Detection of Biomarkers Relating to Quality and Differentiation of Some Commercially Significant Whole Fish Using Spatially Off-Set Raman Spectroscopy
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Raman Measurements
2.3. Spectra Pre-Processing and Chemometrics
3. Results
3.1. Spectra and Band Assignments
3.2. Chemometrics
4. Discussion
4.1. Lipids
4.2. Guanine
4.3. Carotenoids
4.4. Proteins and Amino Acids
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. The State of world fisheries and aquaculture, 2012. Choice Rev. Online 2013, 50, 50. [Google Scholar]
- O’Fallon, J.V.; Busboom, J.R.; Nelson, M.L.; Gaskins, C.T. A direct method for fatty acid methyl ester synthesis: Application to wet meat tissues, oils, and feedstuffs. J. Anim. Sci. 2007, 85, 1511–1521. [Google Scholar] [CrossRef]
- Rasmussen, H.M.; Muzhingi, T.; Eggert, E.M.; Johnson, E.J. Lutein, zeaxanthin, meso-zeaxanthin content in egg yolk and their absence in fish and seafood. J. Food Compos. Anal. 2012, 27, 139–144. [Google Scholar] [CrossRef]
- Skrede, G.; Storebakken, T. Instrumental colour analysis of farmed and wild Atlantic salmon when raw, baked and smoked. Aquaculture 1986, 53, 279–286. [Google Scholar] [CrossRef]
- Skrede, G.; Risvik, E.; Huber, M.; Enersen, G.; Blümlein, L. Developing a Colour Card for Raw Flesh of Astaxanthin-fed Salmon. J. Food Sci. 1990, 55, 361–363. [Google Scholar] [CrossRef]
- Koningstein, J.A. Introduction to the Theory of the Raman Effect; D. Reidel Publishling Company: Dordrecht, the Netherlands, 1972. [Google Scholar]
- Sánchez-Alonso, I.; Carmona, P.; Careche, M. Vibrational spectroscopic analysis of hake (Merluccius merluccius L.) lipids during frozen storage. Food Chem. 2012, 132, 160–167. [Google Scholar]
