Hepatic Bile Acid Reuptake in the Rat Depends on Bile Acid Conjugation but Not on Agonistic Properties towards FXR and TGR5
Abstract
1. Introduction
2. Results
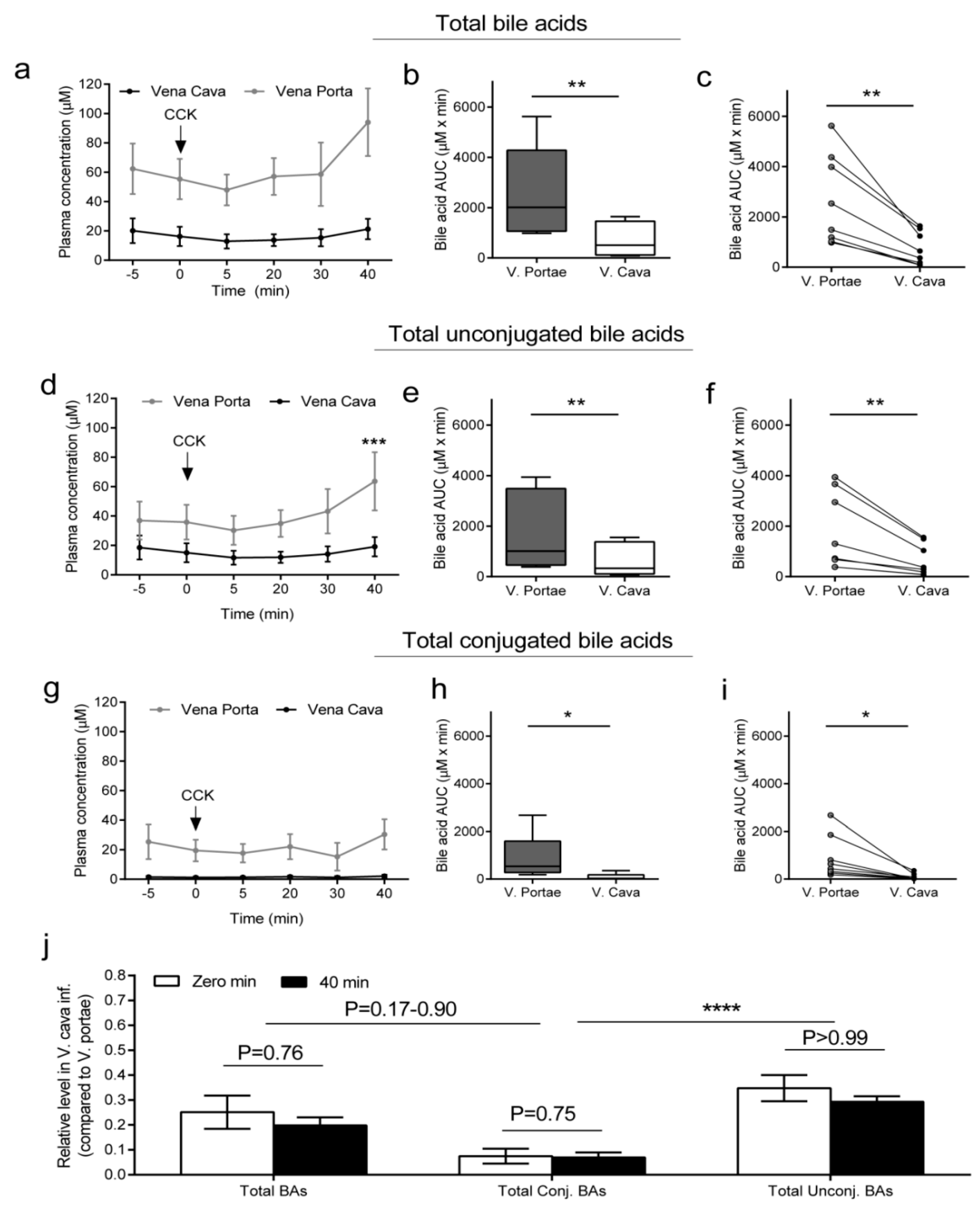
2.1. Total, Total Conjugated, and Total Unconjugated BA Concentrations in the Portal Vein and Vena Cava
2.2. Concentrations of Primary BAs in the Portal Vein and Vena Cava
2.3. Concentrations of Secondary BAs in the Portal Vein and Vena Cava
2.4. Concentrations of Murine Specific BAs in the Portal Vein and Vena Cava
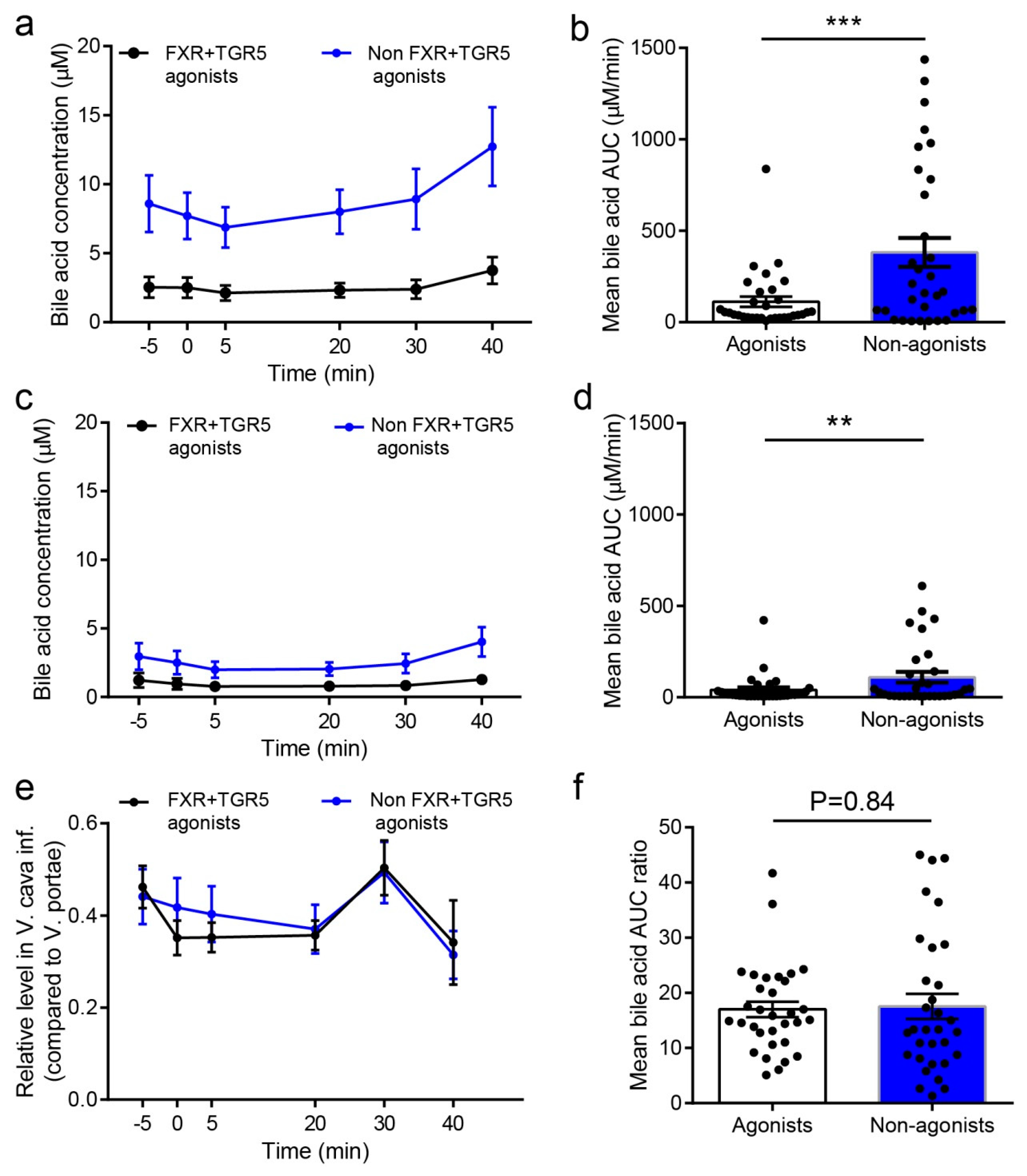
2.5. Concentration of FXR and TGR5 Agonistic BAs in Peripheral Blood is below Activating Concentrations, and Hepatic Extraction of BAs is Independent of Their Potency and Efficacy towards These Receptors
3. Discussion
4. Materials and Methods
4.1. Animal Studies
4.1.1. Ethical Considerations
4.1.2. In Vivo Study
4.2. Biochemical Measurements
4.3. Data Analysis and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cronholm, T.; Sjovall, J. Bile acids in portal blood of fats fed different diets and cholestyramine. Bile acids and steroids 189. Eur. J. Biochem. FEBS 1967, 2, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.W. Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 2009, 50, S120–S125. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y. Bile acid metabolism and signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, F.; Bloks, V.W.; Groen, A.K. Beyond intestinal soap-bile acids in metabolic control. Nat. Rev. Endocrinol. 2014, 10, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, P.; Cariou, B.; Lien, F.; Kuipers, F.; Staels, B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 2009, 89, 147–191. [Google Scholar] [CrossRef] [PubMed]
- Poole, D.P.; Godfrey, C.; Cattaruzza, F.; Cottrell, G.S.; Kirkland, J.G.; Pelayo, J.C.; Bunnett, N.W.; Corvera, C.U. Expression and function of the bile acid receptor GpBAR1 (TGR5) in the murine enteric nervous system. Neurogastroenterol. Motil. Off. J. Eur. Gastrointest. Motil. Soc. 2010, 22, 814–825, e227–e818. [Google Scholar] [CrossRef]
