Flavonoid and Non-Flavonoid Compounds of Autumn Royal and Egnatia Grape Skin Extracts Affect Membrane PUFA’s Profile and Cell Morphology in Human Colon Cancer Cell Lines
Abstract
1. Introduction
2. Results and Discussion
2.1. Total Polyphenolic Content in Autumn Royal and Egnatia GSEs
2.2. Membrane PUFA Profile After GSE Treatment in Human Colon Cancer Cell Lines
2.3. Effects of GSE Treatments on the Gene Expression of 15-LOX-1 and PPAR-γ in Human Colon Cancer Cell Lines
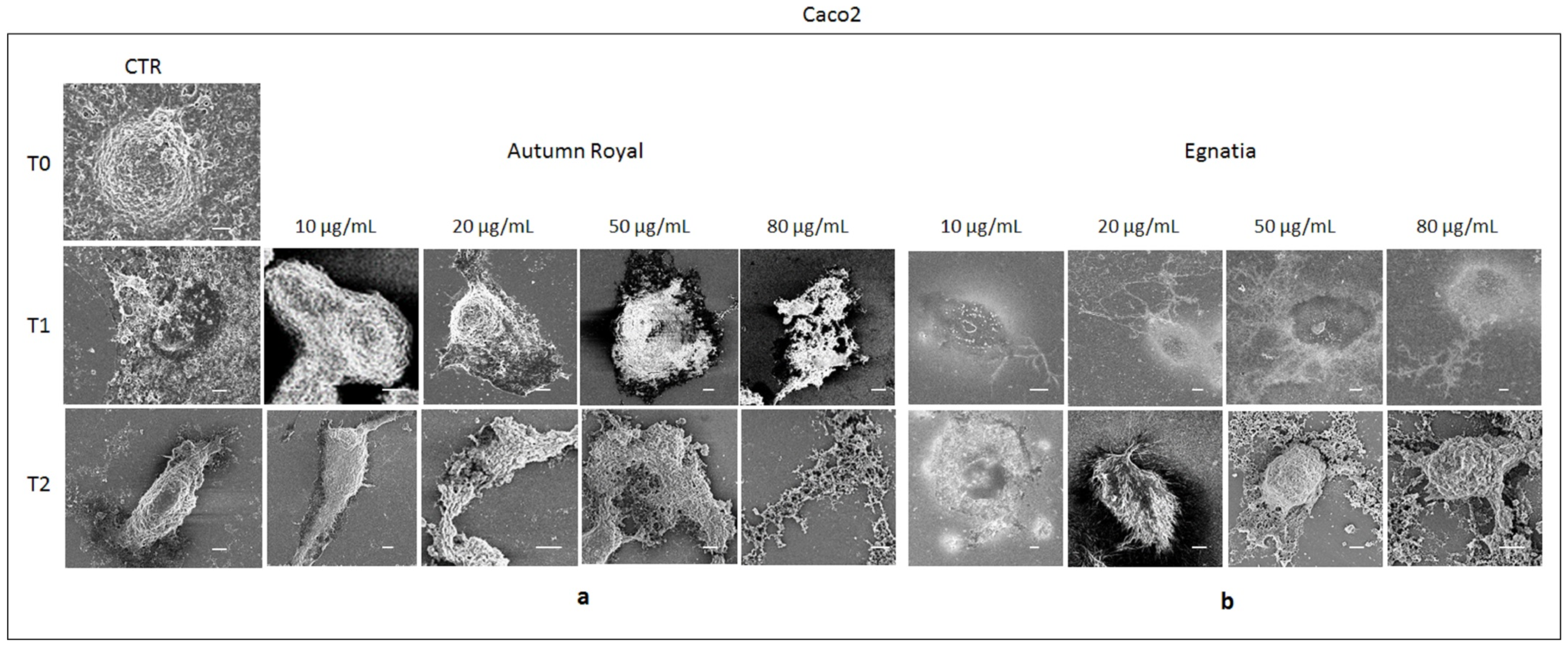
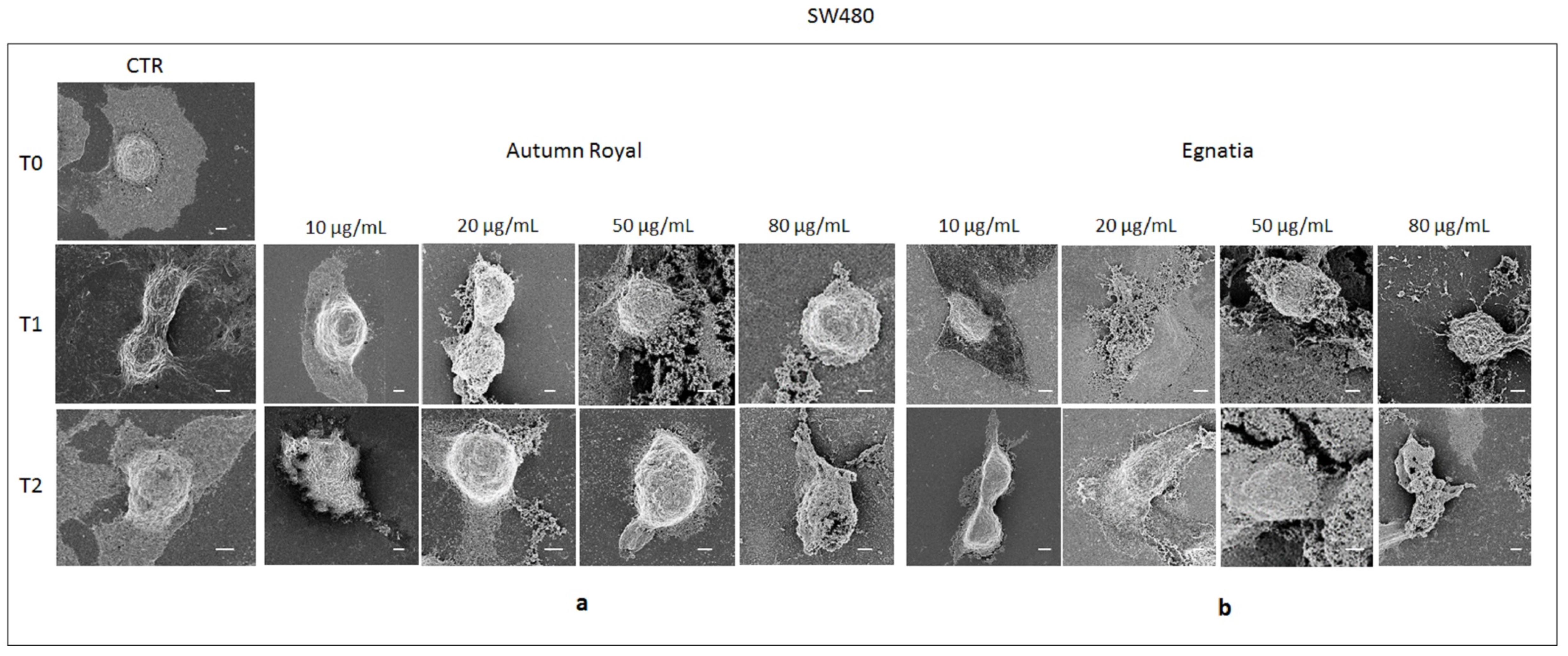
2.4. Autumn Royal and Egnatia GSEs Induce Cell Morphological Changes
3. Materials and Methods
3.1. Preparation of the GSEs
3.2. Total Polyphenolic Content
3.3. UHPLC/QTOF Mass Spectrometry
3.4. Cell Culture and Treatment
3.5. Lipids Extraction and PUFAs Analysis
3.6. RNA Extraction and Quantitative Real-Time PCR
3.7. FE-SEM Investigation
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Magrone, T.; Magrone, M.; Russo, M.A.; Jirillo, E. Magrone Recent Advances on the Anti-Inflammatory and Antioxidant Properties of Red Grape Polyphenols: In Vitro and In Vivo Studies. Antioxidants 2019, 9, 35. [Google Scholar] [CrossRef]
- Mojzer, E.B.; Hrnčič, M.K.; Škerget, M.; Knez, Ž.; Bren, U. Polyphenols: Extraction Methods, Antioxidative Action, Bioavailability and Anticarcinogenic Effects. Molecules 2016, 21, 901. [Google Scholar] [CrossRef] [PubMed]
- Durazzo, A.; Lucarini, M.; Souto, E.B.; Cicala, C.; Caiazzo, E.; Izzo, A.A.; Novellino, E.; Santini, A. Polyphenols: A concise overview on the chemistry, occurrence, and human health. Phytother. Res. 2019, 33, 2221–2243. [Google Scholar] [CrossRef] [PubMed]
- Pereira, D.M.; Valentão, P.; Pereira, J.A.; Andrade, P.B. Phenolics: From Chemistry to Biology. Molecules 2009, 14, 2202–2211. [Google Scholar] [CrossRef]
- Xia, E.-Q.; Deng, G.-F.; Guo, Y.-J.; Li, H.-B. Biological Activities of Polyphenols from Grapes. Int. J. Mol. Sci. 2010, 11, 622–646. [Google Scholar] [CrossRef] [PubMed]
- Hendrich, A.B. Flavonoid-membrane interactions: Possible consequences for biological effects of some polyphenolic compounds1. Acta Pharmacol. Sin. 2006, 27, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xiao, Y.-Y. Grape Phytochemicals and Associated Health Benefits. Crit. Rev. Food Sci. Nutr. 2013, 53, 1202–1225. [Google Scholar] [CrossRef]
