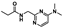Abstract
As fragment-based drug discovery has become mainstream, there has been an increase in various screening methodologies. Protein-observed 19F (PrOF) NMR and 1H CPMG NMR are two fragment screening assays that have complementary advantages. Here, we sought to combine these two NMR-based assays into a new screening workflow. This combination of protein- and ligand-observed experiments allows for a time- and resource-efficient multiplexed screen of mixtures of fragments and proteins. PrOF NMR is first used to screen mixtures against two proteins. Hit mixtures for each protein are identified then deconvoluted using 1H CPMG NMR. We demonstrate the benefit of this fragment screening method by conducting the first reported fragment screens against the bromodomains of BPTF and Plasmodium falciparum (Pf) GCN5 using 467 3D-enriched fragments. The hit rates were 6%, 5% and 4% for fragments binding BPTF, PfGCN5, and fragments binding both proteins, respectively. Select hits were characterized, revealing a broad range of affinities from low µM to mM dissociation constants. Follow-up experiments supported a low-affinity second binding site on PfGCN5. This approach can be used to bias fragment screens towards more selective hits at the onset of inhibitor development in a resource- and time-efficient manner.
1. Introduction
Fragment-based drug discovery (FBDD) is a validated technique to uncover inhibitors for many protein classes and has resulted in four FDA-approved drugs [1,2,3,4]. Compounds in FBDD libraries are typically of low complexity and low molecular weight (average MW ≤ 300 g/mol), allowing these libraries to more effectively sample chemical space, with smaller library sizes than traditional high throughput screening libraries (average MW ≤ 500 g/mol) [5]. Fragments with more complexity via sp3 carbons, saturated ring systems, and stereocenters (i.e., 3D fragments) are also being investigated for providing increased diversity of fragment libraries [6,7,8,9,10,11]. The 3D-enriched fragments offer a potential solution to improve selectivity and physicochemical properties at the beginning of the drug development process [7,12,13,14,15,16,17]. Fragments often have weak affinity for their target protein, making it essential to use screening assays that can detect compounds with affinity values in the high μM to low mM range.
Ligand- and protein-observed NMR binding assays are effective fragment screening techniques. A 2019 poll by Practical Fragments listed ligand- and protein-observed NMR as the second and fourth most popular FBDD techniques, respectively [18]. These two NMR formats have complementary advantages [19]. Ligand-observed NMR assays such as Carr–Purcell–Meiboom–Gill (CPMG), saturation transfer difference (STD), and water–ligand observed via gradient spectroscopy (waterLOGSY) allow for the screening of fragment mixtures without the need for deconvolution experiments due to distinct ligand resonances. In practice, only a single protein is screened in a ligand-observed NMR experiment. For small proteins studied here (<15 kDa), experiment times can take 5–30 min. In contrast, protein-observed NMR techniques such as 1H-15N-heteronuclear single quantum coherence (HSQC) and protein-observed 19F (PrOF) NMR use distinct protein resonances allowing screening of multiple proteins in a single experiment. Although faster than CPMG NMR for small proteins (2–8 min [20]), protein-observed NMR methods require more protein for screening and additional time for resource-heavy deconvolution steps when screening mixtures of fragments.
Here, we take advantage of the benefits of ligand- and protein-observed NMR techniques by combining 1H CPMG NMR and PrOF NMR into a single screening workflow that allows testing fragment mixtures for binding against multiple proteins with minimal deconvolution experiments, which saves time and labeled protein. There are many examples of fragment screens using 1H CPMG NMR [21], STD NMR [21], waterLOGSY NMR [21], PrOF NMR [11,22], dual-protein PrOF NMR [23], and multi-protein HSQC NMR techniques [20,24]. From these NMR methods, we chose 1H CPMG NMR and PrOF NMR for this platform because a previous study showed low discrepancy between the assays [25]. Additionally, PrOF NMR gives significantly less complicated spectra than HSQC NMR, and is 2–3-fold faster [25,26], expediting experiment time and data processing. More than 70 proteins have been studied via PrOF NMR using more than 15 different fluorinated amino acids [27]. PrOF NMR has been used to study proteins of various sizes from the Fyn SH3 domain (7 kDa) [28] to the α7 single-ring of the 20S proteasome core particle from Thermoplasma acidophilum (180 kDa) [29], illustrating its wide applicability to different proteins.
To validate this approach, we screened a library of 467 3D-enriched fragments in mixtures of five against the bromodomains of BPTF and Plasmodium falciparum GCN5 (PfGCN5). PfGCN5 is a homolog of human GCN5, which is in the same bromodomain family (IV) of BPTF allowing early stage assessment of selectivity. A previous study using this 3D-enriched fragment library to target the bromodomain-containing protein BRD4 showed that the inclusion of 3D-enriched fragments into screening decks may be useful in uncovering novel, selective chemical matter for bromodomain inhibitor development [11]. Here, we employ the same library to target two different bromodomains in order to gain a better understanding of the broad applicability of using 3D-enriched fragments for targeting bromodomains.
Bromodomains are epigenetic effector domains that recognize N-ε-acetylated-lysine (Kac) on the N-terminal tails of histone proteins. Widely considered highly druggable [30], bromodomains present a useful set of target proteins to validate this workflow. When dysregulated, bromodomain-containing proteins have been associated with diverse diseases. In particular, the bromodomain of BPTF has been implicated in several cancers including melanoma [31], breast cancer [32,33], and high grade gliomas [33]. While there are two commercially available chemical probes (TP-238, NVS-BPTF-1) and three literature-disclosed BPTF inhibitors [34,35,36,37,38], they all suffer from poor physiochemical properties or poor selectivity against the other 60 human bromodomains. Thus, there is a need for new chemical matter to develop selective inhibitors for BPTF.
Non-human bromodomains have been identified in parasites carrying infectious diseases, including malaria. Epigenetic proteins, including bromodomain-containing proteins, regulate gene expression and replication of the parasitic cells during the sexual and asexual stages [39,40]. Thus, small molecule-mediated disruption of parasitic bromodomain interactions with acetylated lysines on histones is a promising avenue for stopping the disease progression. Plasmodium falciparum (Pf), the most lethal protozoan malarial parasite, has up to eight predicted bromodomains, two of which have been characterized—PfGCN5 and PfBDP1 [41,42,43]. Prior research aimed at targeting these bromodomains has focused on repurposing human bromodomain inhibitors. Consequently, the four lead inhibitors from these studies have <10-fold selectivity for parasitic cells over human cells, limiting their potential as therapeutics [44,45,46]. Given the selectivity challenge for targeting the BPTF and PfGCN5 bromodomains, we rationalized that a dual-protein screen and the use of 3D-fragments would increase the odds of finding selective fragment leads in a time-efficient manner. To our knowledge, we report here the first fragment screens against BPTF and PfGCN5 and conclude our report discussing the time and resource advantages of using this new screening workflow and its applications towards additional bromodomains.
2. Results
2.1. Screening Workflow
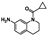
Screen preparation entails expression of unlabeled- and fluorine-labeled proteins and pooling a fragment library into mixtures. Fluorine incorporation is accomplished by metabolic labeling with fluorinated amino acids or amino acid precursors [27,47]. Aromatic amino acids are enriched at bromodomain binding sites and, as such, their fluorinated counterparts are good reporting resonances. Here, we use 5-fluorotryptophan (5FW)-labeled BPTF and PfGCN5 as both proteins have a tryptophan proximal to the native Kac binding site in a region termed the WPF shelf (Trp2824 and Trp1379, respectively, Figure 1). PfGCN5 has an additional tryptophan, Trp1454, distal to the binding site that gives additional surface coverage for binding site analyses and general information on protein behavior during the assay. Unlabeled proteins were used for the 1H CPMG NMR assay.

Figure 1.
Screening workflow. (Left) The location of each 5FW resonance on the proteins and in the 19F NMR spectra (PDB ID = 4QNS, 3UVW). (Middle) Representative binding experiments in which case one or both proteins resonances are affected. (Right) A representative 1H CPMG NMR experiment in which case a decrease in resonance intensity is observed in the ligand + protein vs the ligand alone spectra. To deconvolute the 1H CPMG NMR mixture binding experiments, the 1H NMR spectra of each individual ligand in that mixture are overlaid to identify the corresponding ligand resonances.