- Brighton, C.A.; Rievaj, J.; Kuhre, R.E.; Glass, L.L.; Schoonjans, K.; Holst, J.J.; Gribble, F.M.; Reimann, F. Bile acids trigger GLP-1 release predominantly by accessing basolaterally located g protein-coupled bile acid receptors. Endocrinology 2015, 156, 3961–3970. [Google Scholar] [CrossRef]
- Thomas, C.; Gioiello, A.; Noriega, L.; Strehle, A.; Oury, J.; Rizzo, G.; Macchiarulo, A.; Yamamoto, H.; Mataki, C.; Pruzanski, M.; et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009, 10, 167–177. [Google Scholar] [CrossRef]
- Ullmer, C.; Alvarez Sanchez, R.; Sprecher, U.; Raab, S.; Mattei, P.; Dehmlow, H.; Sewing, S.; Iglesias, A.; Beauchamp, J.; Conde-Knape, K.; et al. Systemic bile acid sensing by G protein-coupled bile acid receptor 1 (GPBAR1) promotes PYY and GLP-1 release. Br. J. Pharmacol. 2013, 169, 671–684. [Google Scholar] [CrossRef]
- Sinal, C.J.; Tohkin, M.; Miyata, M.; Ward, J.M.; Lambert, G.; Gonzalez, F.J. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 2000, 102, 731–744. [Google Scholar] [CrossRef]
- Edwards, P.A.; Kast, H.R.; Anisfeld, A.M. BAREing it all: The adoption of LXR and FXR and their roles in lipid homeostasis. J. Lipid Res. 2002, 43, 2–12. [Google Scholar] [PubMed]
- Davis, R.A.; Attie, A.D. Deletion of the ileal basolateral bile acid transporter identifies the cellular sentinels that regulate the bile acid pool. Proc. Natl. Acad. Sci. USA 2008, 105, 4965–4966. [Google Scholar] [CrossRef] [PubMed]
- Vassileva, G.; Golovko, A.; Markowitz, L.; Abbondanzo, S.J.; Zeng, M.; Yang, S.; Hoos, L.; Tetzloff, G.; Levitan, D.; Murgolo, N.J.; et al. Targeted deletion of Gpbar1 protects mice from cholesterol gallstone formation. Biochem. J. 2006, 398, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Tanaka, K.; Suzuki, J.; Miyoshi, H.; Harada, N.; Nakamura, T.; Miyamoto, Y.; Kanatani, A.; Tamai, Y. Targeted disruption of G protein-coupled bile acid receptor 1 (Gpbar1/M-Bar) in mice. J. Endocrinol. 2006, 191, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Keitel, V.; Gorg, B.; Bidmon, H.J.; Zemtsova, I.; Spomer, L.; Zilles, K.; Haussinger, D. The bile acid receptor TGR5 (Gpbar-1) acts as a neurosteroid receptor in brain. Glia 2010, 58, 1794–1805. [Google Scholar] [CrossRef]