- Lee, H.S.; Ha, A.W.; Kim, W.K. Effect of resveratrol on the metastasis of 4T1 mouse breast cancer cells in vitro and in vivo. Nutr. Res. Pr. 2012, 6, 294–300. [Google Scholar] [CrossRef]
- Wang, L.; Ling, Y.; Chen, Y.; Li, C.-L.; Feng, F.; You, Q.-D.; Lu, N.; Guo, Q. Flavonoid baicalein suppresses adhesion, migration and invasion of MDA-MB-231 human breast cancer cells. Cancer Lett. 2010, 297, 42–48. [Google Scholar] [CrossRef]
- Mantena, S.K.; Baliga, M.S.; Katiyar, S.K. Grape seed proanthocyanidins induce apoptosis and inhibit metastasis of highly metastatic breast carcinoma cells. Carcinogenesis 2006, 27, 1682–1691. [Google Scholar] [CrossRef]
- Ramos, S. Cancer chemoprevention and chemotherapy: Dietary polyphenols and signalling pathways. Mol. Nutr. Food Res. 2008, 52, 507–526. [Google Scholar] [CrossRef]
- Valenzuela, M.; Bastias, L.; Montenegro, I.; Werner, E.; Madrid, A.; Godoy, P.; Párraga, M.; Villena, J. Autumn Royal and Ribier Grape Juice Extracts Reduced Viability and Metastatic Potential of Colon Cancer Cells. Evid.-Based Complement. Altern. Med. 2018, 2018, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Avtanski, D.; Poretsky, L. Phyto-polyphenols as potential inhibitors of breast cancer metastasis. Mol. Med. 2018, 24, 29. [Google Scholar] [CrossRef] [PubMed]
- Hanikoglu, A.; Kucuksayan, E.; Hanikoglu, F.; Ozben, T.; Menounou, G.; Sansone, A.; Chatgilialoglu, C.; Di Bella, G.; Ferreri, C. Effects of somatostatin, curcumin, and quercetin on the fatty acid profile of breast cancer cell membranes. Can. J. Physiol. Pharmacol. 2020, 98, 131–138. [Google Scholar] [CrossRef]
- Ortinau, L.C.; Pickering, R.T.; Nickelson, K.J.; Stromsdorfer, K.L.; Naik, C.Y.; Haynes, R.A.; Bauman, D.E.; Rector, R.S.; Fritsche, K.L.; Ii, J.P. Sterculic Oil, a Natural SCD1 Inhibitor, Improves Glucose Tolerance in Obese ob/ob Mice. ISRN Endocrinol. 2012, 2012, 947323. [Google Scholar] [CrossRef] [PubMed]
- Tutino, V.; Gigante, I.; Scavo, M.P.; Refolo, M.G.; De Nunzio, V.; Milella, R.A.; Caruso, M.G.; Notarnicola, M. Stearoyl-CoA Desaturase-1 Enzyme Inhibition by Grape Skin Extracts Affects Membrane Fluidity in Human Colon Cancer Cell Lines. Nutrients 2020, 12, 693. [Google Scholar] [CrossRef] [PubMed]
