. Not. Bot. Horti Agrobot. Cluj Napoca. 2018, 46, 509–516. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, D.; Fan, S.; Du, L.; Shen, Y.; Xing, L.; Li, Y.; Ma, J.; Han, M. Effect of exogenous GA 3 and its inhibitor paclobutrazol on floral formation, endogenous hormones, and flowering-associated genes in ‘Fuji’ apple (Malus domestica Borkh.). Plant Physiol. Biochem. 2016, 107, 178–186. [Google Scholar] [CrossRef]
- Fuentes, L.; Monsalve, L.; Morales-Quintana, L.; Valdenegro, M.; Martínez, J.P.; Defilippi, B.G.; González-Agüero, M. Differential expression of ethylene biosynthesis genes in drupelets and receptacle of raspberry (Rubus idaeus). J. Plant Physiol. 2015, 179, 100–105. [Google Scholar] [CrossRef]
- Song, Y.; Liu, H.; Zhou, Q.; Zhang, H.J.; Zhang, Z.D.; Li, Y.D.; Wang, H.B.; Liu, F.Z. High-throughput sequencing of highbush bilberry transcriptome and analysis of basic helix-loop-helix transcription factors. J. Integr. Agric. 2017, 16, 591–604. [Google Scholar] [CrossRef]
- Hoang, V.L.T.; Innes, D.J.; Shaw, P.N.; Monteith, G.R.; Gidley, M.J.; Dietzgen, R.G. Sequence diversity and differential expression of major phenylpropanoid-flavonoid biosynthetic genes among three mango varieties. BMC Genom. 2015, 16, 561. [Google Scholar] [CrossRef] [PubMed]
- Ziarovska, J.; Boselova, D.; Bezo, M. Retrotransposon cassandra copies estimating in plums using real-time pcr approach. Emir. J. Food Agric. 2015, 27, 591–604. [Google Scholar] [CrossRef]
- Han, J.; Wu, Y.; Huang, W.; Wang, B.; Sun, C.; Ge, Y.; Chen, Y. PCR and DHPLC methods used to detect juice ingredient from 7 fruits. Food Control 2012, 25, 696–703. [Google Scholar] [CrossRef]
| Fruit | Method | c (ng·µL−1) | A260/A280 | A260/A230 | m (ng) |
|---|---|---|---|---|---|
| Banana | 1 | 5.0 ± 1.7 | 1.40 ± 0.09 | 0.36 ± 0.05 | 500 ± 167 |
| 2 | 3.5 ± 1.0 | 1.95 ± 0.07 | 1.04± 0.06 | 350 ± 101 | |
| 3 | 5.4 ± 0.8 | 2.13 ± 0.15 | 1.46 ± 0.63 | 540 ± 78 | |
| 4 | 4.6 ± 1.5 | 1.23 ± 0.05 | 0.27 ± 0.16 | 460 ± 150 | |
| 5 | 4.1 ± 11.6 | 1.81 ± 0.34 | 0.27 ± 0.16 | 410 ± 1164 | |
| 6 | 227.5 ± 37.5 | 1.59 ± 0.04 | 1.66 ± 0.16 | 22,750 ± 3747 | |
| Bilberry | 1 | 23.2 ± 21.3 | 1.18 ± 0.17 | 0.43 ± 0.10 | 2320 ± 2134 |
| 2 | 1.7 ± 0.2 | 1.65 ± 0.47 | 0.28 ± 0.35 | 170 ± 21 | |