Previously, the replacement of W with 5FW in BPTF has been shown to impart only small structural perturbations and no effect on stability [36,48]. Similarly, circular-dichroism thermal-melt experiments show little structural perturbations and no difference in the protein stability from 5FW incorporation relative to the unlabeled PfGCN5 bromodomain (Tm PfGCN5/5FW-PfGCN5 = 57.1 °C, Figure S2–S5). 1H-15N HSQC NMR chemical shift comparative analysis of 1H-15N-5FW PfGCN5 and 1H-15N-PfGCN5 gave an average Δδ of 0.037 ± 0.024 ppm (see note in Figure S5) and a similar dispersion of resonances supporting a well-folded protein. Additionally, a PrOF NMR titration with bromosporine, a known high-affinity pan-bromodomain inhibitor, showed slow exchange kinetics of the WPF shelf resonance, indicating little perturbation of the fluorinated protein binding function (Figure S21). We further support a lack of significant perturbation from fluorination via comparing the affinity of a new fragment by PrOF NMR and 1H-15N HSQC NMR with the non-19F-labeled protein yielded less than a 2-fold difference in affinity. This analysis is described further in Section 2.3. The observation of only a modest perturbation to structure and function from fluorination is similar to our prior analyses of fluorinated bromodomains [25,49].
A 467-member 3D-enriched fragment library from Life Chemicals was used in this screen. In a previous study, we used this library to demonstrate that the use of 3D fragments is beneficial for bromodomain screening using the first bromodomain of BRD4 (BRD4 D1) [11]. The fragments were pooled into mixtures of 4–5 fragments to increase screening efficiency while minimizing spectral overlap in the 1H NMR spectra for CPMG NMR. We employ this library, and the same screening criteria here, to further understand the implications of 3D fragments for bromodomain screening.
The screening workflow consists of two dependent steps: 1) a PrOF NMR screen of the 5FW-labeled proteins and a cocktail of 4–5 fragments in a single NMR tube; 2) a 1H CPMG NMR binding assay to deconvolute the hit mixtures for each protein (Figure 1). In the first step, a dual-protein PrOF NMR spectrum was obtained in which 5FW-PfGCN5, 5FW-BPTF (~ 50 µM each) and mixtures of fragments (400 µM individual fragment) were present in the same NMR tube. Similar to a previous dual-protein PrOF NMR screen with BRD4 D1 and BPTF [48], the resonances for 5FW-PfGCN5 and 5FW-BPTF are resolved, allowing for monitoring protein-specific resonance perturbations (Figure 1). Chemical shift perturbations indicating a ligand binding event were based on a prior cutoff of Δδ ≥ 0.030 ppm as the definition for a hit mixture. This cutoff was based on a statistical analysis of chemical shift perturbations from a prior BRD4 D1 fragment screen [23] and are described further in the experimental section and in Table S2.
The second step is a 1H CPMG NMR deconvolution of the hit mixtures using the non-fluorine-labeled bromodomains. 1H CPMG NMR employs a T2 filter to take advantage of the difference in transverse relaxation time between protein and small molecules to filter out the bound-state ligand(s). Unlike the PrOF NMR assay, a single protein is assayed in each experiment. A binding event is characterized by a decrease in ligand resonance intensity from the ligand alone spectrum to the ligand + protein spectrum. A hit is defined as a decrease in resonance intensity ≥ 20%. The 1H CPMG NMR assay can be tuned by changing the protein or ligand concentration, filter length (Figure 1), or hit criteria depending on how stringent the desired hit rate is. The number of 1H CPMG NMR deconvolution experiments is directly informed from the first step. If a mixture from the first step was only a hit for one protein, a single 1H CPMG NMR deconvolution experiment is needed. If a mixture from the first step was a hit for both proteins, two separate 1H CPMG NMR deconvolutions experiments are needed to determine the fragments that bind to each protein.
2.2. Screening Results
In the first step, PrOF NMR gave a total of 49 out of 98 (50%) hit mixtures, with Δδ ranging from 0.030 to 0.46 ppm (Table S2). There was an even distribution of hit mixtures binding to BPTF or PfGCN5, and those that bind to both proteins (Table 1). Similarly, the 1H CPMG NMR second deconvolution step gave an even hit rate distribution of fragments that bind selectively to BPTF or PfGCN5. An analysis of the range of PrOF NMR Δδ and 1H CPMG NMR percent resonance decreases showed no major differences between the proteins, indicating a similar responsiveness to binding (Figure S6). Additionally, no obvious trends were observed between the magnitude of the PrOF NMR Δδ and the magnitude of the 1H CPMG NMR response or the number of hits per mixture (Figure S6).

Table 1.
Hit rate for both steps of the screening platform.
There was a 16% (8 mixtures) discrepancy between the steps of the assay where a hit mixture was indicated by PrOF NMR, but no hit fragment(s) was detected via the 1H CPMG NMR deconvolution experiment. A similar low disagreement (15%) between PrOF NMR and 1H CPMG NMR has been described by Urick et al. [25]. Six of eight mixtures leading to the discrepancies are likely due to the lenient cutoff used to define a PrOF NMR hit. These six mixtures had a Δδ < 0.050 ppm, indicating that these could be false positives. More surprising, the remaining two mixtures had a Δδ > 0.100 ppm but gave no 1H CPMG NMR hits. This result may indicate enhanced binding of fragments due to fluorination. An alternative hypothesis is that these mixtures contain high-affinity binders that show a limited signal response in the 1H CPMG NMR assay due to a lack of sufficient chemical exchange, a limitation of the 1H CPMG binding assay [50]. However, this effect was not pursued in this report.
Previously we used the same 3D fragment library in a screen targeting BRD4 D1 [11]. The 34 hits from that screen were also tested for selectivity against BPTF using PrOF NMR, giving 13 fragments that bound BPTF in addition to BRD4 D1. In cross-validation of our data, we identified 11 of these 13 fragment hits in this screen, an 85% agreement between the two screens for fragment hits binding to BPTF (Table S2). The low discrepancy observed between the PrOF NMR and 1H CPMG NMR assays in this screen and the reproducibility of the PrOF NMR data when compared with our prior screen, indicate that the PrOF NMR and 1H CPMG NMR assays are ideal for using in sequential order for a screening workflow.
2.3. Hit Follow Up
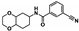
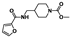
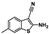
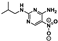
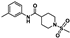
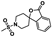
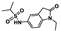
Nine fragment hits were chosen to be characterized—six that selectively bound PfGCN5 (1, 2, 3) or BPTF (4, 5, 6) and three that bound both proteins (7, 8, 9, Table 2). Fragments were selected to represent the broad range of PrOF NMR and 1H CPMG NMR magnitude of responses. Fragment 9 was included in the characterization because its hit mixture (#30) gave the highest and fourth largest Δδ (0.469 ppm, 0.169 ppm) for BPTF and PfGCN5, respectively. Our previous screen identified 9 as binding BRD4 D1 with BPTF as an off target [11]. The single-point PrOF NMR assay with BPTF from the previous screen gave a Δδ of 0.453 ppm (400 µM 9), similar to the Δδ observed with mixture #30, making 9 the most likely ligand from the fragment mixture. While a co-crystal structure of 9 with BPTF has been solved and a Kd for BPTF of 180 µM via PrOF NMR was determined [51], 9 has not been explored for targeting PfGCN5.

Table 2.
Follow up on select fragment hits.
PrOF NMR titrations of the selected fragments allowed for facile affinity determination. Hits that were selective for PfGCN5 all showed a dose dependence; however, only 2 reached saturation, allowing for Kd determination. BPTF selective compounds gave a similar result with 6 but not 5 reaching saturation. While 4 selectively bound weakly to BPTF in the initial PrOF NMR screen and had strong binding response in the 1H CPMG NMR deconvolution assay, it showed no binding during the validation dose-response PrOF NMR experiment. Three other hits were found in mixture #50 containing 4, indicating that this false positive could result from ligand–ligand interactions affecting the screening assays or fluorine effects precluding binding. As expected, 7 and 8 bind BPTF and PfGCN5, albeit with modest affinity. However, 6 is an attractive starting point for PfGCN5 inhibitor development because it binds PfGCN5 > 6-fold better than BPTF and was found previously to not bind BRD4 D1 (Table 2) [11]. Further, 9 is also an attractive target for PfGCN5 inhibitor development because it has a good affinity and high ligand efficiency (Kd = 16 µM, LE = 0.43) and binds PfGCN5 9-fold over BPTF. However, 9 also binds BRD4 D1 with a Kd of 50 µM [11].