- Scaglia, N.; Igal, R.A. Stearoyl-CoA Desaturase Is Involved in the Control of Proliferation, Anchorage-independent Growth, and Survival in Human Transformed Cells. J. Boil. Chem. 2005, 280, 25339–25349. [Google Scholar] [CrossRef] [PubMed]
- Ran, H.; Zhu, Y.; Deng, R.; Zhang, Q.; Liu, X.; Feng, M.; Zhong, J.; Lin, S.; Tong, X.; Su, Q. Stearoyl-CoA desaturase-1 promotes colorectal cancer metastasis in response to glucose by suppressing PTEN. J. Exp. Clin. Cancer Res. 2018, 37, 54. [Google Scholar] [CrossRef]
- Maria, N.; Maria, G.C.; Valeria, T.; Valentina, D.N.; Isabella, G.; Giampiero, D.L.; Nicola, V.; Ornella, R.; Rosa, R.; Elisa, S.; et al. Nutrition and lipidomic profile in colorectal cancers. Acta Bio Med. Atenei Parm. 2018, 89, 87–96. [Google Scholar]
- Long, J.; Zhang, C.-J.; Zhu, N.; Du, K.; Yin, Y.-F.; Tan, X.; Liao, D.-F.; Qin, L. Lipid metabolism and carcinogenesis, cancer development. Am. J. Cancer Res. 2018, 8, 778–791. [Google Scholar]
- Garcia, L.C.; Achón-Tuñón, M.; González-González, M.P. The influence of the polyunsaturated fatty acids in the prevention and promotion of cancer. Nutr. Hosp. 2015, 32, 41–49. [Google Scholar]
- Tutino, V.; De Nunzio, V.; Caruso, M.G.; Veronese, N.; Lorusso, D.; Di Masi, M.; Benedetto, M.L.; Notarnicola, M. Elevated AA/EPA Ratio Represents an Inflammatory Biomarker in Tumor Tissue of Metastatic Colorectal Cancer Patients. Int. J. Mol. Sci. 2019, 20, 2050. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Zuo, X.; Jaoude, J.; Mao, F.; Colby, J.; Shureiqi, I. ALOX15 as a suppressor of inflammation and cancer: Lost in the link. Prostaglandins Other Lipid Mediat. 2017, 132, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N. Pro-resolving lipid mediators are leads for resolution physiology. Nature 2014, 510, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Mathew, G.; Jayne, D.; Pelengaris, S.; Khan, M. 15-Lipoxygenase-1 in Colorectal Cancer: A Review. Tumor Boil. 2009, 30, 185–199. [Google Scholar] [CrossRef]
- Çimen, I.; Astarci, E.; Banerjee, S. 15-lipoxygenase-1 exerts its tumor suppressive role by inhibiting nuclear factor-kappa B via activation of PPAR gamma. J. Cell. Biochem. 2011, 112, 2490–2501. [Google Scholar] [CrossRef]
- Mao, F.; Wang, M.; Wang, J.; Xu, W. The role of 15-LOX-1 in colitis and colitis-associated colorectal cancer. Inflamm. Res. 2015, 64, 661–669. [Google Scholar] [CrossRef]