- Afseth, N.K.; Wold, J.P.; Segtnan, V.H. The potential of Raman spectroscopy for characterisation of the fatty acid unsaturation of salmon. Anal. Chim. Acta 2006, 572, 85–92. [Google Scholar] [CrossRef]
- Hikima, J.-I.; Ando, M.; Hamaguchi, H.-O.; Sakai, M.; Maita, M.; Yazawa, K.; Takeyama, H.; Aoki, T. On-site Direct Detection of Astaxanthin from Salmon Fillet Using Raman Spectroscopy. Mar. Biotechnol. 2017, 19, 157–163. [Google Scholar] [CrossRef]
- Wold, J.P.; Marquardt, B.J.; Dable, B.K.; Robb, D.; Hatlen, B. Rapid Quantification of Carotenoids and Fat in Atlantic Salmon (Salmo Salar L.) by Raman Spectroscopy and Chemometrics. Appl. Spectrosc. 2004, 58, 395–403. [Google Scholar] [CrossRef]
- Velioglu, H.M.; Temiz, H.T.; Boyaci, I.H.; Boyacı, I.H. Differentiation of fresh and frozen-thawed fish samples using Raman spectroscopy coupled with chemometric analysis. Food Chem. 2015, 172, 283–290. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, T.; Xiang, C.; Xu, X.; Tian, X. Rapid Identification of Rainbow Trout Adulteration in Atlantic Salmon by Raman Spectroscopy Combined with Machine Learning. Molecules 2019, 24, 2851. [Google Scholar] [CrossRef] [PubMed]
- Matousek, P.; Clark, I.P.; Draper, E.R.C.; Morris, M.D.; Goodship, A.E.; Everall, N.; Towrie, M.; Finney, W.F.; Parker, A.W. Subsurface Probing in Diffusely Scattering Media Using Spatially Offset Raman Spectroscopy. Appl. Spectrosc. 2005, 59, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Pour, S.O.; Fowler, S.M.; Hopkins, D.L.; Torley, P.; Gill, H.; Blanch, E.W. Investigation of chemical composition of meat using spatially off-set Raman spectroscopy. Analyst 2019, 144, 2618–2627. [Google Scholar] [CrossRef] [PubMed]