2.4. Investigating a Possible Orthogonal Binding Mode to PfGCN5
The two tryptophans of PfGCN5 gave additional biophysical information in the PrOF NMR assay that supported 9, engaging a new low-affinity site on PfGCN5. Typically for the PrOF NMR assays with bromodomains, the resonance displaying the largest response to a ligand binding is the 5FW near the native Kac binding site. Using a W1379F 5FW PfGCN5 mutant, we assigned the downfield resonance in 5FW-PfGCN5 as Trp1379 located on the ZA loop proximal to the Kac binding site in the WPF shelf (Figure 2, Figure S22). The downfield resonance consistently has the largest magnitude of response when tested with ligands known to bind in the native bromodomain binding site. Trp1454 is on the C helix located ~ 22 Å (C5-C5) from Trp1379. The response of both resonances may indicate a conformational change in the protein upon binding or ligands accessing a second binding site near Trp1454. Although rare for bromodomains, recently an additional binding site has been discovered for the second bromodomain of BRD4 [52]. Titration of 9 with PfGCN5 showed significant responses of both 5FW resonances (Figure 2) and warranted additional exploration of a possible second binding site on PfGCN5. Four other mixtures (#32, #53, #51, and #94) had a PfGCN5 upfield Δδ > 0.030 ppm, indicating that other hits may behave like 9. To further explore the binding mode of 9 to PfGCN5, several additional experiments were performed as described below.
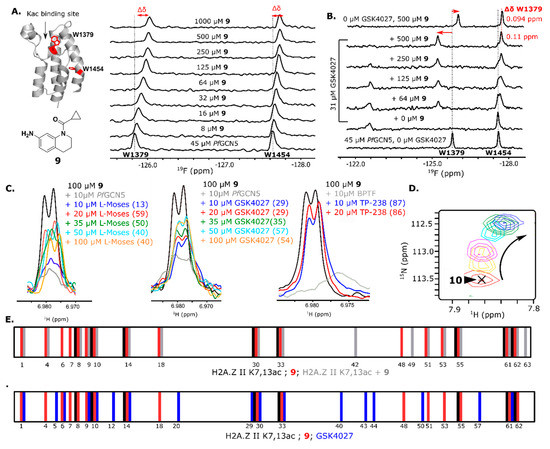
Figure 2.
(A) Fragment 9 and PfGCN5 (PDB 4QNS) with the W locations highlighted. PrOF NMR titration of 9 with PfGCN5. (B) PrOF NMR titration of 9 with GSK4027 present at a saturating concentration. (C) Competition 1H CPMG NMR. The percent recovery for the addition of each competitor is shown in parentheses. (D) 1H-15N HSQC NMR non-linear movement of resonance 10 when titrated with 9. The arrow indicates the direction of movement with increasing concentrations of 9 (0 µM = red, 16 µM = yellow, 32 µM = pink, 64 µM = teal, 125 µM = green, and 250 µM = blue). (E) Strip plots of PfGCN5 1H-15N HSQC NMR resonances with Δδ > x− + 0.60*s at the highest concentration tested (black = H2A.Z II K7,13ac, red = 9, blue = GSK4027, and gray = 9 + H2A.Z II K7,13ac).
The competition 1H CPMG NMR and PrOF NMR assays were employed to further probe the binding mode of 9 to PfGCN5. L-Moses, a potent PCAF/PfGCN5 inhibitor (Kd PfGCN5 = 280 nM [46]), GSK4027, a BPTF/GCN5/PCAF inhibitor [53], and TP-238, a CECR2/BPTF chemical probe [54], were used as competitive molecules. Crystal structures of L-Moses with PfGCN5 (PDB 5TPX), GSK4027 with GCN5 (PDB 5MLJ), and TP-238 with BPTF [51] show these compounds bind the native Kac site. An N-terminal tail peptide of a histone protein, H2A.Z II K7,13ac, was also employed as this peptide was previously shown to bind BPTF (Kd = 310 µM) [55] and also binds PfGCN5 (Kd = 670 µM, Figure S14). Although the affinity for PfGCN5 is unknown, a PrOF NMR titration confirmed that GSK4027 binds PfGCN5 strongly and only perturbs the downfield resonance (Figure S8). In 1H CPMG NMR competition experiments, L-Moses and GSK4027 were unable to compete with 9 and keep it from binding to PfGCN5 at low concentrations. At high concentrations, at or above the protein concentration, the competition maxed out at ~ 50% (Figure 2, Figure S10, Table S5). Conversely, TP-238 was able to compete 9 off from BPTF at low concentrations. This result supports an additional binding interaction with PfGCN5. In a different competition assay, PrOF NMR titrations that were used with a near stoichiometric concentration of GSK4027 while titrating in 9 showed several slow exchange resonances for both 5FW resonances (Figure 2). Interestingly, at high concentrations of 9 in the presence of GSK4027, the WPF shelf resonance, Trp1379, is at a new downfield position relative to the resonance without GSK4027 supporting an additional binding interaction that is stable in the presence of GSK4027 (Figure 2B).
To cross-validate our findings, 1H-15N HSQC NMR was used to further examine the binding mode of 9 to PfGCN5 in the presence and absence of known ligands. The 1H-15N HSQC spectrum of PfGCN5 is unassigned. For ease of spectral interpretation, all resonances that were distinctly visible were assigned as 1-64. Fragment 9, GSK4027, and the acetylated peptide H2A.Z II K7,13ac were titrated against PfGCN5. At the highest ligand concentration, resonances with Δδ > x− + 0.60*s, where x− is the mean and s is the standard deviation at the highest concentration, were defined as significant. The correction factor of 0.60 is to compensate for only 60% of the resonances being usable for analysis. Titration of 9 with PfGCN5 gave linear and non-linear dose-dependent chemical shift perturbations. Of the five resonances with non-linear movement, only one (10) was considered to have significant movement. Nine of the linear dose-dependent resonances were used to calculate a Kd average of 7.5 µM with a standard deviation of 7.0 µM (Figure S13). This affinity with the non-fluorine-labeled protein is similar but higher than the affinity determined by PrOF NMR. The resonances of 9, 30, 33, 55, and 61 are common amongst 9, GSK4027, and H2A.Z II K7,13ac, indicating that these are likely resonances affected by ligand binding the native Kac site. Both GSK4027 and 9 affect unique resonances (9 = 4, 7, 18, 48, 53, and 51; GSK4027 = 5, 12, 20, 40, 43, 44, 50, and 57). It is likely that the unique resonances affected are due to its higher affinity and more protein-specific contacts because GSK4027 does not perturb the upfield resonance in the PrOF NMR assay. For fragment 9, these resonances may be due to interactions outside the binding site. A competition experiment was performed where H2A.Z II K7,13ac was present at a concentration above the Kd (1 mM) and 9 was titrated. In this case, the resonances unique to 9 were affected as well as three additional resonances (42, 49, and 63), supporting a possible ternary complex.
The competition 1H CPMG NMR, PrOF NMR, and 1H-15N HSQC NMR data support an additional low-affinity binding interaction of 9 to PfGCN5. Finally, we used the in silico solvent docking program, FTMap, to identify any additional hot spot residues outside the native histone binding site [56]. This program has been used previously by Olp et al. to identify a new binding site on BRD4 D2 [52]. In this case, whereas several large solvent clusters were found in the histone binding site, only a single cluster of 12 solvent molecules was found near Trp1454. This result supports a potential binding site, but of low affinity (Figure S20). However, more biophysical data are needed to validate the hypothesis.
3. Discussion
3.1. 3D-Enriched Fragment Analysis
Due to drugs and chemical probes having a high degree of 3D character, 3D-enriched fragment libraries are increasingly being explored to generate starting chemical matter for inhibitor development [8,12,15,57,58,59,60]. The main concerns with using 3D-enriched fragments are the potential for lower hit rates due to increased complexity and hits that are biased away from fragments with 3D character limiting the utility of using these higher complexity molecules. Here, we sought to follow up on our previous study using 3D-enriched fragments to target BRD4 D1 [11]. In that screen, we observed a lower hit rate (10%) than a similar 2D-enriched screen (25%); however, the 3D character of the hits matched that of the overall library, indicating that both 2D and 3D fragments should be included in screening decks for increasing library diversity. In this screen, we observed a total hit rate for PfGCN5 and BPTF of 9.8% and 9.2%, respectively, essentially the same hit rate as in the previous screen, indicating a similar ligandability between the three bromodomains, although BRD4 D1 ligands tended to be higher in affinity.
Plane of best fit (PBF) is a common method to quantify the 3D character of a compound [61,62]. While a PBF > 0.25 has been used by others to define a 3D fragment [62], we use a more rigorous cutoff of PBF > 0.30 as being considered highly 3D. The average PBF for all hits (0.42), BPTF selective hits (0.30), and PfGCN5 selective hits (0.55) are above or equal to the 0.30 threshold (Figure 3A). Interestingly, PfGCN5 selective hits are more 3D than BPTF selective hits. The non-parametric Mann–Whitney–Wilcoxon statistical test was used to analyze the PBF distributions. The overall hits and PfGCN5 selective hits fall within the same PBF distribution of the library, indicating that the hits are not enriched for 2D or 3D character (Table S4). Despite having an average PBF above the 3D character threshold, the distribution of BPTF selective hits is statistically different from the overall library, indicating that these hits are biased towards flatter fragments relative to the content of the overall library. In agreement with our previous conclusion, these results support including both 2D- and 3D-enriched fragments in screening decks for capturing more diverse screening hits which may improve the selectivity of fragment hits over structurally related bromodomains.