| 3 | 48.5 ± 14.7 | 0.63 ± 0.26 | 0.14 ± 0.06 | 4850 ± 1468 | |
| 4 | 3.0 ± 1.0 | 0.99 ± 0.07 | 0.30 ± 0.03 | 300 ± 103 | |
| 5 | 4.3 ± 6.7 | 1.96 ± 0.25 | 0.33 ± 0.82 | 430 ± 668 | |
| 6 | 80.5 ± 26.4 | 1.39 ± 0.03 | 1.66 ± 0.22 | 8050 ± 2644 | |
| Pear | 1 | 41.3 ± 23.5 | 1.39 ± 0.14 | 0.57 ± 0.19 | 4130 ± 2,353 |
| 2 | 5.6 ± 1.2 | 2.60 ± 0.14 | 0.55 ± 0.07 | 560 ± 121 | |
| 3 | 5.7 ± 0.4 | 1.54 ± 0.07 | 0.59 ± 0.04 | 570 ± 36 | |
| 4 | 4.1 ± 0.5 | 1.10 ± 0.13 | 0.28 ± 0.03 | 410 ± 50 | |
| 5 | 5.6 ± 3.0 | 1.73 ± 0.31 | 0.32 ± 0.77 | 560 ± 303 | |
| 6 | 54.9 ± 11.0 | 1.36 ± 0.01 | 1.61 ± 0.15 | 5490 ± 1104 | |
| Apple | 1 | 11.8 ± 3.1 | 1.21 ± 0.04 | 0.37 ± 0.06 | 1180 ± 305 |
| 2 | 1.6 ± 1.3 | 2.37 ± 2.32 | 0.22 ± 0.06 | 160 ± 126 | |
| 3 | 3.9 ± 0.7 | 1.43 ± 0.03 | 0.51 ± 0.04 | 390 ± 66 | |
| 4 | 15.7 ± 1.7 | 1.19 ± 0.02 | 0.21 ± 0.03 | 1570 ± 168 | |
| 5 | 3.3 ± 1.9 | 1.59 ± 0.24 | 0.53 ± 2.97 | 330 ± 193 | |
| 6 | 67.5 ± 7.4 | 1.39 ± 0.02 | 1.48 ± 0.08 | 6750 ± 745 | |
| Strawberry | 1 | 7.0 ± 0.4 | 1.25 ± 0.10 | 0.39 ± 0.04 | 700 ± 35 |
| 2 | 6.0 ± 2.9 | 2.08 ± 0.13 | 1.11 ± 0.21 | 600 ± 295 | |
| 3 | 19.0 ± 24.9 | 0.60 ± 0.12 | 0.14 ± 0.04 | 1900 ± 2491 | |
| 4 | 3.1 ± 7.8 | 1.17 ± 0.05 | 0.41 ± 0.05 | 310 ± 783 | |
| 5 | 2.6 ± 0.6 | 1.53 ± 0.69 | 1.98 ± 5,18 | 260 ± 56 | |
| 6 | 298.5 ± 159.1 | 1.59 ± 0.13 | 1.82 ± 0.05 | 29,850 ± 15,914 | |
| Raspberry | 1 | 241.9 ± 44.2 | 1.50 ± 0.05 | 0.37 ± 0.03 | 24,190 ± 4423 |
| 2 | 4.0 ± 2.1 | 2.09 ± 0.08 | 1.02 ± 0.20 | 400 ± 211 | |
| 3 | 6.3 ± 1.3 | 1.39 ± 0.10 | 1.17 ± 0.10 | 630 ± 125 | |
| 4 | 1.7 ± 0.9 | 1.25 ± 0.27 | 0.33 ± 0.07 | 170 ± 86 | |
| 5 | 9.3 ± 19.4 | 1.99 ± 0.04 | 0.38 ± 2.15 | 930 ± 1938 | |
| 6 | 650.3 ± 65.2 | 1.43 ± 0.02 | 1.78 ± 0.18 | 65,030 ± 6524 | |
| Apricot | 1 | 48.2 ± 16.4 | 1.24 ± 0.06 | 0.43 ± 0.04 | 4820 ± 1638 |
| 2 | 0.9 ± 0.4 | 1.20 ± 0.35 | 0.19 ± 0.14 | 90 ± 40 | |
| 3 | 33.3 ± 14.6 | 0.76 ± 0.19 | 0.16 ± 0.04 | 3330 ± 1456 | |
| 4 | 2.2 ± 0.3 | 1.11 ± 0.09 | 0.23 ± 0.03 | 220 ± 30 | |
| 5 | 4.6 ± 2.0 | 1.93 ± 0.16 | 0.27 ± 0.06 | 460 ± 203 | |
| 6 | 75.9 ± 23.1 | 1.43 ± 0.03 | 1.53 ± 0.06 | 7590 ± 2308 | |
| Plum | 1 | 50.4 ± 21.9 | 1.25 ± 0.06 | 0.34 ± 0.04 | 5040 ± 2187 |
| 2 | 1.7 ± 0.3 | 2.29 ± 0.20 | 0.41 ± 0.07 | 170 ± 30 | |