- Afseth, N.K.; Bloomfield, M.; Wold, J.P.; Matousek, P. A Novel Approach for Subsurface Through-Skin Analysis of Salmon Using Spatially Offset Raman Spectroscopy (SORS). Appl. Spectrosc. 2014, 68, 255–262. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Mathlouthi, M.; Seuvre, A.M.; Koenig, J.L. F.T.-I.R. and laser-raman spectra of guanine and guanosine. Carbohydr. Res. 1986, 146, 15–27. [Google Scholar] [CrossRef]
- Lafleur, L.; Rice, J.; Thomas, G.J. Raman Studies of Nucleic Acids. VII. Poly A, Poly U, and Poly G and Poly C. Biopolymers 1972, 11, 2423–2437. [Google Scholar] [CrossRef]
- Zhu, F.; Peng, J.; Gao, J.; Zhao, Y.; Yu, K.; He, Y. Determination and visualization of fat contents in salmon fillets based on visible and near-infrared imagery. Trans. Chin. Soc. Agric. Eng. 2012, 30, 314–323. [Google Scholar]
- Zhol, S.; Ackman, R.G.; Morrison, C. Storage of lipids in the myosepta of Atlantic salmon (Salmo salar). Fish. Physiol. Biochem. 1995, 14, 171–178. [Google Scholar] [CrossRef]
- Kaneko, G.; Shirakami, H.; Hirano, Y.; Oba, M.; Yoshinaga, H.; Khieokhajonkhet, A.; Nagasaka, R.; Kondo, H.; Hirono, I.; Ushio, H.; et al. Diversity of Lipid Distribution in Fish Skeletal Muscle. Zool. Sci. 2016, 33, 170–178. [Google Scholar] [CrossRef]
- Taşbozan, O.; Gökçe, M.A. Fatty Acids in Fish. In Fatty Acids; Catala, A., Ed.; IntechOpen: College Park, MD, USA, 2017. [Google Scholar]
- Bekhit, M.Y.; Grung, B.; Mjøs, S.A. Determination of Omega-3 Fatty Acids in Fish Oil Supplements Using Vibrational Spectroscopy and Chemometric Methods. Appl. Spectrosc. 2014, 68, 1190–1200. [Google Scholar] [CrossRef] [PubMed]
