Biodegradation of Organophosphorus Compounds Predicted by Enzymatic Process Using Molecular Modelling and Observed in Soil Samples Through Analytical Techniques and Microbiological Analysis: A Comparison
Abstract
1. Introduction
2. Results and Discussion
2.1. Molecular Docking Studies
2.2. Molecular Dynamics Study
2.3. Total Cell Count and Recovering from Soil Sample
2.4. Methodological Analytical Parameters
2.5. Microbial Degradation Study
3. Materials and Methods
3.1. Molecular Docking Studies
3.2. Molecular Dynamics Studies
3.3. Spiking Chemicals
3.4. Soil Samples
3.4.1. Sampling and Initial Preparation of Soil Samples
3.4.2. Analysis of the Physicochemical Characteristics
3.4.3. Sterilization Process
3.4.4. Contamination with Spiking Chemicals
3.5. Time Degradation Experiment
3.6. Microbial Viability in Soil
3.7. Gas Chromatography and Mass Spectrometry Analyses
3.7.1. Soil Sample Preparation for Analysis
3.7.2. Instrumentation and Analysis
3.7.3. Internal Verification for the Methodology of Analysis
3.7.4. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Singh, B.K.; Walker, A. Microbial degradation of organophosphorus compounds. FEMS Microbiol. Rev. 2006, 30, 428–471. [Google Scholar] [CrossRef] [PubMed]
- Sogorb, M.A.; Vilanova, E. Enzymes involved in the detoxification of organophosphorus, carbamate and pyrethroid insecticides through hydrolysis. Toxicol. Lett. 2002, 128, 215–228. [Google Scholar] [CrossRef]
- De Paula, R.L.; De Almeida, J.S.F.D.; Cavalcante, S.F.A.; Gonçalves, A.S.; Simas, A.B.C.; Franca, T.C.C.; Valis, M.; Kuca, K.; Nepovimova, E.; Granjeiro, J.M. Molecular Modeling and In Vitro Studies of a Neutral Oxime as a Potential Reactivator for Acetylcholinesterase Inhibited by Paraoxon. Molecules 2018, 23, 2954. [Google Scholar] [CrossRef]
- Schecter, W.P. Cholinergic symptoms due to nerve agent attack: A strategy for management. Anesthesiol. Clin. N. Am. 2004, 22, 579–590. [Google Scholar] [CrossRef]
- Deng, S.; Chen, Y.; Wang, D.; Shi, T.; Wu, X.; Ma, X.; Li, X.; Hua, R.; Tang, X.; Li, Q.X. Rapid biodegradation of organophosphorus pesticides by Stenotrophomonas sp: G1. J. Hazard. Mater. 2015, 297, 17–24. [Google Scholar] [CrossRef]
- Arias-Estévez, M.; López-Periago, E.; Martínez-Carballo, E.; Simal-Gándara, J.; Mejuto, J.C.; García-Río, L. The mobility and degradation of pesticides in soils and the pollution of groundwater resources. Agric. Ecosyst. Environ. 2008, 123, 247–260. [Google Scholar] [CrossRef]
- Da Rocha Voris, D.G.; dos Santos Dias, L.; Lima, J.A.; Lima, K.D.; Lima, J.B.; dos Santos Lima, A.L. Evaluation of larvicidal, adulticidal, and anticholinesterase activities of essential oils of Illicium verum Hook. f., Pimenta dioica (L.) Merr., and Myristica fragrans Houtt. against Zika virus vectors. Environ. Sci. Pollut. Res. 2018, 25, 22541–22551. [Google Scholar] [CrossRef]
- Kitagawa, D.A.S.; de Cavalcante, S.F.; de Paula, R.L.; Rodrigues, R.B.; Bernardo, L.B.; da Silva, M.C.J.; da Silva, T.N.; dos Santos, W.V.; Granjeiro, J.M.; de Almeida, J.S.F.D.; et al. In Vitro Evaluation of Neutral Aryloximes as Reactivators for Electrophorus eel Acetylcholinesterase Inhibited by Paraoxon. Biomolecules 2019, 9, 583. [Google Scholar] [CrossRef]
- Ramalho, T.C.; de Castro, A.C.; Silva, D.R.; Silva, M.C.; Franca, T.C.C.; Bennion, B.J.; Kuca, K. Computational Enzymology and Organophosphorus Degrading Enzymes: Promising Approaches Toward Remediation Technologies of Warfare Agents and Pesticides. Curr. Med. Chem. 2016, 23, 1041–1061. [Google Scholar] [CrossRef]
- OPCW. Preventing the Re-Emergence of Chemical Weapons. Available online: https://www.opcw.org/our-work/preventing-re-emergence-chemical-weapons (accessed on 10 September 2019).