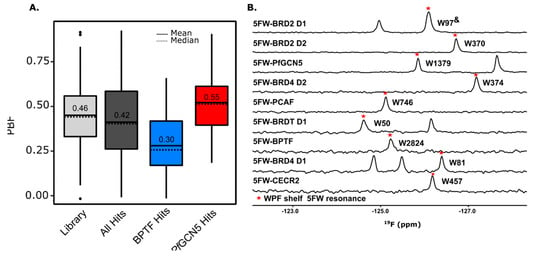
Figure 3.
(A) Boxplot of the PBF of the library, total hits, and hits selective for BPTF and PfGCN5. (B) PrOF NMR overlay of various 5FW-labeled bromodomains. The W in the WPF shelf near the binding site is denoted with a red star and label. & The 5FW resonances have not been assigned but this W is the one that shows the largest response with known ligands [55]. Also see Figure S9.
3.2. Initial Assessment of the Broader Applicability of the Screening Workflow
The dual-protein PrOF NMR, 1H CPMG NMR deconvolution screening workflow described herein can be broadly applicable to screening different targets. Although 19F is hypersensitive to its environment, making it a useful NMR active reporting nucleus, the main concern in screening two proteins simultaneously in PrOF NMR is the overlap of 19F resonances. We have screened pairs of proteins that are not members of the same bromodomain family (BRD4 D1 and BPTF [48]) and from different species (BPTF and PfGCN5) in the same NMR tube with resolved resonances. Importantly for gaining selectivity information at the early stages in development, bromodomains that are members of the same family (IV), CECR2 and PCAF, have resolved 5FW resonances and can be used in a dual-protein assay (Figure S9). Moreover, a survey of nine 5FW-labeled bromodomains (BRD4 D1, BRD4 D2, BRDT D1, BRD2 D1, BRD2 D2, CECR2, PCAF, PfGCN5, and BPTF) illustrates that 32 out of the 36 (89%) possible pair combinations of proteins have resolved resonances including different bromodomains on the same protein (Figure 3B, S9). If the desired target proteins have overlapping 5FW resonances, other 19F-labeled amino acids such as 3-fluorotyrosine, 4-fluorophenalanine, trifluoromethyl probes, or other isomers of fluorinated tryptophan (e.g., 6-fluorotryptophan) could be employed, as these resonances fall in a different region of the 19F NMR spectrum [63,64,65]. Additionally, if PrOF NMR cannot be used to screen multiple proteins at once due to overlapping resonances or perturbation of structure or function due to fluorinated amino acid incorporation, the RAMPED UP HSQC NMR [24] can be substituted for PrOF NMR in the protein-observed step of this workflow.
A concern with screening multiple proteins in the same solution is the possibility of non-specific protein–protein interactions that can influence ligand binding interactions. To explore this, we examined the chemical shift and line width at half height of the resonances for the three sets of proteins that have been tested in the same PrOF NMR experiment (BPTF/BRD4 D1, PfGCN5/BPTF, CECR2/PCAF). The average change in chemical shift and change in line width of the resonances when the proteins were tested individual or in pairs was 0.04 ppm (0.0–0.08 ppm) and 4.3 Hz (0.4–9.2 Hz), respectively (Table S5). If significant protein–protein interactions were occurring, we would expect a substantial change in chemical shift and line width of the resonances. Given the sensitivity of the 19F nucleus, low micromolar concentration of fluorinated protein can be used (25–50 μM), which may help to further mitigate non-specific interactions.
In comparison to similar screening methods, the dual-protein PrOF NMR screen and subsequent 1H CPMG NMR deconvolution is time and resource efficient while giving more biophysical information. There are several variations on the screening workflow described here to accomplish a fragment screen against two proteins—two separate 1H CPMG NMR screens, two separate PrOF NMR screens with PrOF NMR deconvolution experiments, or a dual-protein PrOF NMR screen with PrOF NMR deconvolution (Table 3). Conducting two separate 1H CPMG NMR screens is the most material efficient and the second most time efficient. However, all ligand-observed methods do not provide information on protein behavior during the assay or binding site location without additional competition experiments increasing time and material consumption. This added structural information is a general advantage of protein-observed NMR experiments. Because alternate PrOF NMR screening formats require multiple experiments for deconvolution of hit mixtures, this method is ~ 2.5-fold faster, uses 1.5–2-fold less ligand, and 1.3–1.8-fold less fluorine-labeled protein. In the case where a mixture only binds one protein in the PrOF NMR screen, the same NMR tube could be used for the 1H CPMG deconvolution, further reducing time and resource consumption.

Table 3.
Comparison of the resources used in different screening methods based on the results of this screen.
In silico screening could be incorporated into this workflow in order to increase the amount of chemical matter explored. Virtual screening has been used to support and conduct fragment screens [66,67]. In particular, virtual fragment screens have been conducted against various bromodomains including BAZ2A [68], BET family bromodomains [69,70], and CREBBP [71]. In this workflow, large fragment libraries could be first screened against the two proteins of interests filtering the libraries into smaller libraries. The initial in silico hit libraries can then be subjected to this workflow.
4. Materials and Methods
4.1. Materials
The 3D-enriched fragment library was purchased from Life Chemicals and divided into mixtures of 4–5 fragments as previously described [11]. Amino acids, uracil, thiamine-HCl, LB broth, biotin, and nicotinic acid were purchased from RPI Corp. The 5-fluoroindole, magnesium sulfate, succinic acid, calcium chloride, Iron(III) chloride, 15N ammonium chloride, L-Moses, and imidazole were purchased from Millipore Sigma. Potassium diphosphate, potassium monophosphate, sodium phosphate, manganese (II) chloride, zinc(I) chloride, and sodium chloride were purchased from Fischer Scientific. Cobalt (II) chloride, Ethylenediaminetetraacetic acid, and copper (II) chloride was purchased from Acros Organics. Boric acid was purchased from Mallinckrodt. Deuterium oxide and dimethyl sulfoxide-d6 were purchased from Cambridge Isotope Laboratories. GSK4027 was purchased from Cayman Chemicals and TP238 was synthesized previously [55].
4.2. Protein Expression and Purification
5FW-labeled proteins. The 5FW-PfGCN5-his6 and 5FW-BPTF-his6 were expressed as previously described [36]. The protein expression plasmid for expressing W1379F 5F2-PfGCN5-his6 was purchased from Genesrcript.
Unlabeled PfGCN5 and BPTF. Unlabeled PfGCN5 and BPTF were expressed as follows. The pNIC28-BSA4 (kanomycin resistance) encoding BPTF and pET15-MHL (ampicillin resistance) encoding PfGCN5 were gifts from Stefan Knapp (Nuffield Department of Medicine, University of Oxford) and Ray Hui (University of Toronto), respectively. E.coli strain BL21(DE3) was transformed with the plasmid of the desired protein sequence and plated onto an agar plates containing the respective antibiotics. The plated cells were incubated at 37 °C overnight. A single colony was selected from the agar plate and inoculated in 5 mL LB media and the appropriate antibiotic (100 mg /L). The primary culture was grown overnight at 25 °C with shaking at 250 RPM. The primary culture was transferred into 1 L of LB containing the appropriate antibiotic (100 mg/L). The secondary culture was allowed to grow at 37 °C with shaking at 250 RPM until the optical density at 600 nm reached 0.6–0.8. The temperature was reduced to 20 °C for 30 min before inducing protein expression with 1 mM isopropyl β-D-1-thiogalactopyranoside. Cells were harvested after 12–16 h.
15N and 15N, 5FW-labeled PfGCN5. Following the procedure for unlabeled PfGCN5, when the secondary reached an optical density at 600 nm of 0.6–0.8, cells were harvested via centrifugation and resuspended in minimal media (33.7 mM disodium phosphate, 22.0 mM potassium phosphate, 8.55 mM sodium chloride, and 1g/L 15N ammonium chloride) with trace elements (134 µM ethylenediaminetetraacetic acid, 31 µM iron(III) chloride, 6.2 µM zinc(III) chloride, 0.72 µM copper(II) chloride, 0.42 µM cobalt(II) chloride, 1.62 µM boric acid, and 0.081 µM manganese(II) chloride), magnesium chloride (1 mM), calcium chloride (0.3 mM), biotin (1 µg/L), thiamine (µg/L), and glucose (0.4% w/v). For 15N/5FW labeling, 100 mg/L of 5-fluoroindole was added. Cells were allowed to equilibrate at 37 °C, 250 RPM for 1 h. The temperature was reduced to 20 °C for 30 min before protein induction with 1 mM isopropyl β-D-1-thiogalactopyranosid. Cells were harvested after 12–16 h.