- Alemi, F.; Kwon, E.; Poole, D.P.; Lieu, T.; Lyo, V.; Cattaruzza, F.; Cevikbas, F.; Steinhoff, M.; Nassini, R.; Materazzi, S.; et al. The TGR5 receptor mediates bile acid-induced itch and analgesia. J. Clin. Investig. 2013, 123, 1513–1530. [Google Scholar] [CrossRef]
- Duboc, H.; Tache, Y.; Hofmann, A.F. The bile acid TGR5 membrane receptor: From basic research to clinical application. Dig. Liver Dis. Off. J. Ital. Soc. Gastroenterol. Ital. Assoc. Study Liver 2014, 46, 302–312. [Google Scholar] [CrossRef]
- Bookout, A.L.; Jeong, Y.; Downes, M.; Yu, R.T.; Evans, R.M.; Mangelsdorf, D.J. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 2006, 126, 789–799. [Google Scholar] [CrossRef]
- Bishop-Bailey, D.; Walsh, D.T.; Warner, T.D. Expression and activation of the farnesoid X receptor in the vasculature. Proc. Natl. Acad. Sci. USA 2004, 101, 3668–3673. [Google Scholar] [CrossRef]
- Sonne, D.P.; van Nierop, F.S.; Kulik, W.; Soeters, M.R.; Vilsboll, T.; Knop, F.K. Postprandial plasma concentrations of individual bile acids and FGF-19 in patients with type 2 diabetes. J. Clin. Endocrinol. Metab. 2016, 101, 3002–3009. [Google Scholar] [CrossRef]
- Angelin, B.; Bjorkhem, I.; Einarsson, K.; Ewerth, S. Hepatic uptake of bile acids in man. Fasting and postprandial concentrations of individual bile acids in portal venous and systemic blood serum. J. Clin. Investig. 1982, 70, 724–731. [Google Scholar] [CrossRef] [PubMed]
- Kuhre, R.E.; Wewer Albrechtsen, N.J.; Larsen, O.; Jepsen, S.L.; Balk-Moller, E.; Andersen, D.B.; Deacon, C.F.; Schoonjans, K.; Reimann, F.; Gribble, F.M.; et al. Bile acids are important direct and indirect regulators of the secretion of appetite- and metabolism-regulating hormones from the gut and pancreas. Mol. Metab. 2018. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Miyamoto, Y.; Nakamura, T.; Tamai, Y.; Okada, H.; Sugiyama, E.; Nakamura, T.; Itadani, H.; Tanaka, K. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 2002, 298, 714–719. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.; Hollister, K.; Sowers, L.C.; Forman, B.M. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 1999, 3, 543–553. [Google Scholar] [CrossRef]