- Bryce, D. CBRN: Latest Incidents around the world. Def. IQ. 2019. Available online: http://www.defenceiq.com/events-cbrn-conference/downloads/cbrn-threats-latest-incidents-around-the-globe (accessed on 27 September 2019).
- OPCW. OPCW by the Numbers. Available online: https://www.opcw.org/media-centre/opcw-numbers (accessed on 9 September 2019).
- OPCW. Eliminating Chemical Weapons Committed to Complete and Verifiable Destruction. Available online: https://www.opcw.org/our-work/eliminating-chemical-weapons (accessed on 9 September 2019).
- Karigar, C.S.; Rao, S.S. Role of microbial enzymes in the bioremediation of pollutants: A review. Enzyme Res. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Copley, S.D. Evolution of Efficient Pathways for Degradation of Anthropogenic Chemicals. Nat. Chem. Biol. 2009, 5, 559–566. [Google Scholar] [CrossRef]
- Souza, R.C.; Cantão, M.E.; Nogueira, M.A.; Vasconcelos, A.T.R.; Hungria, M. Outstanding impact of soil tillage on the abundance of soil hydrolases revealed by a metagenomic approach. Braz. J. Microbiol. 2018, 49, 723–730. [Google Scholar] [CrossRef]
- Benning, M.M.; Kuo, J.M.; Raushel, F.M.; Holden, H.M. Three-Dimensional Structure of the Binuclear Metal Center of Phosphotriesterase. Biochemistry 1995, 34, 7973–7978. [Google Scholar] [CrossRef]
- Iyer, R.; Iken, B.; Damania, A. A comparison of organophosphate degradation genes and bioremediation applications. Environ. Microbiol. Rep. 2013, 5, 787–798. [Google Scholar] [CrossRef]
- Sikora, L.J.; Kaufman, D.D.; Horng, L.C. Enzyme activity in soils showing enhanced degradation of organophosphate insecticides. Biol. Fertil. Soils 1990, 9, 14–18. [Google Scholar] [CrossRef]
- Zheng, Y.; Long, L.; Fan, Y.; Gan, J.; Fang, J.; Jin, W. A review on the detoxification of organophosphorus compounds by microorganisms. Afr. J. Microbiol. Res. 2013, 7, 2127–2134. [Google Scholar]
- Theriot, C.M.; Grunden, A.M. Hydrolysis of organophosphorus compounds by microbial enzymes. Appl. Microbiol. Biotechnol. 2011, 89, 35–43. [Google Scholar] [CrossRef]
- Firozjaei, S.A.A.; Latifi, A.M.; Khodi, S.; Abolmaali, S.; Choopani, A. A review on biodegradation of toxic organophosphate compounds. J. Appl. Biotechnol. Rep. 2015, 2, 215–224. [Google Scholar]
- De Castro, A.A.; Assis, L.C.; Silva, D.R.; Corrêa, S.; Assis, T.M.; Gajo, G.C.; Soares, F.V.; Ramalho, T.C. Computational enzymology for degradation of chemical warfare agents: Promising technologies for remediation processes. AIMS Microbiol. 2017, 3, 108–135. [Google Scholar] [CrossRef]
- Omburo, G.A.; Mullins, L.S.; Raushel, F.M. Structural Characterization of the Divalent Cation Sites of Bacterial Phosphotriesterase by 113Cd NMR Spectroscopy. Biochemistry 1993, 32, 9148–9155. [Google Scholar] [CrossRef]
- Omburo, G.A.; Kuo, J.M.; Mullins, L.S.; Raushels, F.M. Characterization of the Zinc Binding Site of Bacterial Phosphotriesterase. J. Biol. Chem. 1992, 267, 13278–13283. [Google Scholar]
- Zhang, X.; Wu, R.; Song, L.; Lin, Y.; Lin, M.; Cao, Z.; Wu, W.; Mo, Y. Molecular Dynamics Simulations of the Detoxification of Paraoxon Catalyzed by Phosphotriesterase (PTE). J. Comput. Chem. 2009, 30, 2388–2401. [Google Scholar] [CrossRef]