Protein Purification and Characterization. Protein purification was accomplished as previously described. Quadrupole time-of-flight (Q-TOF) LC/MS was used to confirm the identity of the protein and determine percent fluorine incorporation. The accuracy of the instrument is 0.001% or an error in mass of ±0.001% (±1.5 Da for a 15 kDa protein). Percent fluorine incorporation was calculated using Equation (1), where 1 F is the peak intensity for the mass corresponding to the incorporation of 1 fluorine, etc.
Calculated and observed masses for 5FW-PfGCN5his6, 5FW-BPTFhis6, unlabeled PfGCN5 and unlabeled BPTF: 16922 and 16921 kDa (85–95% 19F incorporation); 14719 and 14720 kDa (85–95% 19F incorporation); 14740 and 14739 kDa; and 16904 and 16092 kDa, respectively For 15N, 5FW PfGCN5 mass spectral analysis proved difficult for accurate percent labeling determination, but PrOF NMR shows moderate incorporation (Figure S3) and the lack of Trp peaks in the 1H-15N HSQC spectrum (Figure S4) indicate that it is sufficient for comparing the secondary structure of unlabeled of 5FW-labeled PfGCN5.
4.3. NMR Methods
PrOF NMR. PrOF NMR spectra were obtained on a Bruker Avance III HD 500 with the 5 mm Prodigy TCI inverse cryoprobe (19F S:N 2000:1). All PrOF NMR experiments had 5% (v/v) D2O, 0.42% trifluoroacetic acid (TFA), and the desired fragment(s) dissolved in DMSO. The DMSO concentration for all experiments was kept constant at 1%. TFA (−76.55 ppm) was used to reference the spectra. For the PrOF NMR screen, 5FW-BPTF and 5FW-PfGCN5 (35–40 µM in 50 mM phosphate, 100 mM NaCl, pH 7.4) were tested with mixtures of 4–5 fragments, where each individual fragment was 400 µM. Parameters values were: relaxation delay = 0.7 s, acquisition time = 0.05 s, sweep width = 10, and transmitter frequency offset = −125 (for binding experiment) and −75 (for TFA reference experiment). A 10 Hz line broadening was applied. Titrations were fit to Equation (2), where p is the protein concentration and L is the ligand concentration.
A PrOF NMR binding event is characterized as a change in chemical shift (Δδ) from the protein-only spectrum to the protein + fragment mixture spectrum. We used a prior cutoff of Δδ ≥ 0.030 ppm as the definition of a hit mixture, based on a statistical analysis of chemical shift perturbations from a prior BRD4 D1 fragment screen [22]. This cutoff was determined to be reasonable given the average Δδ of the non-WPF shelf resonance of 0.017 ppm for PfGCN5 observed in this screen. Although the binding site residues are more significantly perturbed, the average Δδ of the WPF shelf resonance was close to 0.030 ppm (BPTF = 0.049 ppm, PfGCN5 = 0.043 ppm, Table S2).
1H CPMG NMR. 1H CPMG NMR screen data were acquired on Bruker Avance III HD 500 with the 5 mm Prodigy TCI cryoprobe inverse cryoprobe (1H 2500:1 S/N). Competition 1H CPMG NMR data were acquired on Bruker 700 MHz Avance with a CryoProbe 5 mm TXI (1H S:N with H2O suppression (2mM sucrose) - 900:1) and excitation sculpting water suppression. Proteins were present at 10 µM in 50 mM phosphate, 100 mM NaCl, pH 7.4 in 100% D2O. In separate NMR tubes, a 1H CPMG NMR pulse was applied to a 1) mixture of ligands (individual ligands present at 100 µM for the screen and 50 µM for competition experiments) and 2) ligands + protein. For competition experiments, the ligands were used individually, and the competitor ligands were spiked into tube 2. 1H CPMG NMR parameters for the screen: NS = 128, acquisition time = 2 s, τ = 2.5 ms, and L = 120. For the competition experiments, τ was increased to 4.0 ms.
1H-15N SOFAST HSQC.1H-15N SOFAST HSQC spectra were obtained on a Bruker 600 MHz Avance NEO with a CryoProbe 5 mm TCI (1H, 13C, 15N, 2H) w/ Z-gradient using Bruker’s IBS_SOFAST.x method with 32 scans, 0.05 s acquisition time, and 0.2 s delay. Data were acquired at 3015.15 K and 50–52 µM protein in 50 mM phosphate, 100 mM NaCl, pH 7.4 and 5% (v/v) D2O. Data were processed with nmrDraw, nmrPipe, and NFRAM Sparky. Δδ1H-15N was calculated using Equation (3).
4.4. Peptide Synthesis
The H2A.Z II K7ac, K13ac peptide was synthesized using standard N-9- fluorenylmethoxycarbonyl (Fmoc) solid-phase synthesis methods on NovaSyn TGR resin (Novabiochem, 0.25 mmol/g) using a Liberty Blue automated microwave synthesizer and N,N- diisopropylcarbodiimide (DIC) and Oxyma for amino acid activation. The peptide was cleaved from the solid support in a mixture of 95/2.5/2.5 trifluoroacetic acid (TFA)/triisopropylsilane/water for 2–5 h followed by evaporation of solvent under a nitrogen stream. The crude peptide was precipitated into cold diethyl ether and purified by reverse-phase HPLC on a C-18 column using 0.1% TFA water and CH3CN as solvents (4–24% CH 3CN gradient over 30 min). Peptide molecular weight was confirmed using an Ab-Sciex 5800 matrix assisted laser desorption ionization (MALDI) time-of-flight mass spectrometer. Peptide theoretical and observed masse and analytical HPLC trace can be found in Figure S7.
4.5. Docking
Open-source FTMap [72] was used to probe possible binding sites on PfGCN5 (PDB ID = 4QNS).
4.6. Circular Dichroism
A Peltier-equipped temperature-controlled Jasco J-815 spectropolarimeter was used to probe the secondary structure of 5FW and unlabeled PfGCN5 by scanning the far-UV range (200–260 nm) at 25 °C. For measurements, 10.4 µM unlabeled PfGCN5 and 8.3 µM 5FW-PfGCN5 (8.8 mM NaCl, 3.3 mM phosphate at pH 7.4) at a pathlength of 1 mm were used. Spectra were collected at a rate of 50 nm/min over–260 nm. Thermal melting temperatures were determined by monitoring the ellipticity at 222 nm though a temperature scan from 20 to 95 °C (60 °C/h).
4.7. FW PfGCN5 Resonance Assignment
The W1379F PfGCN5 mutant was purchased from GenScript. The W1379F 5FW-PfGCN5 mutant was expressed using the method described above.
5. Conclusions
We presented a time- and resource-efficient screening workflow that uses PrOF NMR followed by 1H CPMG NMR deconvolution to screen mixtures of ligand against two proteins, completing two screens simultaneously. To demonstrate the applicability of this workflow, we completed the first reported fragment screens against the bromodomains of BPTF and PfGCN5 using a library of 3D-enriched fragments. Moderate hit rates were observed for both proteins and hit follow up of a select set led to molecules with moderate selectivity in some cases and, in the case of 9, a potential new low-affinity binding site on PfGCN5. This workflow is broadly applicable to different protein targets and could serve to gain selectivity knowledge at the onset of inhibitor development.