- Gigante, I.; Milella, R.A.; Tutino, V.; DeBiase, G.; Notarangelo, L.; Giannandrea, M.A.; De Nunzio, V.; Orlando, A.; D’Alessandro, R.; Caruso, M.; et al. Autumn Royal and Egnatia Grape Extracts differently modulate Cell Proliferation in Human Colorectal Cancer Cells. Endocr. Metab. Immune Disord. Drug Targets 2020, 20, 1–19. [Google Scholar] [CrossRef]
- Lin, B.-W.; Gong, C.-C.; Song, H.-F.; Cui, Y.-Y. Effects of anthocyanins on the prevention and treatment of cancer. Br. J. Pharmacol. 2016, 174, 1226–1243. [Google Scholar] [CrossRef]
- Hou, D.-X.; Kai, K.; Li, J.-J.; Lin, S.; Terahara, N.; Wakamatsu, M.; Fujii, M.; Young, M.R.; Colburn, N. Anthocyanidins inhibit activator protein 1 activity and cell transformation: Structure-activity relationship and molecular mechanisms. Carcinogenesis 2004, 25, 29–36. [Google Scholar] [CrossRef]
- Unusan, N. Proanthocyanidins in grape seeds: An updated review of their health benefits and potential uses in the food industry. J. Funct. Foods 2020, 67, 103861. [Google Scholar] [CrossRef]
- Nandakumar, V.; Singh, T.; Katiyar, S.K. Multi-targeted prevention and therapy of cancer by proanthocyanidins. Cancer Lett. 2008, 269, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-Y.; Li, Q.; Bi, K.-S. Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian J. Pharm. Sci. 2018, 13, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Oteiza, P.I.; Erlejman, A.G.; Verstraeten, S.V.; Keen, C.L.; Fraga, C.G. Flavonoid-membrane Interactions: A Protective Role of Flavonoids at the Membrane Surface? Clin. Dev. Immunol. 2005, 12, 19–25. [Google Scholar] [CrossRef]
- Mandić, L.; Sadžak, A.; Strasser, V.; Baranović, G.; Jurašin, D.D.; Sikirić, M.D.; Šegota, S. Enhanced Protection of Biological Membranes during Lipid Peroxidation: Study of the Interactions between Flavonoid Loaded Mesoporous Silica Nanoparticles and Model Cell Membranes. Int. J. Mol. Sci. 2019, 20, 2709. [Google Scholar] [CrossRef]
- Pakiet, A.; Kobiela, J.; Stepnowski, P.; Sledzinski, T.; Mika, A. Changes in lipids composition and metabolism in colorectal cancer: A review. Lipids Health Dis. 2019, 18, 29. [Google Scholar] [CrossRef]