- Strobel, C.; Jahreis, G.; Kuhnt, K. Survey of n-3 and n-6 polyunsaturated fatty acids in fish and fish products. Lipids Heal. Dis. 2012, 11, 144. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.D. The Effects of Intermedian on the Ultrastructure of Amphibian Iridophores. Gen. Comp. Endrocrinol. 1969, 12, 405–416. [Google Scholar] [CrossRef]
- Harris, J.; Hunt, S. The fine structure of iridophores in the skin of the Atlantic salmon (Salmo salar L.). Tissue Cell 1973, 5, 479–488. [Google Scholar] [CrossRef]
- Torrissen, O.J.; Naevdal, G. Pigmentation of salmonids—Variation in flesh carotenoids of Atlantic salmon. Aquaculture 1988, 68, 305–310. [Google Scholar] [CrossRef]
- Orona-Navar, A.; Aguilar-Hernández, I.A.; Cerdán-Pasarán, A.; López-Luke, T.; Rodriguez-Delgado, M.; Cárdenas-Chávez, D.; Cepeda-Pérez, E.; Ornelas-Soto, N. Astaxanthin from Haematococcus pluvialis as a natural photosensitizer for dye-sensitized solar cell. Algal Res. 2017, 26, 15–24. [Google Scholar] [CrossRef]
- Rajasingh, H.; Øyehaug, L.; Våge, D.I.; Omholt, S.W. Carotenoid dynamics in Atlantic salmon. BMC Biol. 2006, 4, 10. [Google Scholar] [CrossRef]
- Alam, S.; Teshima, S.I.; Yaniharto, D.; Sumule, O.; Ishikawa, M.; Koshio, S. Assessment of reference dietary amino acid pattern for juvenile red sea bream, Pagrus major. Aquac. Int. 2005, 13, 369–379. [Google Scholar] [CrossRef]
- Mohanty, B.P.; Mahanty, A.; Ganguly, S.; Sankar, T.V.; Chakraborty, K.; Rangasamy, A.; Paul, B.; Sarma, D.; Mathew, S.; Asha, K.K.; et al. Amino Acid Compositions of 27 Food Fishes and Their Importance in Clinical Nutrition. J. Amino Acids 2014, 2014, 1–7. [Google Scholar] [CrossRef]