Supplementary Materials
The following are available online at https://www.mdpi.com/1420-3049/25/17/3949/s1, Table S1. List of fragments in each mixture purchased from Life Chemicals; Table S2. PrOF NMR results from part one of the screen; Table S3. Screen hits; Table S4. PBF comparison; Table S5. Competition 1H CPMG NMR of 9; Figure S1. PMI plots; Figure S2. Circular dichroism and thermal melts of 5FW- and unlabeled PfGCN5; Figure S3. Overlay of 19F spectra of 5FW-PfGCN5 and 15N,5FW-PfGCN5; Figure S4. 1H-15N HSQC comparison of 5FW and unlabeled PfGCN5; Figure S5. Δδ comparison of 15N,5FW PfGCN5 and 15N PfGCN5; Figure S6. Analysis of the two steps of the screening platform; Figure S7. HPLC trace and MALDI mass spectrometry data for H2A.ZII K7,13ac; Figure S8. Titration of GSK4027 with PfGCN5; Figure S9. Stacked spectra of various 5FW-labeled bromodomains; Figure S10. Competition 1H CPMG NMR of 9 raw data; Figure S11. 1H-15N HSQC titration of 9 with PfGCN5; Figure S12. Δδ for each residue for the 1H-15N HSQC titration of 9 with PfGCN5; Figure S13. Titration isotherms for the 1H-15N HSQC titration of 9 with PfGCN5; Figure S14. 1H-15N HSQC titration of H2A.Z II K7,13ac with PfGCN5; Figure S15. Δδ for each residue for the 1H-15N HSQC titration of H2A.Z II K7,13ac with PfGCN5; Figure S16. 1H-15N HSQC titration of 9 with a saturating concentration of H2A.Z II K7,13ac with PfGCN5; Figure S17. Δδ for each residue for the 1H-15N HSQC titration of 9 with a saturating concentration of H2A.Z II K7,13ac with PfGCN5; Figure S18. 1H-15N HSQC titration of GSK4207 with PfGCN5; Figure S19. Δδ for each residue for the 1H-15N HSQC titration of GSK4027 with PfGCN5; Figure S20. FT map of PfGCN5; Figure S21. PrOF NMR of5FW PfGCN5 with bromosporine; Figure S22. PrOF NMR of W1379F 5FW-PfGCN5; Table S6. Chemical shifts and resonance width of 5FW-labeled bromodomain when tested alone or in the presence of another 5FW-labeled bromodomain. PrOF NMR titrations of 1–9.
Author Contributions
Conceptualization, J.A.J. and W.C.K.P.; methodology, J.A.J., N.M.O., M.J.T., and S.K.B; validation, J.A.J., N.M.O. and S.K.B.; formal analysis, J.A.J.; investigation, J.A.J.; resources, J.A.J., N.M.O., and S.K.B.; data curation, J.A.J.; writing—original draft preparation, J.A.J.; writing—review and editing, J.A.J., N.M.O., S.K.B., and W.C.K.P.; supervision, W.C.K.P.; project administration, W.C.K.P.; funding acquisition, S.K.B. and W.C.K.P. All authors have read and agreed to the published version of the manuscript.
Funding
Research reported in this publication was supported by National Institute of General Medical Sciences Grant R01GM121414, National Science Foundation RUI1806228, and CHE-1904071. J.A.J. and N.M.O. were supported by National Institutes of Health Biotechnology Training Grants 5T32GM008347-27 and 5T32GM008347-29, respectively.
Acknowledgments
The authors would like to acknowledge the Minnesota NMR Center and Michelle Miller for her help in acquiring 1H-15N HSQC NMR data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tap, W.D.; Wainberg, Z.A.; Anthony, S.P.; Ibrahim, P.N.; Zhang, C.; Healey, J.H.; Chmielowski, B.; Staddon, A.P.; Cohn, A.L.; Shapiro, G.I.; et al. Structure-Guided Blockade of CSF1R Kinase in Tenosynovial Giant-Cell Tumor. New Engl. J. Med. 2015, 373, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.W.; Newell, D.R.; Angibaud, P. A successful collaboration between academia, biotech and pharma led to discovery of erdafitinib, a selective FGFR inhibitor recently approved by the FDA. MedChemComm 2019, 10, 1509–1511. [Google Scholar] [CrossRef]
- Souers, A.J.; Leverson, J.D.; Boghaert, E.R.; Ackler, S.L.; Catron, N.D.; Chen, J.; Dayton, B.D.; Ding, H.; Enschede, S.H.; Fairbrother, W.J.; et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013, 19, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.; Lee, J.T.; Wang, W.; Zhang, J.; Cho, H.; Mamo, S.; Bremer, R.; Gillette, S.; Kong, J.; Haass, N.K.; et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl. Acad. Sci. USA 2008, 105, 3041–3046. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.J.; Mortenson, P.N.; Murray, C.W. Efficient exploration of chemical space by fragment-based screening. Prog. Biophys. Mol. Biol. 2014, 116, 82–91. [Google Scholar] [CrossRef]
- Di Micco, S.; Vitale, R.; Pellecchia, M.; Rega, M.F.; Riva, R.; Basso, A.; Bifulco, G. Identification of Lead Compounds As Antagonists of Protein Bcl-xL with a Diversity-Oriented Multidisciplinary Approach. J. Med. Chem. 2009, 52, 7856–7867. [Google Scholar] [CrossRef]
- Foley, D.J.; Craven, P.G.E.; Collins, P.M.; Doveston, R.G.; Aimon, A.; Talon, R.; Churcher, I.; von Delft, F.; Marsden, S.P.; Nelson, A. Synthesis and Demonstration of the Biological Relevance of sp3-rich Scaffolds Distantly Related to Natural Product Frameworks. Chem. Eur. J. 2017, 23, 15227–15232. [Google Scholar] [CrossRef]
- Grädler, U.; Schwarz, D.; Blaesse, M.; Leuthner, B.; Johnson, T.L.; Bernard, F.; Jiang, X.; Marx, A.; Gilardone, M.; Lemoine, H.; et al. Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies. Bioorg. Med. Chem. Lett. 2019, 29, 126717. [Google Scholar] [CrossRef]
- Vu, H.; Pedro, L.; Mak, T.; McCormick, B.; Rowley, J.; Liu, M.; Di Capua, A.; Williams-Noonan, B.; Pham, N.B.; Pouwer, R.; et al. Fragment-Based Screening of a Natural Product Library against 62 Potential Malaria Drug Targets Employing Native Mass Spectrometry. Acs Infect. Dis. 2018, 4, 431–444. [Google Scholar] [CrossRef]
- Zhang, R.; McIntyre, P.J.; Collins, P.M.; Foley, D.J.; Arter, C.; vonDelft, F.; Bayliss, R.; Warriner, S.; Nelson, A. Construction of a Shape-Diverse Fragment Set: Design, Synthesis and Screen against Aurora-A Kinase. Chem. Euro. J. 2019, 25, 6831–6839. [Google Scholar] [CrossRef]
- Johnson, J.A.; Nicolaou, C.A.; Kirberger, S.E.; Pandey, A.K.; Hu, H.; Pomerantz, W.C.K. Evaluating the Advantages of Using 3D-Enriched Fragments for Targeting BET Bromodomains. Acs Med. Chem. Lett. 2019, 10, 1648–1654. [Google Scholar] [CrossRef] [PubMed]
- Morley, A.D.; Pugliese, A.; Birchall, K.; Bower, J.; Brennan, P.; Brown, N.; Chapman, T.; Drysdale, M.; Gilbert, I.H.; Hoelder, S.; et al. Fragment-based hit identification: Thinking in 3D. Drug Discov. Today 2013, 18, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Clemons, P.A.; Bodycombe, N.E.; Carrinski, H.A.; Wilson, J.A.; Shamji, A.F.; Wagner, B.K.; Koehler, A.N.; Schreiber, S.L. Small molecules of different origins have distinct distributions of structural complexity that correlate with protein-binding profiles. Proc. Natl. Acad. Sci. USA 2010, 107, 18787–18792. [Google Scholar] [CrossRef]
- Dobson, C.M. Chemical space and biology. Nature 2004, 432, 824. [Google Scholar] [CrossRef] [PubMed]
- Lovering, F.; Bikker, J.; Humblet, C. Escape from Flatland: Increasing Saturation as an Approach to Improving Clinical Success. J. Med. Chem. 2009, 52, 6752–6756. [Google Scholar] [CrossRef]
- Sauer, W.H.B.; Schwarz, M.K. Molecular Shape Diversity of Combinatorial Libraries: A Prerequisite for Broad Bioactivity. J. Chem. Inf. Comput. Sci. 2003, 43, 987–1003. [Google Scholar] [CrossRef]
- Schuffenhauer, A.; Brown, N.; Selzer, P.; Ertl, P.; Jacoby, E. Relationships between Molecular Complexity, Biological Activity, and Structural Diversity. J. Chem. Inf. Model. 2006, 46, 525–535. [Google Scholar] [CrossRef]
- Erlanson, D. Poll results: Affiliation and fragment-finding methods in 2019. Practicle Fragments. Erlanson, D., Ed.; 2019. Available online: https://practicalfragments.blogspot.com/ (accessed on 25 July 2020).