| 3 | 18.9 ± 3.2 | 1.18 ± 0.05 | 0.39 ± 0.04 | 1890 ± 323 | |
| 4 | 2.1 ± 0.2 | 1.05 ± 0.13 | 0.27 ± 0.12 | 210 ± 15 | |
| 5 | 4.1 ± 0.4 | 1.56 ± 0.44 | 0.49 ± 0.11 | 410 ± 42 | |
| 6 | 101.2 ± 12.0 | 1.38 ± 0.02 | 1.14 ± 0.06 | 10,120 ± 1196 | |
| Peach | 1 | 4.3 ± 2.1 | 1.30 ± 0.48 | 0.23 ± 0.05 | 430 ± 206 |
| 2 | 15.4 ± 0.8 | 2.25 ± 0.05 | 1.82 ± 0.21 | 1540 ± 75 | |
| 3 | 2.7 ± 1.1 | 1.28 ± 1.43 | 0.32 ± 0.41 | 270 ± 108 | |
| 4 | 14.3 ± 3.9 | 1.25 ± 0.05 | 0.26 ± 0.05 | 1430 ± 391 | |
| 5 | 3.1 ± 0.2 | 1.80 ± 0.15 | 1.67 ± 1.61 | 310 ± 21 | |
| 6 | 184.2 ± 30.5 | 1.65 ± 0.01 | 1.45 ± 0.08 | 18,420 ± 3054 | |
| Mango | 1 | 2.6 ± 7.0 | 1.75 ± 0.58 | 0.27 ± 0.11 | 260 ± 696 |
| 2 | 8.5 ± 1.7 | 2.68 ± 0.10 | 1.43 ± 0.44 | 850 ± 170 | |
| 3 | 1.9 ± 0.7 | 1.55 ± 3,8 | 1.69 ± 3,41 | 190 ± 66 | |
| 4 | 4.1 ± 0.5 | 1.54 ± 0.02 | 0.59 ± 0.01 | 410 ± 47 | |
| 5 | 4.7 ± 2.2 | 1.81 ± 0.22 | 0.66 ± 32,58 | 470 ± 217 | |
| 6 | 123.8 ± 12.5 | 1.57 ± 0.02 | 1.63 ± 0.08 | 12,380 ± 1246 |
| Fruit | Method | Ct—Plant Specific Primers | Ct—Species Specific Primers | Figure | ||||
|---|---|---|---|---|---|---|---|---|
| Strawberry | Kit 1 (Qiagen) | 21.64 | 21.24 | 26.91 | 25.03 | 24.77 | S9, 1,2 13, 14 | |
| Kit 2 (Elisabeth Pharmacon) | 19.52 | 20.21 | 33.54 | 23.56 | 25.13 | 29.28 | S9, 3, 4, 15, 16 | |
| Kit 3 (Invitrogen) | 18.18 | 18.48 | 19.07 | 21.11 | 22.22 | 22.88 | S9, 5, 6, 17, 18 | |
| Kit 4 (PerkinElmer) | 22.48 | 21.98 | 21.56 | 26.46 | 32.49 | 26.34 | S9, 7, 8, 19, 20 | |
| Kit 5 (Tools) | 18.19 | 18.19 | 17.25 | 20.16 | 24.03 | 23.01 | S9, 9, 10, 21, 22 | |
| 6 (CTAB protocol) | 32.15 | 24.0 | 21.17 | 29.81 | 31.47 | S9, 11, 12, 23, 24 | ||
| Raspberry | Kit 1 (Qiagen) | 33.25 | 31.18 | S10, 1, 2, 13, 14 | ||||
| Kit 2 (Elisabeth Pharmacon) | 16.62 | 22.15 | 18.33 | 28.14 | 23.79 | 22.66 | S10, 3, 4, 15, 16 | |
| Kit 3 (Invitrogen) | 18.05 | 17.31 | 19.28 | 25.08 | 24.91 | 25.47 | S10, 5, 6, 17, 18 | |
| Kit 4 (PerkinElmer) | 19.21 | 19.13 | 17.33 | 27.05 | 25.68 | 24.9 | S10, 7, 8, 19, 20 | |
| Kit 5 (Tools) | 15.01 | 16.57 | 18.32 | 21.98 | 24.11 | 25.3 | S10, 9, 10, 21, 22 | |
| 6 (CTAB protocol) | 20.6 | 17.62 | 24.1 | 25.25 | 23.88 | 29.58 | S10, 11, 12, 23, 24 | |