- Lew, J.L.; Zhao, A.; Yu, J.; Huang, L.; De Pedro, N.; Pelaez, F.; Wright, S.D.; Cui, J. The farnesoid X receptor controls gene expression in a ligand- and promoter-selective fashion. J. Biol. Chem. 2004, 279, 8856–8861. [Google Scholar] [CrossRef]
- Parks, D.J.; Blanchard, S.G.; Bledsoe, R.K.; Chandra, G.; Consler, T.G.; Kliewer, S.A.; Stimmel, J.B.; Willson, T.M.; Zavacki, A.M.; Moore, D.D.; et al. Bile acids: Natural ligands for an orphan nuclear receptor. Science (New York, NY) 1999, 284, 1365–1368. [Google Scholar] [CrossRef]
- Chiang, J.Y.L. Bile acids: Regulation of synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef]
- Kawamata, Y.; Fujii, R.; Hosoya, M.; Harada, M.; Yoshida, H.; Miwa, M.; Fukusumi, S.; Habata, Y.; Itoh, T.; Shintani, Y.; et al. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 2003, 278, 9435–9440. [Google Scholar] [CrossRef]
- Jorgensen, N.B.; Dirksen, C.; Bojsen-Moller, K.N.; Kristiansen, V.B.; Wulff, B.S.; Rainteau, D.; Humbert, L.; Rehfeld, J.F.; Holst, J.J.; Madsbad, S.; et al. Improvements in glucose metabolism early after gastric bypass surgery are not explained by increases in total bile acids and fibroblast growth factor 19 concentrations. J. Clin. Endocrinol. Metab. 2015, 100, E396–E406. [Google Scholar] [CrossRef]
- Patti, M.E.; Houten, S.M.; Bianco, A.C.; Bernier, R.; Larsen, P.R.; Holst, J.J.; Badman, M.K.; Maratos-Flier, E.; Mun, E.C.; Pihlajamaki, J.; et al. Serum bile acids are higher in humans with prior gastric bypass: Potential contribution to improved glucose and lipid metabolism. Obesity (Silver Spring, MD) 2009, 17, 1671–1677. [Google Scholar] [CrossRef]
- Ahlberg, J.; Angelin, B.; Bjorkhem, I.; Einarsson, K. Individual bile acids in portal venous and systemic blood serum of fasting man. Gastroenterology 1977, 73, 1377–1382. [Google Scholar] [CrossRef]