- Raymond, J.W.; Rogers, T.N.; Shonnard, D.R.; Kline, A.A. A review of structure-based biodegradation estimation methods. J. Hazard. Mater. 2001, 84, 189–215. [Google Scholar] [CrossRef]
- Loos, M.; Krauss, M.; Fenner, K. Pesticide nonextractable residue formation in soil: Insights from inverse modeling of degradation time series. Environ. Sci. Technol. 2012, 46, 9830–9837. [Google Scholar] [CrossRef]
- OECD. Revised Introduction to the OECD Guidelines for Testing of Chemicals, Section 3; OECD Guidelines for the Testing of Chemicals, Section 3; OECD Publishing: Paris, France, 2006. [Google Scholar]
- Kontoyianni, M.; Mcclellan, L.M.; Sokol, G.S. Evaluation of Docking Performance: Comparative Data on Docking Algorithms. J. Med. Chem. 2004, 47, 558–565. [Google Scholar] [CrossRef]
- Pal, R.; Chakrabarti, K.; Chakraborty, A.; Chowdhury, A. Degradation and Effects of Pesticides on Soil Microbiological Parameters-a Review. Int. J. Agric. Res. 2006, 1, 240–258. [Google Scholar]
- EPA Predictive Models and Tools for Assessing Chemicals under the Toxic Substances Control Act (TSCA). Available online: https://www.epa.gov/tsca-screening-tools (accessed on 17 September 2019).
- Gianfreda, L.; Rao, M.A. Interactions Between Xenobiotics and Microbial and Enzymatic Soil Activity. Crit. Rev. Environ. Sci. Technol. 2008, 38, 269–310. [Google Scholar] [CrossRef]
- Thomsen, R.; Christensen, M.H. MolDock: A New Technique for High-Accuracy Molecular Docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef]
- Hehre, W.; Ohlinger, S.; Klunzinger, P.; Deppmeier, B.; Driessen, A.; Johnson, J.; Ohsan, P. Spartan’08 Tutorial and User’s Guide; Q-CHEM, INC.: Irvine, CA, USA, 2006; ISBN 978-1-890661-38-4. [Google Scholar]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J. Am. Chem. Soc. 1996, 7863, 11225–11236. [Google Scholar] [CrossRef]
- Sousa, A.W.; Vranken, W.F. Open Access ACPYPE—AnteChamber PYthon Parser interface. BMC Res. Notes 2012, 5, 367. [Google Scholar]
- James, M.; Murtola, T.; Schulz, R.; Smith, J.C.; Hess, B.; Lindahl, E. ScienceDirect GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Cavalcante, S.F.A.; Kitagawa, D.A.S.; Rodrigues, R.B.; Silva, T.C.; Bernardo, L.B.; Correa, A.B.A.; Simas, A.B.C. One-Pot Synthesis of NEMP, a VX Surrogate, and Reactivation of NEMP-Inhibited Electrophorus Eel Acetylcholinesterase by Current Antidotes. J. Braz. Chem. Soc. 2019, 30, 1095–1102. [Google Scholar] [CrossRef]
- Salgado, J.R.S. Sistematização, avaliação e teste de protocolo para detecção de esporos de Bacillus anthracis em solo, Universidade Federal do Rio de Janeiro. 2018. Available online: http://www.sibi.ufrj.br (accessed on 28 August 2019).
- Paixão, M.S.G. Analyses of Accuracy of Estimates of Water Table Positioning and of Soil Water Contents by Using Shallow Seismic Refraction and Georadar from a Study at USP’s Campus, São Paulo/SP, Universidade de São Paulo. 2005. Available online: http://www.teses.usp.br/teses/disponiveis/14/ 14132/tde-31032006-192248/pt-br.php (accessed on 22 August 2019).