- Keserű, G.M.; Erlanson, D.A.; Ferenczy, G.G.; Hann, M.M.; Murray, C.W.; Pickett, S.D. Design Principles for Fragment Libraries: Maximizing the Value of Learnings from Pharma Fragment-Based Drug Discovery (FBDD) Programs for Use in Academia. J. Med. Chem. 2016, 59, 8189–8206. [Google Scholar] [CrossRef]
- Harner, M.J.; Chauder, B.A.; Phan, J.; Fesik, S.W. Fragment-Based Screening of the Bromodomain of ATAD2. J. Med. Chem. 2014, 57, 9687–9692. [Google Scholar] [CrossRef]
- Mashalidis, E.H.; Śledź, P.; Lang, S.; Abell, C. A three-stage biophysical screening cascade for fragment-based drug discovery. Nat. Protoc. 2013, 8, 2309–2324. [Google Scholar] [CrossRef]
- Norton, R.S.; Leung, E.W.W.; Chandrashekaran, I.R.; MacRaild, C.A. Applications of (19)F-NMR in Fragment-Based Drug Discovery. Molecules 2016, 21, 860. [Google Scholar] [CrossRef] [PubMed]
- Urick, A.K.; Hawk, L.M.L.; Cassel, M.K.; Mishra, N.K.; Liu, S.; Adhikari, N.; Zhang, W.; dos Santos, C.O.; Hall, J.L.; Pomerantz, W.C.K. Dual Screening of BPTF and Brd4 Using Protein-Observed Fluorine NMR Uncovers New Bromodomain Probe Molecules. Acs Chem. Biol. 2015, 10, 2246–2256. [Google Scholar] [CrossRef] [PubMed]
- Zartler, E.R.; Hanson, J.; Jones, B.E.; Kline, A.D.; Martin, G.; Mo, H.; Shapiro, M.J.; Wang, R.; Wu, H.; Yan, J. RAMPED-UP NMR: Multiplexed NMR-Based Screening for Drug Discovery. J. Am. Chem. Soc. 2003, 125, 10941–10946. [Google Scholar] [CrossRef]
- Urick, A.K.; Calle Jiménez, L.P.; Espinosa, J.F.; Hu, H.; Pomerantz, W.C.K. Protein-Observed Fluorine NMR is a Complementary Ligand Discovery Method to 1H CPMG Ligand-Observed NMR. Acs Chem. Biol. 2016, 11, 3154–3164. [Google Scholar] [CrossRef] [PubMed]
- Stadmiller, S.S.; Aguilar, J.S.; Waudby, C.A.; Pielak, G.J. Rapid Quantification of Protein-Ligand Binding via (19)F NMR Lineshape Analysis. Biophys. J. 2020, 118, 2537–2548. [Google Scholar] [CrossRef] [PubMed]
- Arntson, K.E.; Pomerantz, W.C.K. Protein-Observed Fluorine NMR: A Bioorthogonal Approach for Small Molecule Discovery. J. Med. Chem. 2016, 59, 5158–5171. [Google Scholar] [CrossRef] [PubMed]
- Evanics, F.; Kitevski, J.L.; Bezsonova, I.; Forman-Kay, J.; Prosser, R.S. 19F NMR studies of solvent exposure and peptide binding to an SH3 domain. Biochim. Biophys. Acta. (Bba)-Gen. Subj. 2007, 1770, 221–230. [Google Scholar] [CrossRef]
- Boeszoermenyi, A.; Chhabra, S.; Dubey, A.; Radeva, D.L.; Burdzhiev, N.T.; Chanev, C.D.; Petrov, O.I.; Gelev, V.M.; Zhang, M.; Anklin, C.; et al. Aromatic 19F-13C TROSY: A background-free approach to probe biomolecular structure, function, and dynamics. Nat. Methods 2019, 16, 333–340. [Google Scholar] [CrossRef]
- Vidler, L.R.; Brown, N.; Knapp, S.; Hoelder, S. Druggability Analysis and Structural Classification of Bromodomain Acetyl-lysine Binding Sites. J. Med. Chem. 2012, 55, 7346–7359. [Google Scholar] [CrossRef]
- Dar, A.A.; Nosrati, M.; Bezrookove, V.; de Semir, D.; Majid, S.; Thummala, S.; Sun, V.; Tong, S.; Leong, S.P.L.; Minor, D.; et al. The Role of BPTF in Melanoma Progression and in Response to BRAF-Targeted Therapy. J. Natl. Cancer Ins. 2015, 107, djv034. [Google Scholar] [CrossRef]
- Landry, J.; Sharov, A.A.; Piao, Y.; Sharova, L.V.; Xiao, H.; Southon, E.; Matta, J.; Tessarollo, L.; Zhang, Y.E.; Ko, M.S.H.; et al. Essential Role of Chromatin Remodeling Protein Bptf in Early Mouse Embryos and Embryonic Stem Cells. Plos Genet. 2008, 4, e1000241. [Google Scholar] [CrossRef]
- Frey, W.D.; Chaudhry, A.; Slepicka, P.F.; Ouellette, A.M.; Kirberger, S.E.; Pomerantz, W.C.K.; Hannon, G.J.; dos Santos, C.O. BPTF Maintains Chromatin Accessibility and the Self-Renewal Capacity of Mammary Gland Stem Cells. Stem Cell Rep. 2017, 9, 23–31. [Google Scholar] [CrossRef] [PubMed]
- TP-238 A chemical probe for CECR2/BPTF bromodomains. Available online: https://www.thesgc.org/chemical-probes/TP-238 (accessed on 5 September 2018).
- NVS-BPTF-1 A chemical probe for BPTF. Available online: https://www.thesgc.org/chemical-probes/NVS-BPTF-1 (accessed on 19 February 2019).
- Kirberger, S.E.; Ycas, P.D.; Johnson, J.A.; Chen, C.; Ciccone, M.F.; Woo, R.W.L.; Urick, A.K.; Zahid, H.; Shi, K.; Aihara, H.; et al. Selectivity, ligand deconstruction, and cellular activity analysis of a BPTF bromodomain inhibitor. Org. Biomol. Chem. 2019, 17, 2020–2027. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, Q.; Leung, E.L.H.; Li, Y.; Fan, X.; Wu, Q.; Yao, X.; Liu, L. Compound C620-0696, a new potent inhibitor targeting BPTF, the chromatin-remodeling factor in non-small-cell lung cancer. Front. Med. 2019, 14, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Han, J.; Lu, W.; Lian, F.; Wang, J.; Lu, T.; Tao, H.; Xiao, S.; Zhang, F.; Liu, Y.-C.; et al. Discovery of alkoxy benzamide derivatives as novel BPTF bromodomain inhibitors via structure-based virtual screening. Bioorg. Chem. 2019, 86, 494–500. [Google Scholar] [CrossRef]
- Saraf, A.; Cervantes, S.; Bunnik, E.M.; Ponts, N.; Sardiu, M.E.; Chung, D.-W.D.; Prudhomme, J.; Varberg, J.M.; Wen, Z.; Washburn, M.P.; et al. Dynamic and Combinatorial Landscape of Histone Modifications during the Intraerythrocytic Developmental Cycle of the Malaria Parasite. J. Proteome Res. 2016, 15, 2787–2801. [Google Scholar] [CrossRef] [PubMed]
- Trelle, M.B.; Salcedo-Amaya, A.M.; Cohen, A.M.; Stunnenberg, H.G.; Jensen, O.N. Global Histone Analysis by Mass Spectrometry Reveals a High Content of Acetylated Lysine Residues in the Malaria Parasite Plasmodium falciparum. J. Proteome Res 2009, 8, 3439–3450. [Google Scholar] [CrossRef] [PubMed]
- Josling Gabrielle, A.; Petter, M.; Oehring Sophie, C.; Gupta Archna, P.; Dietz, O.; Wilson Danny, W.; Schubert, T.; Längst, G.; Gilson Paul, R.; Crabb Brendan, S.; et al. A Plasmodium Falciparum Bromodomain Protein Regulates Invasion Gene Expression. Cell Host Microbe. 2015, 17, 741–751. [Google Scholar] [CrossRef]
- Fan, Q.; An, L.; Cui, L. Plasmodium falciparum Histone Acetyltransferase, a Yeast GCN5 Homologue Involved in Chromatin Remodeling. Eukaryot. Cell 2004, 3, 264–276. [Google Scholar] [CrossRef]
- Jeffers, V.; Yang, C.; Huang, S.; Sullivan, W.J. Bromodomains in Protozoan Parasites: Evolution, Function, and Opportunities for Drug Development. Microbiol. Mol. Biol. Rev. 2017, 81. [Google Scholar] [CrossRef]