- Eggink, H.M.; van Nierop, F.S.; Schooneman, M.G.; Boelen, A.; Kalsbeek, A.; Koehorst, M.; Ten Have, G.A.M.; de Brauw, L.M.; Groen, A.K.; Romijn, J.A.; et al. Transhepatic bile acid kinetics in pigs and humans. Clin. Nutr. (Edinb. Scotl.) 2018, 37, 1406–1414. [Google Scholar] [CrossRef] [PubMed]
- Portman, O.W.; Shah, S. The determination of concentrations of bile acids in peripheral, portal and hepatic blood of Cebus monkeys. Arch. Biochem. Biophys. 1962, 96, 516–523. [Google Scholar] [CrossRef]
- Dawson, P.A.; Lan, T.; Rao, A. Bile acid transporters. J. Lipid Res. 2009, 50, 2340–2357. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, K.; Ichimiya, H.; Higashijima, H.; Yamashita, H.; Kuroki, S.; Chijiiwa, K.; Tanaka, M. Regulation of bile acid synthesis in the rat: Relationship between hepatic cholesterol 7 alpha-hydroxylase activity and portal bile acids. J. Lipid Res. 1995, 36, 315–321. [Google Scholar]
- Takahashi, S.; Fukami, T.; Masuo, Y.; Brocker, C.N.; Xie, C.; Krausz, K.W.; Wolf, C.R.; Henderson, C.J.; Gonzalez, F.J. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 2016, 57, 2130–2137. [Google Scholar] [CrossRef]
- Steiner, C.; Othman, A.; Saely, C.H.; Rein, P.; Drexel, H.; von Eckardstein, A.; Rentsch, K.M. Bile acid metabolites in serum: Intraindividual variation and associations with coronary heart disease, metabolic syndrome and diabetes mellitus. PLoS ONE 2011, 6, e25006. [Google Scholar] [CrossRef]
- LaRusso, N.F.; Hoffman, N.E.; Korman, M.G.; Hofmann, A.F.; Cowen, A.E. Determinants of fasting and postprandial serum bile acid levels in healthy man. Am. J. Dig. Dis. 1978, 23, 385–391. [Google Scholar] [CrossRef]
- Eggink, H.M.; Oosterman, J.E.; de Goede, P.; de Vries, E.M.; Foppen, E.; Koehorst, M.; Groen, A.K.; Boelen, A.; Romijn, J.A.; la Fleur, S.E.; et al. Complex interaction between circadian rhythm and diet on bile acid homeostasis in male rats. Chronobiol. Int. 2017, 34, 1339–1353. [Google Scholar] [CrossRef]
- Lee, H.B.; Blaufox, M.D. Blood volume in the rat. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 1985, 26, 72–76. [Google Scholar]