- Wilson, R.; Cowey, C. Amino acid composition of whole body tissue of rainbow trout and Atlantic salmon. Aquaculture 1985, 48, 373–376. [Google Scholar] [CrossRef]
- Benjakul, S.; Nalinanon, S.; Shahidi, F. Fish Collagen. In Food Biochemistry and Food Processing; Simpson, B.K., Nollet, L., Toldrá, F., Benjakul, S., Paliyath, G., Hui, Y.H., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Fowler, S.M.; Schmidt, H.; Van De Ven, R.; Wynn, P.; Hopkins, D.L. Predicting meat quality traits of ovine m. semimembranosus, both fresh and following freezing and thawing, using a hand held Raman spectroscopic device. Meat Sci. 2015, 108, 138–144. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

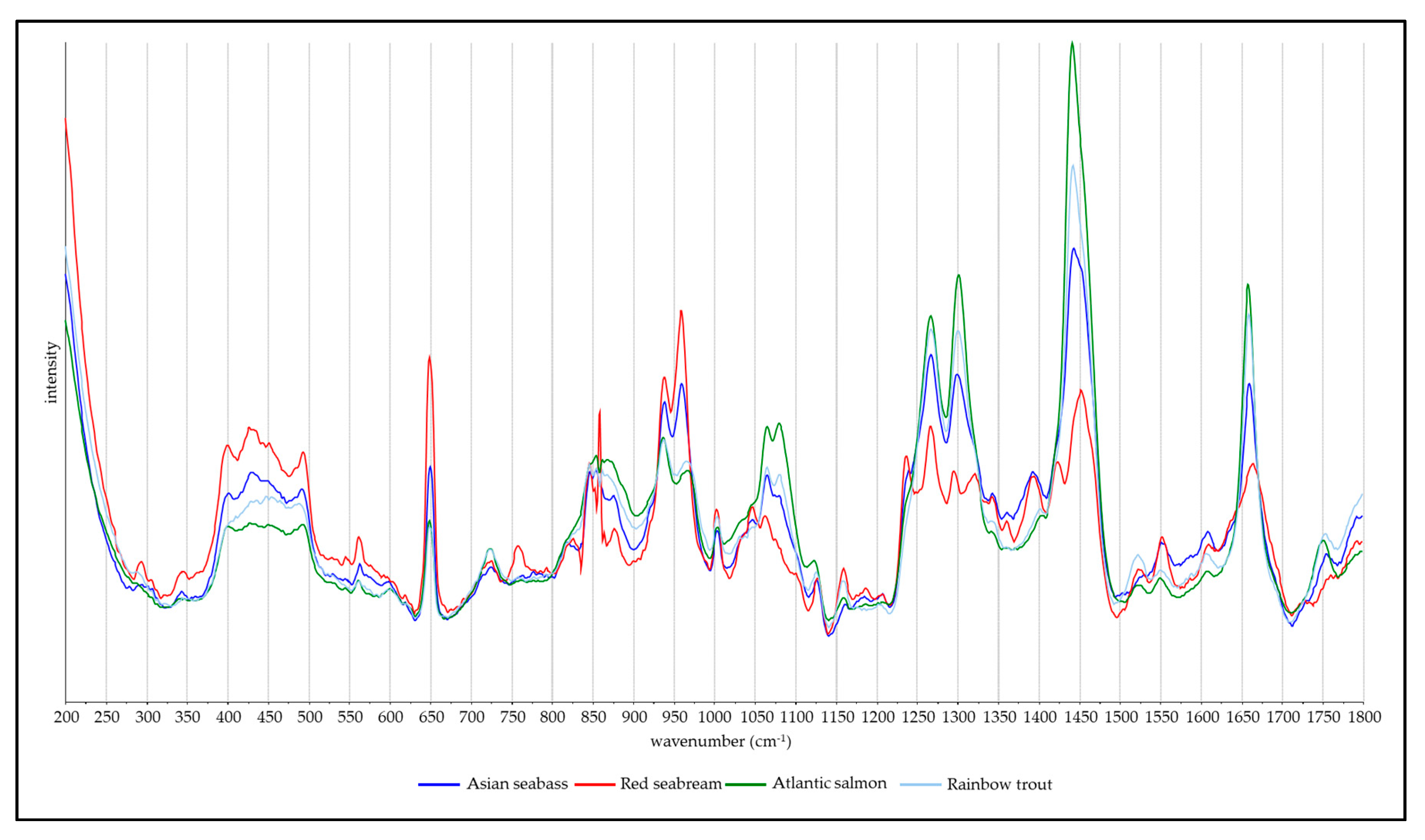
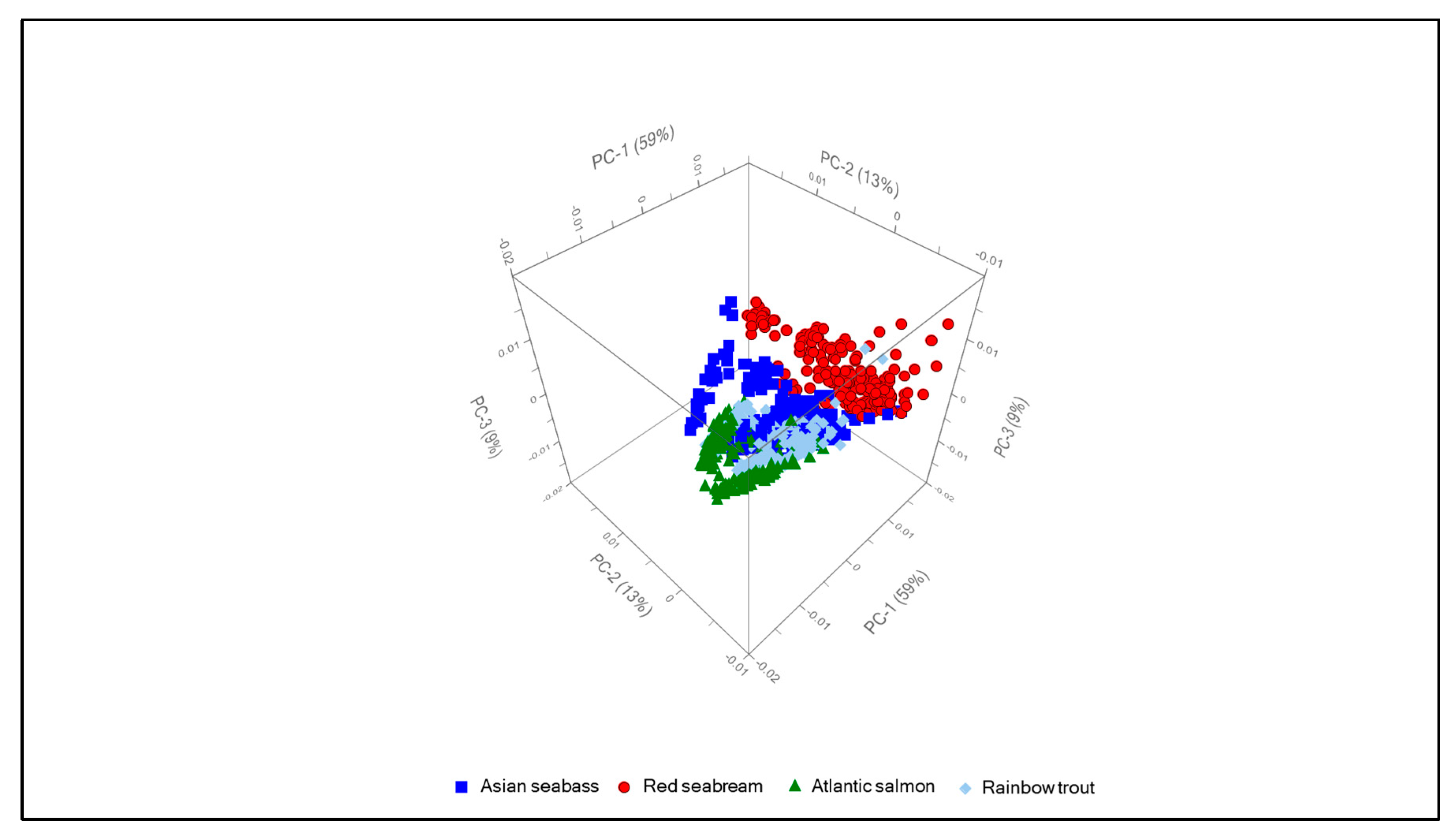
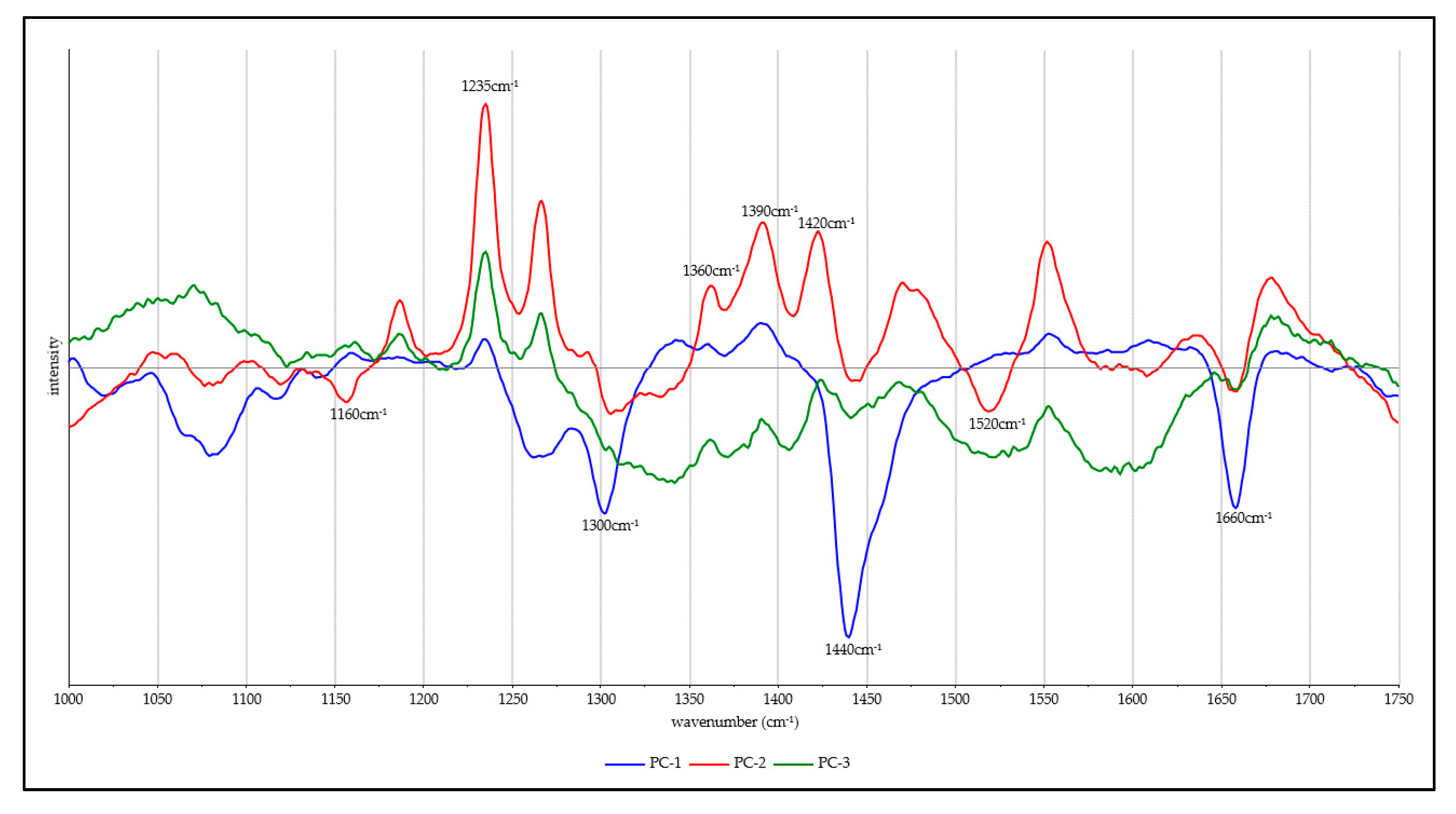
| Wavenumber (cm−1) | Vibration | Component | Reference |
|---|---|---|---|
| 339 | δ(C=O) | Guanine | [17,18] |
| 400 | δ(C=O) | Guanine | |
| 493 | δ(N-9-C-4=C-5) and δ(N-7-C-=C-4) | Guanine | |
| 560 | δ(C-C=C) | Guanine | |
| 650 | Ring breathing | Guanine | |
| 710 | δ(ring) | Guanine | |
| 724 | C-C stretching | Collagen | [16] |
| 760 | Ring breathing | Tryptophan | |
| 825 | - | Collagen | |
| 850 | Single bond stretching | Amino acids | |
| 860 | ν(C-C) | Collagen | |
| 868 | ν(C-C) stretching | Collagen | |
| 930 | δ(N-C=N) | Guanine | [17,18] |
| 968 | δ(=C-H) trans RHC=CHR | Lipid | [7,8] |
| 1004 | In plane CH3 rock Aromatic ring breathing | Carotenoids Phenylalanine | [16] |
| 1048 | ν(C-H) | Lipids | [7,8] |
| 1065 | ν(C-C) stretching | Lipids | |
| 1079 | Asymmetric CCC stretching | Lipids | |
| 1118 | C-C stretching | Lipids | |
| 1126 | δ(C-N=C) | Guanine | [17,18] |
| 1160 | C-C polyene chain stretch | Carotenoids | [8,9] |