| Bilberry | Kit 1 (Qiagen) | 19.71 | 18.83 | 19.1 | 26.72 | 24.77 | 24.09 | S5, 1, 2, 13, 14 |
| Kit 2 (Elisabeth Pharmacon) | 19.94 | 20.55 | 14.38 | 28.87 | 31.31 | 28.16 | S5, 3, 4, 15, 16 | |
| Kit 3 (Invitrogen) | 17.45 | 16.93 | 17.73 | 22.84 | 22.61 | 23.49 | S5, 5, 6, 17, 18 | |
| Kit 4 (PerkinElmer) | 22.6 | 23.91 | 21.07 | 28.27 | 28.78 | 26.42 | S5, 7, 8, 19, 20 | |
| Kit 5 (Tools) | 16.75 | 18.08 | 16.09 | 22.36 | 24.37 | 23.56 | S5, 9, 10, 21, 22 | |
| 6 (CTAB protocol) | 31.01 | 29.32 | 27.87 | 30.36 | S5, 11, 12, 23, 24 | |||
| Fruit | Method | Ct—Plant Specific Primers | Ct—Species Specific Primers | Figure | ||||
|---|---|---|---|---|---|---|---|---|
| Plum | Kit 1 (Qiagen) | 30.96 | 33.41 | 30.06 | 29.56 | 29.62 | S13, 1, 2, 13, 14 | |
| Kit 2 (Elisabeth Pharmacon) | 22.91 | 21.94 | 21.74 | 20.78 | 22.71 | 22.56 | S13, 3, 4. 15, 16 | |
| Kit 3 (Invitrogen) | 31.12 | 30.39 | 35.6 | 21.91 | 30.5 | 29.98 | S13, 5, 6, 17, 18 | |
| Kit 4 (Perkin-Elmer) | 23.75 | 24.05 | 21.44 | 25.82 | 25.79 | 24.32 | S13, 7, 8, 19, 20 | |
| Kit 5 (Tools) | 18.61 | 20.06 | 19.78 | 21.37 | 23.53 | 22.26 | S13, 9, 10, 21, 22 | |
| 6 (CTAB protocol) | 29.36 | 31.69 | 28.25 | 31.24 | 25.34 | 28.73 | S13, 23, 24 | |
| Peach | Kit 1 (Qiagen) | 16.09 | 18.53 | 17.48 | 23.96 | 28.22 | 27.43 | S6, 1, 2, 13, 14 |
| Kit 2 (Elisabeth Pharmacon) | 19.43 | 20.49 | 19.54 | 28.46 | 26.64 | 27.27 | S6, 3, 4, 15, 16 | |
| Kit 3 (Invitrogen) | 18.85 | 17.98 | 18.45 | 26.59 | 26.36 | 26.13 | S6, 5, 6, 17, 18 | |
| Kit 4 (Perkin-Elmer) | 20.08 | 19.66 | 21.11 | 27.93 | 27.09 | 26.36 | S6, 7, 8, 19, 20 | |
| Kit 5 (Tools) | 15.68 | 18.09 | 17.06 | 25.25 | 26.43 | 25.51 | S6, 9, 10, 21, 22 | |
| 6 (CTAB protocol) | 20.61 | 20.5 | 20.27 | 26.06 | 24.61 | 27.04 | S6, 11, 12, 23, 24 | |
| Apricot | Kit 1 (Qiagen) | 31.38 | 32.25 | 31.85 | S12, 1, 2, 13, 14 | |||
| Kit 2 (Elisabeth Pharmacon) | 25.07 | 23.51 | 30.76 | 30.42 | 30.96 | S12, 3, 4, 15, 16 | ||
| Kit 3 (Invitrogen) | 17.94 | 15.61 | 17.26 | 25.78 | 24.27 | 26.16 | S12, 5, 6, 17, 18 | |
| Kit 4 (Perkin-Elmer) | 24.65 | 23.54 | 24.48 | 32.28 | 29.5 | S6, 7, 8, 19, 20 | ||
| Kit 5 (Tools) | 18.51 | 19.05 | 18.28 | 27.17 | 27.93 | 28.99 | S6, 9, 10, 21, 22 | |
| 6 (CTAB protocol) | 29.76 | 28.82 | 24.92 | 32.0 | S6, 11, 12, 23, 24 | |||