- Else, P.L. The highly unnatural fatty acid profile of cells in culture. Prog. Lipid Res. 2020, 77, 101017. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, F.; Todo, S.; Imashuku, S. Antitumor effect of gamma-linolenic acid on cultured human neuroblastoma cells. Prostaglandins Leukot. Med. 1986, 23, 311–320. [Google Scholar] [CrossRef]
- Das, U.N. Essential fatty acids and their metabolites and cancer. Nutrition 1999, 15, 239–240. [Google Scholar]
- Ramchurren, N.; Karmali, R. Effects of gamma-linolenic and dihomo-gamma-linolenic acids on 7,12-dimethylbenz(α)anthracene-induced mammary tumors in rats. Prostaglandins Leukot. Essent. Fat. Acids 1995, 53, 95–101. [Google Scholar] [CrossRef]
- Xu, Y.; Qian, S.Y. Anti-cancer activities of omega-6 polyunsaturated fatty acids. Biomed. J. 2014, 37, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Çimen, I.; Tunçay, S.; Banerjee, S. 15-Lipoxygenase-1 expression suppresses the invasive properties of colorectal carcinoma cell lines HCT-116 and HT-29. Cancer Sci. 2009, 100, 2283–2291. [Google Scholar] [CrossRef] [PubMed]
- Derry, M.; Raina, K.; Agarwal, R.; Agarwal, C. Differential effects of grape seed extract against human colorectal cancer cell lines: The intricate role of death receptors and mitochondria. Cancer Lett. 2012, 334, 69–78. [Google Scholar] [CrossRef] [PubMed]
- DiNicola, S.; Cucina, A.; Pasqualato, A.; Proietti, S.; D’Anselmi, F.; Pasqua, G.; Santamaria, A.R.; Coluccia, P.; Lagana, A.; Antonacci, D.; et al. Apoptosis-inducing factor and caspase-dependent apoptotic pathways triggered by different grape seed extracts on human colon cancer cell line Caco-2. Br. J. Nutr. 2010, 104, 824–832. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kaur, M.; Mandair, R.; Agarwal, R.; Agarwal, C. Grape Seed Extract Induces Cell Cycle Arrest and Apoptosis in Human Colon Carcinoma Cells. Nutr. Cancer 2008, 60, 2–11. [Google Scholar] [CrossRef] [PubMed]
- DiNicola, S.; Cucina, A.; Pasqualato, A.; D’Anselmi, F.; Proietti, S.; Lisi, E.; Pasqua, G.; Antonacci, D.; Bizzarri, M. Antiproliferative and Apoptotic Effects Triggered by Grape Seed Extract (GSE) versus Epigallocatechin and Procyanidins on Colon Cancer Cell Lines. Int. J. Mol. Sci. 2012, 13, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Tollefsbol, T.O. Combinational Proanthocyanidins and Resveratrol Synergistically Inhibit Human Breast Cancer Cells and Impact Epigenetic–Mediating Machinery. Int. J. Mol. Sci. 2018, 19, 2204. [Google Scholar] [CrossRef]