- Miao, J.; Fan, Q.; Cui, L.; Li, J.; Li, J.; Cui, L. The malaria parasite Plasmodium falciparum histones: Organization, expression, and acetylation. Gene. 2006, 369, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Chua, M.J.; Robaa, D.; Skinner-Adams, T.S.; Sippl, W.; Andrews, K.T. Activity of bromodomain protein inhibitors/binders against asexual-stage Plasmodium falciparum parasites. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Moustakim, M.; Clark, P.G.K.; Trulli, L.; Fuentes de Arriba, A.L.; Ehebauer, M.T.; Chaikuad, A.; Murphy, E.J.; Mendez-Johnson, J.; Daniels, D.; Hou, C.-F.D.; et al. Discovery of a PCAF Bromodomain Chemical Probe. Angew. Chem. Int. Ed. 2017, 56, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Gee, C.T.; Arntson, K.E.; Urick, A.K.; Mishra, N.K.; Hawk, L.M.; Wisniewski, A.J.; Pomerantz, W.C. Protein-Observed (19)F-NMR for Fragment Screening, Affinity Quantification and Druggability Assessment. Nat. Protc. 2016, 11, 1414–1427. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.K.; Urick, A.K.; Ember, S.W.J.; Schönbrunn, E.; Pomerantz, W.C. Dual Screening of BPTF and Brd4 Using Protein-Observed Fluorine NMR Uncovers New Bromodomain Probe Molecules. Acs Chem. Biol. 2014, 9, 2755–2760. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.K.; Urick, A.K.; Ember, S.W.J.; Schönbrunn, E.; Pomerantz, W.C. Fluorinated Aromatic Amino Acids Are Sensitive 19F NMR Probes for Bromodomain-Ligand Interactions. Acs Chem. Biol. 2014, 9, 2755–2760. [Google Scholar] [CrossRef] [PubMed]
- Gossert, A.D.; Jahnke, W. NMR in drug discovery: A practical guide to identification and validation of ligands interacting with biological macromolecules. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 97, 82–125. [Google Scholar] [CrossRef]
- Ycas, P.D.; Zahid, H.; Chan, A.; Olson, N.M.; Johnson, J.A.; Talluri, S.K.; Schonbrunn, E.; Pomerantz, W.C.K. New inhibitors for the BPTF bromodomain enabled by structural biology and biophysical assay. Org. Biomol. Chem. 2020, 18, 5174–5182. [Google Scholar] [CrossRef]
- Olp, M.D.; Sprague, D.J.; Goetz, C.J.; Kathman, S.G.; Wynia-Smith, S.L.; Shishodia, S.; Summers, S.B.; Xu, Z.; Statsyuk, A.V.; Smith, B.C. Covalent-Fragment Screening of BRD4 Identifies a Ligandable Site Orthogonal to the Acetyl-Lysine Binding Sites. Acs Chem. Biol. 2020, 15, 1036–1049. [Google Scholar] [CrossRef]
- Humphreys, P.G.; Bamborough, P.; Chung, C.W.; Craggs, P.D.; Gordon, L.; Grandi, P.; Hayhow, T.G.; Hussain, J.; Jones, K.L.; Lindon, M.; et al. Discovery of a Potent, Cell Penetrant, and Selective p300/CBP-Associated Factor (PCAF)/General Control Nonderepressible 5 (GCN5) Bromodomain Chemical Probe. J. Med. Chem. 2017, 60, 695–709. [Google Scholar] [CrossRef]
- Structural Genomics Consortium. Available online: https://www.thesgc.org/chemical-probes/TP-238 (accessed on 15 May 2004).
- Olson, N.M.; Kroc, S.; Johnson, J.A.; Zahid, H.; Ycas, P.D.; Chan, A.; Kimbrough, J.R.; Kalra, P.; Schönbrunn, E.; Pomerantz, W.C.K. NMR Analyses of Acetylated H2A.Z Isoforms Identify Differential Binding Interactions with the Bromodomain of the NURF Nucleosome Remodeling Complex. Biochemistry 2020, 59, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.; Morcos, F.; Cheng, R.R.; Jiang, H.; Onuchic, J.N. Elucidating the druggable interface of protein−protein interactions using fragment docking and coevolutionary analysis. Proc. Natl. Acad. Sci. USA 2016, 113, E8051–E8058. [Google Scholar] [CrossRef] [PubMed]
- Feher, M.; Schmidt, J.M. Property Distributions: Differences between Drugs, Natural Products, and Molecules from Combinatorial Chemistry. J. Chem. Inf. Comput. Sci. 2003, 43, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Garner, P.; Cox, P.B.; Rathnayake, U.; Holloran, N.; Erdman, P. Design and Synthesis of Pyrrolidine-based Fragments That Sample Three-dimensional Molecular Space. Acs Med. Chem. Lett. 2019, 10, 811–815. [Google Scholar] [CrossRef] [PubMed]
- Hanby, A.R.; Troelsen, N.S.; Osberger, T.J.; Kidd, S.L.; Mortensen, K.T.; Spring, D.R. Fsp(3)-rich and diverse fragments inspired by natural products as a collection to enhance fragment-based drug discovery. Chemcomm (Camb. Engl. ) 2020, 56, 2280–2283. [Google Scholar] [CrossRef]
- Liu, M.; Quinn, R.J. Fragment-based screening with natural products for novel anti-parasitic disease drug discovery. Expert Opin. Drug Discov. 2019, 14, 1283–1295. [Google Scholar] [CrossRef]
- Firth, N.C.; Brown, N.; Blagg, J. Plane of Best Fit: A Novel Method to Characterize the Three-Dimensionality of Molecules. J. Chem. Inf. Model. 2012, 52, 2516–2525. [Google Scholar] [CrossRef]
- Fuller, N.; Spadola, L.; Cowen, S.; Patel, J.; Schonherr, H.; Cao, Q.; McKenzie, A.; Edfeldt, F.; Rabow, A.; Goodnow, R. An Improved Model for Fragment-Based Lead Generation at AstraZeneca. Drug Discov Today 2016. [Google Scholar] [CrossRef]
- Divakaran, A.; Kirberger, S.E.; Pomerantz, W.C.K. SAR by (Protein-Observed) 19F NMR. Acc. Chem. Res. 2019, 52, 3407–3418. [Google Scholar] [CrossRef]
- Ye, L.; Larda, S.T.; Frank Li, Y.F.; Manglik, A.; Prosser, R.S. A comparison of chemical shift sensitivity of trifluoromethyl tags: Optimizing resolution in ¹⁹F NMR studies of proteins. J. Biomol Nmr 2015, 62, 97–103. [Google Scholar] [CrossRef]
- Lu, M.; Ishima, R.; Polenova, T.; Gronenborn, A.M. 19F NMR relaxation studies of fluorosubstituted tryptophans. J. Biomol. Nmr 2019, 73, 401–409. [Google Scholar] [CrossRef] [PubMed]
- de Souza Neto, L.R.; Moreira-Filho, J.T.; Neves, B.J.; Maidana, R.L.B.R.; Guimarães, A.C.R.; Furnham, N.; Andrade, C.H.; Silva, F.P., Jr. In silico Strategies to Support Fragment-to-Lead Optimization in Drug Discovery. Front Chem 2020, 8, 93. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Ranganathan, A.; Ijzerman, A.P.; Siegal, G.; Carlsson, J. Complementarity between in Silico and Biophysical Screening Approaches in Fragment-Based Lead Discovery against the A2A Adenosine Receptor. J. Chem. Inf. Model. 2013, 53, 2701–2714. [Google Scholar] [CrossRef] [PubMed]
- Spiliotopoulos, D.; Wamhoff, E.-C.; Lolli, G.; Rademacher, C.; Caflisch, A. Discovery of BAZ2A bromodomain ligands. Eur. J. Med. Chem. 2017, 139, 564–572. [Google Scholar] [CrossRef]
- Zhao, H.; Gartenmann, L.; Dong, J.; Spiliotopoulos, D.; Caflisch, A. Discovery of BRD4 bromodomain inhibitors by fragment-based high-throughput docking. Bioorganic Med. Chem. Lett. 2014, 24, 2493–2496. [Google Scholar] [CrossRef] [PubMed]
- Speck-Planche, A.; Scotti, M.T. BET bromodomain inhibitors: Fragment-based in silico design using multi-target QSAR models. Mol. Divers. 2019, 23, 555–572. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Unzue, A.; Dong, J.; Spiliotopoulos, D.; Nevado, C.; Caflisch, A. Discovery of CREBBP Bromodomain Inhibitors by High-Throughput Docking and Hit Optimization Guided by Molecular Dynamics. J. Med. Chem. 2016, 59, 1340–1349. [Google Scholar] [CrossRef]
- Kozakov, D.; Grove, L.E.; Hall, D.R.; Bohnuud, T.; Mottarella, S.E.; Luo, L.; Xia, B.; Beglov, D.; Vajda, S. The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Protoc. 2015, 10, 733–755. [Google Scholar] [CrossRef]
Sample Availability: All small molecule fragments are commercially available and can be purchased from Life Chemicals. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).