| 1192 | Carotenoids | ||
| 1235 | ν(C-2-NH2) | Guanine | [17,18] |
| 1266 | C-H rocking | Lipid | |
| 1300 | CH2 twist | Methylene-Lipid | |
| 1360 | δ(C-8-N-H), δ(C-8-H), ν(C-8-N) | Guanine | [17,18] |
| 1393 | Guanine | ||
| 1420 | δ(N-7=C-8-H) | Guanine | |
| 1440 | CH2 scissoring | Methylene-Lipid | [7,8] |
| 1520 | In phase C=C stretch | Carotenoids | [8] |
| 1550 | ν(C=C) | Tryptophan | [16] |
| 1608 | C=C | Phenylalanine | |
| 1660 | cis C=C stretch/amide I | Lipid/Protein | [7,8] |
| 1750 | C=O | Lipid |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landry, J.D.; Torley, P.J.; Blanch, E.W. Detection of Biomarkers Relating to Quality and Differentiation of Some Commercially Significant Whole Fish Using Spatially Off-Set Raman Spectroscopy. Molecules 2020, 25, 3776. https://doi.org/10.3390/molecules25173776
Landry JD, Torley PJ, Blanch EW. Detection of Biomarkers Relating to Quality and Differentiation of Some Commercially Significant Whole Fish Using Spatially Off-Set Raman Spectroscopy. Molecules. 2020; 25(17):3776. https://doi.org/10.3390/molecules25173776
Chicago/Turabian StyleLandry, Jeremy D., Peter J. Torley, and Ewan W. Blanch. 2020. "Detection of Biomarkers Relating to Quality and Differentiation of Some Commercially Significant Whole Fish Using Spatially Off-Set Raman Spectroscopy" Molecules 25, no. 17: 3776. https://doi.org/10.3390/molecules25173776
APA StyleLandry, J. D., Torley, P. J., & Blanch, E. W. (2020). Detection of Biomarkers Relating to Quality and Differentiation of Some Commercially Significant Whole Fish Using Spatially Off-Set Raman Spectroscopy. Molecules, 25(17), 3776. https://doi.org/10.3390/molecules25173776