- Wang, S.; Tian, Q.; An, F. Growth inhibition and apoptotic effects of total flavonoids from Trollius chinensis on human breast cancer MCF-7 cells. Oncol. Lett. 2016, 12, 1705–1710. [Google Scholar] [CrossRef]
- Subramanian, A.P.; Jaganathan, S.K.; Mandal, M.; Supriyanto, E.; Muhamad, I.I. Gallic acid induced apoptotic events in HCT-15 colon cancer cells. World J. Gastroenterol. 2016, 22, 3952–3961. [Google Scholar] [CrossRef]
- Milella, R.A.; Basile, T.; Alba, V.; Gasparro, M.; Giannandrea, M.A.; DeBiase, G.; Genghi, R.; Antonacci, D. Optimized ultrasonic-assisted extraction of phenolic antioxidants from grape (Vitis vinifera L.) skin using response surface methodology. J. Food Sci. Technol. 2019, 56, 4417–4428. [Google Scholar] [CrossRef]
- Flamini, R.; De Rosso, M.; De Marchi, F.; Vedova, A.D.; Panighel, A.; Gardiman, M.; Maoz, I.; Bavaresco, L. An innovative approach to grape metabolomics: Stilbene profiling by suspect screening analysis. Metabolomics 2013, 9, 1243–1253. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Boil. Chem. 1957, 226, 497–509. [Google Scholar]
- DePalo, N.; Fanizza, E.; Vischio, F.; Denora, N.; Laquintana, V.; Cutrignelli, A.; Striccoli, M.; Giannelli, G.; Agostiano, A.; Curri, M.L.; et al. Imaging modification of colon carcinoma cells exposed to lipid based nanovectors for drug delivery: A scanning electron microscopy investigation. RSC Adv. 2019, 9, 21810–21825. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
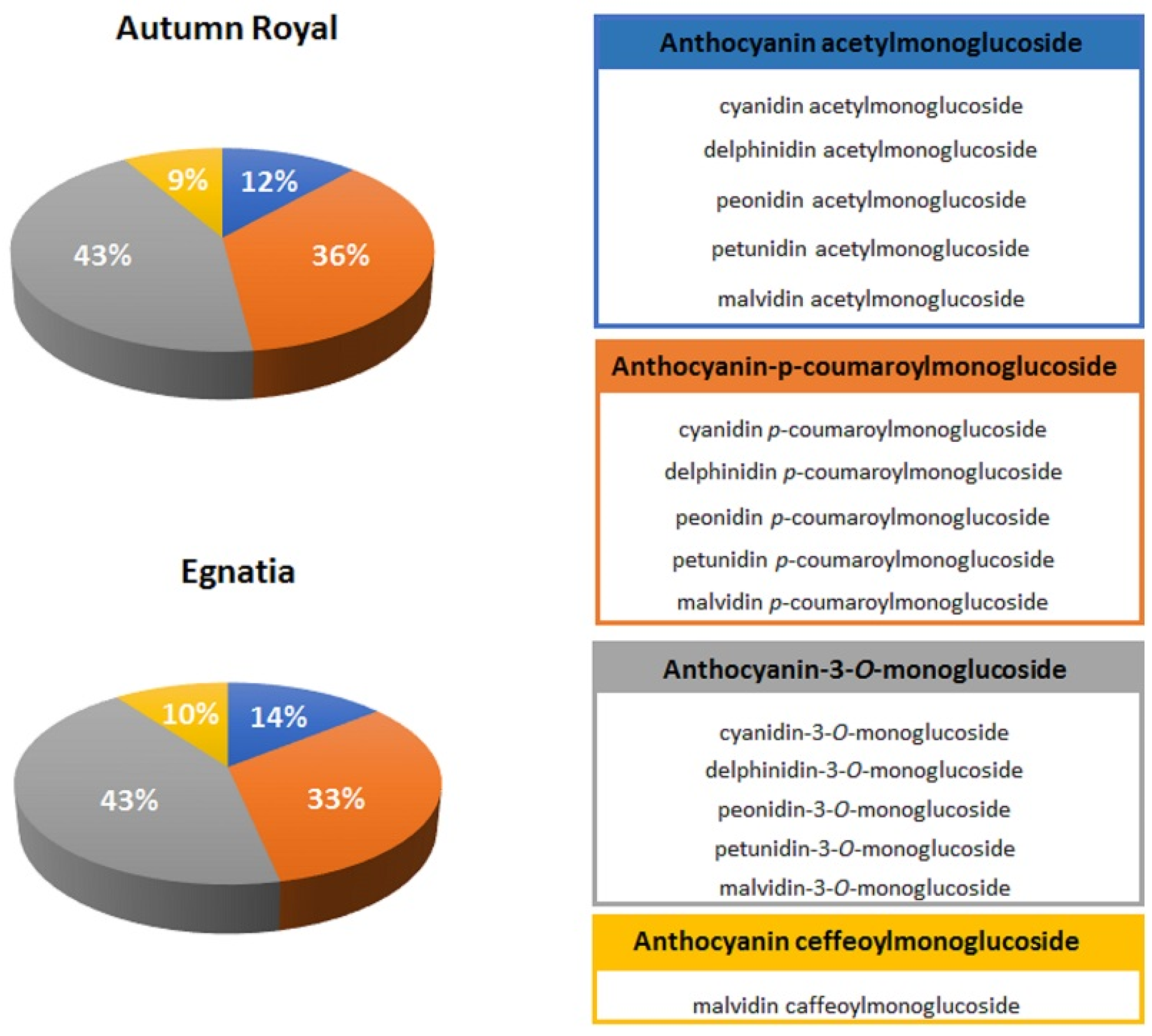
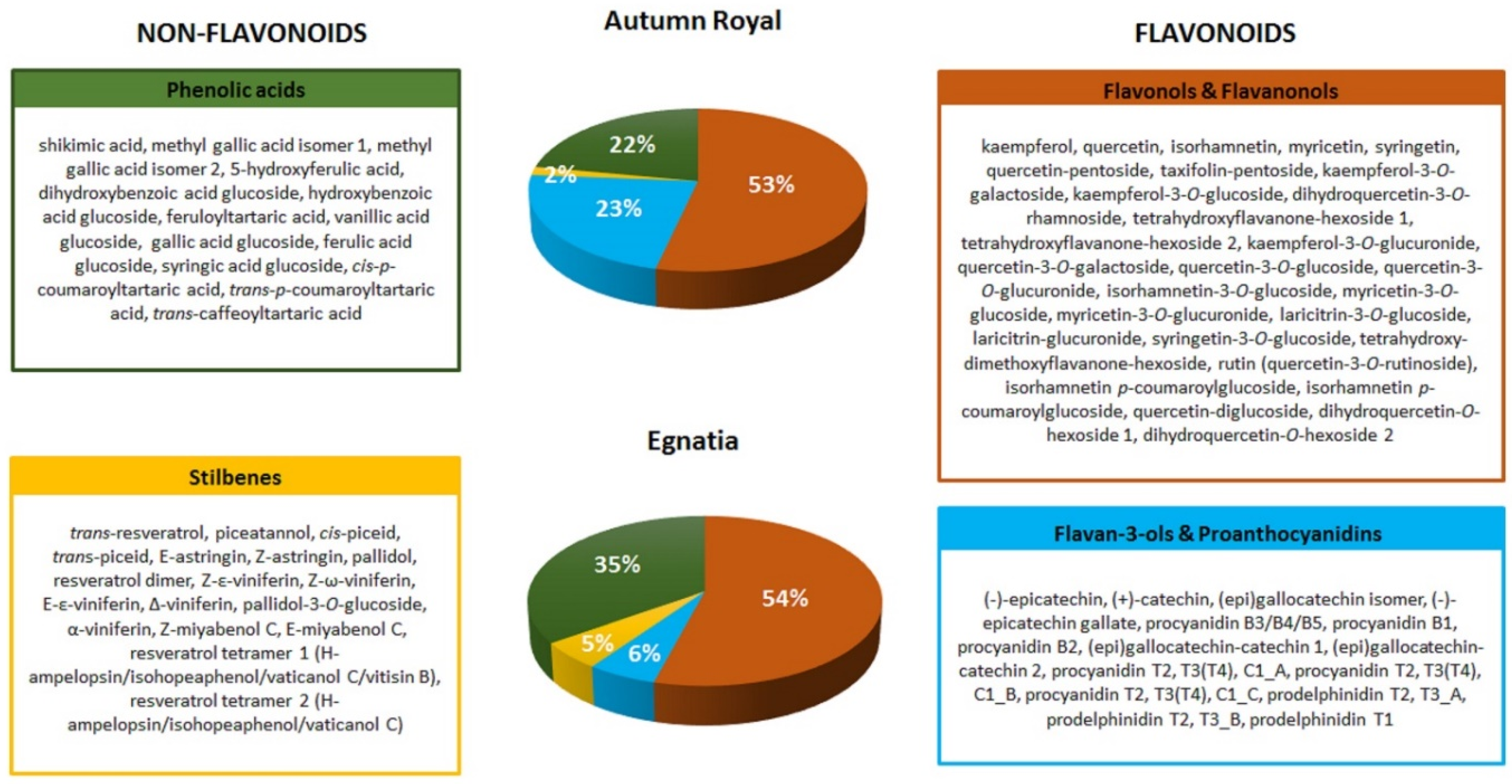

| o-Diphenol Compounds | |
|---|---|
| Phenolic acids | shikimic acid, methyl gallic acid, 5-hydroxy-ferulic acid, gallic acid glucoside, trans-caffeoyltartaric acid |
| Flavonols and Flavanonols | quercetin glycosides, myricetin glycosides, laricitrin glycosides, taxifolin-pentoside, dihydroquercetin-3-O-rhamnoside |
| Flavan-3-ols and Proanthocyanidins | (+)-catechin/(−)-epicatechin, (epi)gallocatechins, (−)-epicatechin gallate, procyanidins/prodelphinidins |
| Anthocyanins | cyanidin glycosides, delphinidin glycosides, petunidin glycosides |
| a | Caco2 PUFAs (%) | Autumn Royal | Egnatia | |||||