| Fruit | Method | Ct—Plant Specific Primers | Ct—Species Specific Primers | Figure | ||||
|---|---|---|---|---|---|---|---|---|
| Banana | Kit 1 (Qiagen) | 16.51 | 16.97 | 18.13 | 21.04 | 21.63 | 23.06 | S4, 1, 2, 13, 14 |
| Kit 2 (Elisabeth Pharmacon) | 17.38 | 19.24 | 19.06 | 24.2 | 22.31 | 23.45 | S4, 3, 4, 15, 16 | |
| Kit 3 (Invitrogen) | 20.09 | 20.38 | 19.61 | 23.03 | 24.2 | 23.85 | S4, 5, 6, 17, 18 | |
| Kit 4 (Perkin-Elmer) | 22.25 | 23.52 | 23.76 | 25.69 | 25.3 | 24.42 | S4, 7, 8, 19. 20 | |
| Kit 5 (Tools) | 18.77 | 17.4 | 18.56 | 20.77 | 17.87 | 20.75 | S4, 9, 10, 21, 22 | |
| 6 (CTAB protocol) | 20.82 | 29.96 | 28.24 | 25.24 | 32.1 | 29.47 | S4, 11, 12, 23, 24 | |
| Mango | Kit 1 (Qiagen) | 22.43 | 19.25 | 18.52 | 21.98 | 20.72 | 20.86 | S11, 1, 2, 13, 14 |
| Kit 2 (Elisabeth Pharmacon) | 23.26 | 23.75 | 22.83 | 30.92 | 30.0 | S11, 3, 4, 15, 16 | ||
| Kit 3 (Invitrogen) | 21.83 | 21.92 | 22.05 | 25.03 | 25.77 | 25.94 | S11, 5, 6, 17, 18 | |
| Kit 4 (Perkin-Elmer) | 21.54 | 22.39 | 21.7 | 25.11 | 26.2 | 25.17 | S11, 7, 8, 19, 20 | |
| Kit 5 (Tools) | 16.91 | 18.02 | 17.33 | 21.18 | 24.01 | 21.98 | S11, 9, 10, 21, 22 | |
| 6 (CTAB protocol) | 21.15 | 21.78 | 25.03 | 25.62 | 26.64 | 28.02 | S11, 11, 12, 23, 24 | |
| Fruit | Method | Ct—Plant Specific Primers | Ct—Species Specific Primers | Figure | ||||
|---|---|---|---|---|---|---|---|---|
| Pear | Kit 1 (Qiagen) | 25.23 | 24.17 | 26.28 | 29.03 | 28.36 | 28.91 | S7, 1, 2, 13, 14 |
| Kit 2 (Elisabeth Pharmacon) | 21.89 | 20.94 | 19.35 | 26.08 | 24.97 | 23.88 | S7, 3, 4, 15, 16 | |
| Kit 3 (Invitrogen) | 26.04 | 26.27 | 26.29 | 29.06 | 29.71 | 29.11 | S7, 5, 6, 17, 18 | |
| Kit 4 (Perkin-Elmer) | 25.27 | 27.43 | 26.34 | 26.77 | 28.73 | 26.54 | S7, 7, 8, 19, 20 | |
| Kit 5 (Tools) | 25.92 | 24.63 | 24.35 | 27.84 | 28.48 | 29.93 | S7, 9, 10, 21, 22 | |
| 6 (CTAB protocol) | 29.58 | 31.65 | 32.42 | 28.27 | 26.65 | 27.97 | S7, 11, 12, 23, 24 | |
| Apple | Kit 1 (Qiagen) | 28.24 | 27.25 | 27.89 | 32.27 | 35.9 | 32.84 | S8, 1, 2, 13, 14 |
| Kit 2 (Elisabeth Pharmacon) | 29.34 | 27.93 | 29.11 | 34.72 | 32.68 | S8, 3, 4, 15, 16 | ||
| Kit 3 (Invitrogen) | 27.38 | 27.52 | 25.88 | 30.76 | 30.87 | 30.79 | S8, 5, 6, 17, 18 | |