- Morville, T.; Sahl, R.E.; Trammell, S.A.; Svenningsen, J.S.; Gillum, M.P.; Helge, J.W.; Clemmensen, C. Divergent effects of resistance and endurance exercise on plasma bile acids, FGF19, and FGF21 in humans. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Sayin, S.I.; Wahlstrom, A.; Felin, J.; Jantti, S.; Marschall, H.U.; Bamberg, K.; Angelin, B.; Hyotylainen, T.; Oresic, M.; Backhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
| Plasma from Vena Portae | Plasma from Vena Cava Inferior | ||||||
|---|---|---|---|---|---|---|---|
| Bile acid | Baseline (µM) | 40 min (µM) | AUC (µM x min) | Baseline (µM) | 40 min (µM) | AUC (µM x min) | Ratio: V. Cava/V. Portae |
| Collected | |||||||
| Total | 58.9 ± 10.7 | 87.4 ± 25.1 | 2650 ± 634 | 18.3 ± 5.18 | 28.0 ± 8.68 | 726 ± 231 ** | 0.25 ± 0.07 (0 min) 0.20 ± 0.03 (40 min) |
| Total conjugated | 22.5 ± 9.32 | 30.4 ± 10.1 | 899 ± 317 | 1.43 ± 0.57 | 2.18 ± 0.91 | 92.4 ± 45.1 * | 0.08 ± 0.03 (0 min) 0.07 ± 0.02 (40 min) |
| Total unconjugated | 36.4 ± 12.1 | 63.7 ± 19.8 | 1752 ± 535 | 16.8 ± 7.30 | 19.1 ± 6.56 | 636 ± 222 ** | 0.35 ± 0.05 (0 min) 0.29 ± 0.02 (40 min) |
| Primary BAs | |||||||
| CA | 13.6 ± 4.10 | 21.9 ± 5.87 | 190 ± 50.4 | 4.67 ± 1.73 | 5.54 ± 1.35 | 658 ± 168 ** | 0.30 ± 0.03 (0 min) 0.28 ± 0.02 (40 min) |
| TCA | 10.5 ± 4.39 | 12.6 ± 4.38 | 404 ± 153 | 0.56 ± 0.19 | 0.81 ± 0.32 | 37.4 ± 17.2 * | 0.08 ± 0.03 (0 min) 0.07 ± 0.02 (40 min) |
| CDCA | 4.41 ± 2.67 | 6.50 ± 3.21 | 186 ± 99.4 | 2.89 ± 1.18 | 2.34 ± 1.01 | 93.3 ± 50.2 | 0.61 ± 0.06 (0 min) 0.43 ± 0.03 (40 min) |
| TCDCA | 2.30 ± 0.91 | 3.34 ± 0.96 | 95.1 ± 25.8 | 0.31 ± 0.08 | 0.37 ± 0.09 | 16.0 ± 5.31 ** | 0.19 ± 0.03 (0 min) 0.13 ± 0.01 (40 min) |
| Secondary BAs | |||||||
| DCA | 2.45 ± 0.85 | 4.87 ± 1.66 | 128 ± 43.4 | 0.97 ± 0.35 | 1.14 ± 0.35 | 43.2 ± 12.9 * | 0.43 ± 0.03 (0 min) 0.33 ± 0.08 (40 min) |
| TDCA | 0.92 ± 0.18 | 1.34 ± 0.39 | 39.4 ± 10.5 | 0.18 ± 0.17 | 0.20 ± 0.035 | 8.86 ± 1.20 | 0.25 ± 0.04 (0 min) 0.19 ± 0.03 (40 min) |
| UDCA | 7.02 ± 3.28 | 15.7 ± 7.60 | 387 ± 179 | 5.44 ± 2.89 | 6.43 ± 3.285 | 197 ± 91.3 * | 0.25 ± 0.04 (0 min) 0.19 ± 0.03 (40 min) |
| TUDCA | 1.58 ± 0.81 | 3.69 ± 1.80 | 387 ± 179 | 0.26 ± 0.07 | 0.40 ± 0.19 | 197 ± 91.3 | 0.47 ± 0.13 (0 min) 0.39 ± 0.13 (40 min) |