| CTR | 20 µg/mL | 50 µg/mL | 80 µg/mL | 20 µg/mL | 50 µg/mL | 80 µg/mL | ||
| n-6 PUFAs | linoleic acid (LA) | 2.20 ± 0.16 | 2.89 ± 0.37 * | 3.20 ± 0.33 * | 4.60 ± 0.39 * | 3.58 ± 0.18 * | 4.14 ± 0.32 * | 6.40 ± 0.39 * |
| γ-linolenic acid (GLA) | 0.14 ± 0.04 | 0.22 ± 0.07 | 0.12 ± 0.05 | 0.21 ± 0.03 | 0.18 ± 0.11 | 0.13 ± 0.04 | 0.15 ± 0.05 | |
| dihomo-γ-linoleic acid (DGLA) | 0.38 ± 0.10 | 0.44 ± 0.04 | 0.30 ± 0.05 | 0.29 ± 0.10 | 0.37 ± 0.08 | 0.34 ± 0.10 | 0.30 ± 0.14 | |
| arachidonic acid (AA) | 3.87 ± 0.51 | 4.27 ± 0.26 | 4.26 ± 0.42 | 4.25 ± 0.33 | 4.60 ± 0.64 | 3.52 ± 0.63 | 3.65 ± 0.61 | |
| n-3 PUFAs | α-linolenic acid (ALA) | 0.19 ± 0.04 | 0.57 ± 0.09 * | 0.81 ± 0.07 * | 1.34 ± 0.21 * | 0.64 ± 0.06 * | 0.93 ± 0.07 * | 1.74 ± 0.03 * |
| eicosapentaenoic acid (EPA) | 0.93 ± 0.06 | 1.03 ± 0.14 | 0.78 ± 0.08 | 0.89 ± 0.11 | 0.92 ± 0.13 | 0.90 ± 0.08 | 0.94 ± 0.11 | |
| docosaepentaenoic acid (DPA) | 1.32 ± 0.10 | 1.52 ± 0.06 | 1.26 ± 0.10 | 1.29 ± 0.09 | 1.58 ± 0.12 | 1.53 ± 0.13 | 1.52 ± 0.09 | |
| docosaehenanoic acid (DHA) | 3.21 ± 0.50 | 3.64 ± 0.37 | 3.06 ± 0.21 | 3.26 ± 0.41 | 3.54 ± 0.95 | 3.20 ± 0.67 | 3.63 ± 0.77 | |
| Total PUFAs | 12.89 ± 0.24 | 15.74 ± 0.51 * | 14.24 ± 0.71 * | 16.34 ± 0.44 * | 15.73 ± 0.50 * | 15.29 ± 0.70 * | 18.88 ± 0.42 * | |
| b | SW480 PUFAs (%) | Autumn Royal | Egnatia | |||||
| CTR | 20 µg/mL | 50 µg/mL | 80 µg/mL | 20 µg/mL | 50 µg/mL | 80 µg/mL | ||
| n-6 PUFAs | linoleic acid (LA) | 3.00 ± 0.36 | 3.91 ± 0.37 * | 4.20 ± 0.41 * | 4.36 ± 0.35 * | 3.82 ± 0.26 | 4.69 ± 0.51 * | 5.64 ± 0.63 * |
| γ-linolenic acid (GLA) | 0.20 ± 0.02 | 0.18 ± 0.01 | 0.21 ± 0.03 | 0.23 ± 0.02 | 0.16 ± 0.03 | 0.21 ± 0.03 | 0.22 ± 0.06 | |
| dihomo-γ-linoleic acid (DGLA) | 0.06 ± 0.02 | 0.04 ± 0.02 | 0.04 ± 0.02 | 0.06 ± 0.01 | 0.03 ± 0.01 | 0.11 ± 0.04 | 0.06 ± 0.06 | |
| arachidonic acid (AA) | 6.28 ± 0.36 | 5.92 ± 0.45 | 4.68 ± 0.32 * | 4.41 ± 0.26 * | 5.69 ± 0.37 | 4.73 ± 0.70 * | 4.37 ± 0.38 * | |
| n-3 PUFAs | α-linolenic acid (ALA) | 0.15 ± 0.06 | 0.12 ± 0.07 | 0.15 ± 0.02 | 0.52 ± 0.06 * | 0.32 ± 0.05 * | 0.46 ± 0.09 * | 0.75 ± 0.10 * |
| eicosapentaenoic acid (EPA) | 0.63 ± 0.14 | 0.51 ± 0.22 | 0.65 ± 0.20 | 0.73 ± 0.07 | 0.54 ± 0.06 | 0.77 ± 0.02 | 0.79 ± 0.02 | |
| docosaepentaenoic acid (DPA) | 2.21 ± 0.41 | 2.89 ± 0.11 | 2.53 ± 0.11 | 2.55 ± 0.11 | 2.82 ± 0.17 | 2.64 ± 0.35 | 2.57 ± 0.60 | |