| Kit 4 (Perkin-Elmer) | 27.4 | 28.08 | 27.56 | 32.75 | S8, 7, 8, 19, 20 | |||
| Kit 5 (Tools) | 28.05 | 27.84 | 27.69 | 29.48 | 30.62 | S8, 9, 10, 21, 22 | ||
| 6 (CTAB protocol) | 32.99 | 29.13 | 31.26 | 30.96 | 32.32 | S8, 11, 12, 23, 24 | ||
| Method No. | Manufacturer | Kit/Method | Sample Homogenization | DNA Capture and Purification |
|---|---|---|---|---|
| 1 | Qiagen | DNEasy PowerPlant Pro Kit | Bead-beating of small pieces of plant tissue at room temperature | Spin column |
| 2 | Elisabeth Pharmacon | EliGene Plant DNA Isolation Kit | Mechanical (pestle and sand) and chemical (detergent), at room temperature | Capture on a spin filter in the presence of a chaotropic detergent |
| 3 | Invitrogen | ChargeSwitch gDNA Plant Kit | Grinding with mortar and pestle in liquid nitrogen | Magnetic beads positively charged in acidic pH and neutral in pH above 8.5 |
| 4 | Perkin- Elmer | Chemagic DNA Plant Kit | Grinding with mortar and pestle in liquid nitrogen | Magnetic beads with chaotropic salt |
| 5 | Tools | Easy Prep Polyphenol Plant DNA Extraction Kit | Grinding with mortar and pestle in liquid nitrogen | Spin column |
| 6 | - | CTAB protocol | Grinding with mortar and pestle in liquid nitrogen | Isopropanol and ethanol precipitation [12,13] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fialova, L.; Romanovska, D.; Marova, I. A Comparative Study of Some Procedures for Isolation of Fruit DNA of Sufficient Quality for PCR-Based Assays. Molecules 2020, 25, 4317. https://doi.org/10.3390/molecules25184317
Fialova L, Romanovska D, Marova I. A Comparative Study of Some Procedures for Isolation of Fruit DNA of Sufficient Quality for PCR-Based Assays. Molecules. 2020; 25(18):4317. https://doi.org/10.3390/molecules25184317
Chicago/Turabian StyleFialova, Lenka, Denisa Romanovska, and Ivana Marova. 2020. "A Comparative Study of Some Procedures for Isolation of Fruit DNA of Sufficient Quality for PCR-Based Assays" Molecules 25, no. 18: 4317. https://doi.org/10.3390/molecules25184317
APA StyleFialova, L., Romanovska, D., & Marova, I. (2020). A Comparative Study of Some Procedures for Isolation of Fruit DNA of Sufficient Quality for PCR-Based Assays. Molecules, 25(18), 4317. https://doi.org/10.3390/molecules25184317