| Murine specific BAs | |||||||
| MCA alpha | 1.95 ± 0.87 | 4.23 ± 1.77 | 97.4 ± 39.4 | 0.93 ± 0.44 | 1.00 ± 0.42 | 34.8 ± 13.6 * | 0.46 ± 0.04 (0 min) 0.37 ± 0.06 (40 min) |
| MCA beta | 2.65 ± 0.94 | 6.42 ± 2.02 | 144 ± 46.3 | 1.26 ± 0.49 | 1.85 ± 0.58 | 60.0 ± 15.9 * | 0.42 ± 0.04 (0 min) 0.69 ± 0.37 (40 min) |
| TMCA alpha | 2.01 ± 0.82 | 2.98 ± 0.91 | 81.9 ± 25.7 | 0.29 ± 0.08 | 0.36 ± 0.10 | 14.9 ± 5.09 | 0.25 ± 0.05 (0 min) 0.16 ± 0.03 (40 min) |
| TMCA beta | 5.29 ± 2.60 | 6.52 ± 2.22 | 203 ± 78.1 | 0.25 ± 0.07 | 0.39 ± 0.14 | 20.7 ± 10.9 ** | 0.16 ± 0.06 (0 min) 0.08 ± 0.01 (40 min) |
| TGR5/FXR | |||||||
| Agonists | 2.52 ± 0.74 | 3.75 ± 0.97 | 112 ± 28.3 | 1.09 ± 0.48 | 1.27 ± 0.33 | 40.4 ± 13.7 ** | 0.38 ± 0.04 (0 min) 0.25 ± 0.03 (40 min) |
| Non-agonists | 8.73 ± 1.64 | 12.7 ± 2.85 | 382 ± 78.6 | 2.74 ± 0.91 | 4.03 ± 1.06 | 110 ± 29.4 *** | 0.25 ± 0.03 (0 min) 0.31 ± 0.05 (40 min) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trammell, S.A.J.; Svenningsen, J.S.; Holst, J.J.; Gillum, M.P.; Kuhre, R.E. Hepatic Bile Acid Reuptake in the Rat Depends on Bile Acid Conjugation but Not on Agonistic Properties towards FXR and TGR5. Molecules 2020, 25, 2371. https://doi.org/10.3390/molecules25102371
Trammell SAJ, Svenningsen JS, Holst JJ, Gillum MP, Kuhre RE. Hepatic Bile Acid Reuptake in the Rat Depends on Bile Acid Conjugation but Not on Agonistic Properties towards FXR and TGR5. Molecules. 2020; 25(10):2371. https://doi.org/10.3390/molecules25102371
Chicago/Turabian StyleTrammell, Samuel A. J., Jens S. Svenningsen, Jens J. Holst, Matthew P. Gillum, and Rune E. Kuhre. 2020. "Hepatic Bile Acid Reuptake in the Rat Depends on Bile Acid Conjugation but Not on Agonistic Properties towards FXR and TGR5" Molecules 25, no. 10: 2371. https://doi.org/10.3390/molecules25102371
APA StyleTrammell, S. A. J., Svenningsen, J. S., Holst, J. J., Gillum, M. P., & Kuhre, R. E. (2020). Hepatic Bile Acid Reuptake in the Rat Depends on Bile Acid Conjugation but Not on Agonistic Properties towards FXR and TGR5. Molecules, 25(10), 2371. https://doi.org/10.3390/molecules25102371