| docosaehenanoic acid (DHA) | 4.60 ± 0.47 | 5.31 ± 1.03 | 5.04 ± 0.09 | 4.92 ± 0.78 | 5.05 ± 0.80 | 4.89 ± 0.53 | 4.60 ± 0.39 | |
| Total PUFAs | 18.58 ± 0.62 | 19.87 ± 0.14 * | 18.00 ± 0.15 | 18.52 ± 0.49 | 19.51 ± 0.59 | 18.49 ± 0.49 | 19.30 ± 0.29 | |
| Target Genes | Gene Symbol | Gene Aliases | ID Assay |
|---|---|---|---|
| Arachidonate 15-lipoxygenase | ALOX15 | 15-LOX-1, 15LOX-1 | qHsaCED0045954 |
| Peroxisome proliferator-activated receptor gamma | PPARG | CIMT1, GLM1, NR1C3, PPARG1, PPARG2, PPARgamma | qHsaCID0011718 |
| Actin, beta | ACTB | PS1TP5BP1 | qHsaCED0036269 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tutino, V.; Gigante, I.; Milella, R.A.; De Nunzio, V.; Flamini, R.; De Rosso, M.; Scavo, M.P.; Depalo, N.; Fanizza, E.; Caruso, M.G.; et al. Flavonoid and Non-Flavonoid Compounds of Autumn Royal and Egnatia Grape Skin Extracts Affect Membrane PUFA’s Profile and Cell Morphology in Human Colon Cancer Cell Lines. Molecules 2020, 25, 3352. https://doi.org/10.3390/molecules25153352
Tutino V, Gigante I, Milella RA, De Nunzio V, Flamini R, De Rosso M, Scavo MP, Depalo N, Fanizza E, Caruso MG, et al. Flavonoid and Non-Flavonoid Compounds of Autumn Royal and Egnatia Grape Skin Extracts Affect Membrane PUFA’s Profile and Cell Morphology in Human Colon Cancer Cell Lines. Molecules. 2020; 25(15):3352. https://doi.org/10.3390/molecules25153352
Chicago/Turabian StyleTutino, Valeria, Isabella Gigante, Rosa Anna Milella, Valentina De Nunzio, Riccardo Flamini, Mirko De Rosso, Maria Principia Scavo, Nicoletta Depalo, Elisabetta Fanizza, Maria Gabriella Caruso, and et al. 2020. "Flavonoid and Non-Flavonoid Compounds of Autumn Royal and Egnatia Grape Skin Extracts Affect Membrane PUFA’s Profile and Cell Morphology in Human Colon Cancer Cell Lines" Molecules 25, no. 15: 3352. https://doi.org/10.3390/molecules25153352
APA StyleTutino, V., Gigante, I., Milella, R. A., De Nunzio, V., Flamini, R., De Rosso, M., Scavo, M. P., Depalo, N., Fanizza, E., Caruso, M. G., & Notarnicola, M. (2020). Flavonoid and Non-Flavonoid Compounds of Autumn Royal and Egnatia Grape Skin Extracts Affect Membrane PUFA’s Profile and Cell Morphology in Human Colon Cancer Cell Lines. Molecules, 25(15), 3352. https://doi.org/10.3390/molecules25153352