- García-Gaines, R.A.; Frankenstein, S. USCS and the USDA Soil Classification System, ERDC/CRREL TR-15-4; ERDC/CRREL: Vicksburg, MS, USA, 2015. [Google Scholar]
- ANVISA Farmacopeia Brasileira, Quinta Edição. Available online: http://portal.anvisa.gov.br/documents/33832/260079/5a+edição+-+Volume+1/4c530f86-fe83-4c4a-b907-6a96b5c2d2fc (accessed on 24 September 2018).
- Vieira, F.C.S.; Nahas, E. Quantificação de bactérias totais e esporuladas no solo. Sci. Agric. 2000, 57, 539–545. [Google Scholar] [CrossRef]
- Gravett, M.R.; Hopkins, F.B.; Main, M.J.; Self, A.J.; Timperley, C.M.; Webba, A.J.; Baker, M.J. Detection of the organophosphorus nerve agent VX and its hydrolysis products in white mustard plants grown in contaminated soil. Anal. Methods 2013, 5, 50–53. [Google Scholar] [CrossRef]
- Gravett, M.R.; Hopkins, F.B.; Self, A.J.; Webb, A.J.; Timperley, C.M.; Baker, M.J. Evidence of VX nerve agent use from contaminated white mustard plants. Proc. R. Soc. A 2014, 470, 1–14. [Google Scholar] [CrossRef]
- Ribani, M.; Beatriz, C.; Bottoli, G.; Collins, C.H.; Jardim, I.C.S.F. Validação em Métodos Cromatográficos e Eletroforéticos. Quim. Nova 2004, 27, 771–780. [Google Scholar] [CrossRef]
- Witkiewicz, Z.; Sliwka, E.; Neffe, S. Chromatographic analysis of chemical compounds related to the Chemical Weapons Convention. Trends Anal. Chem. 2016, 85, 21–33. [Google Scholar] [CrossRef]
Sample Availability: None. |
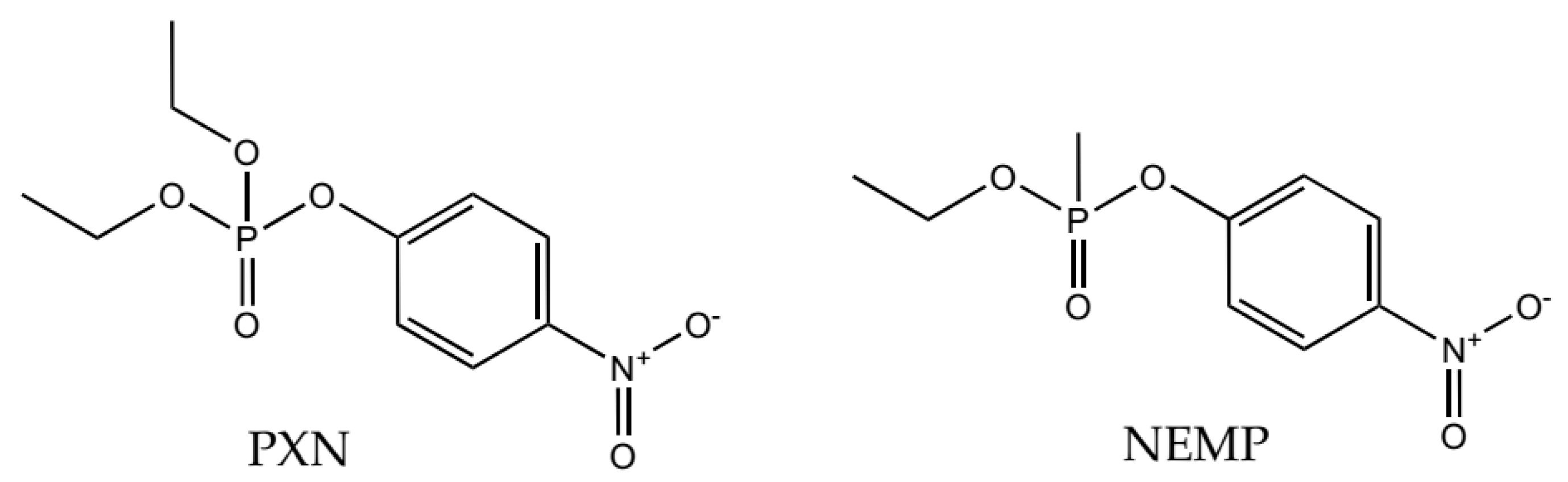

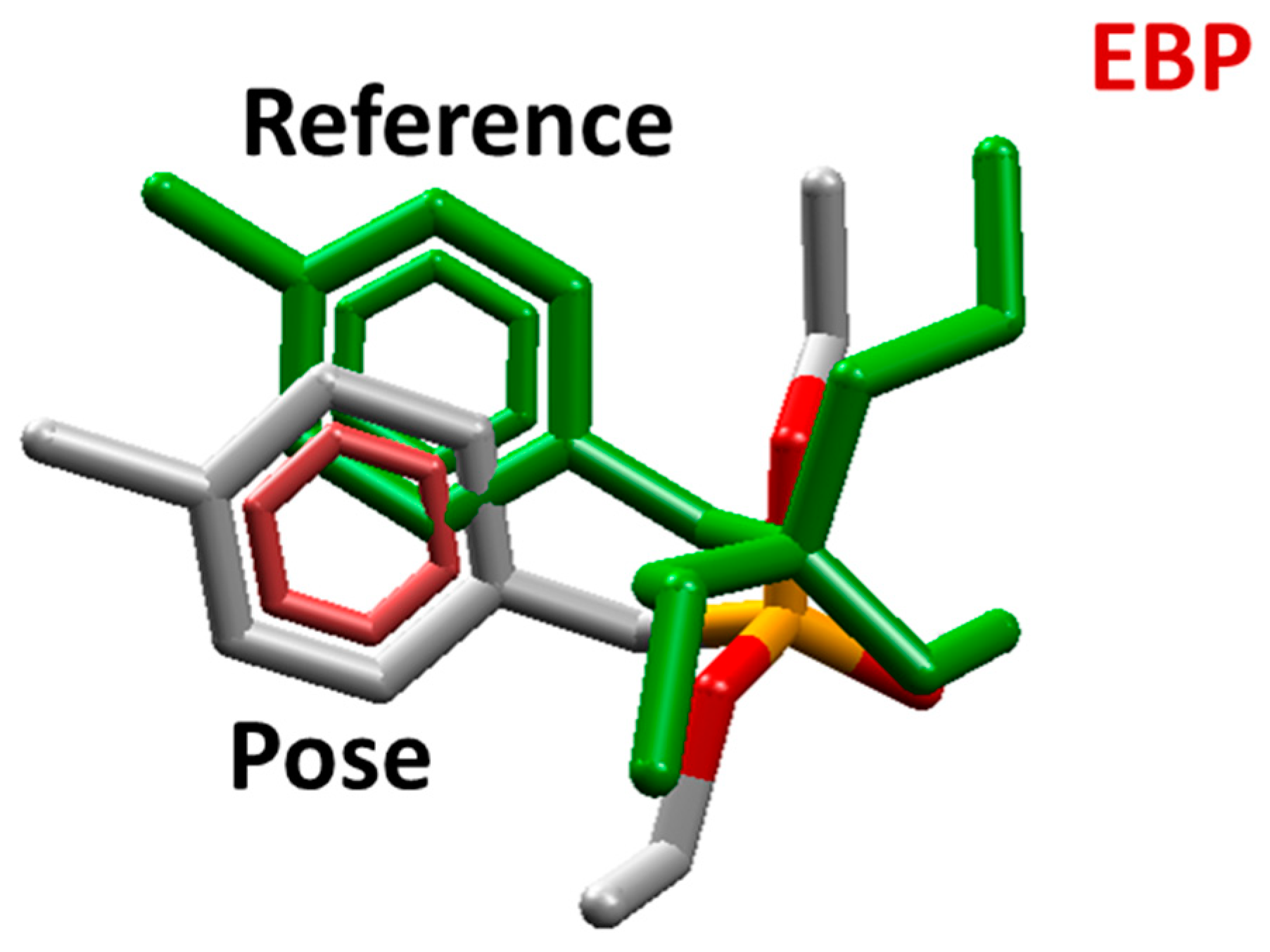
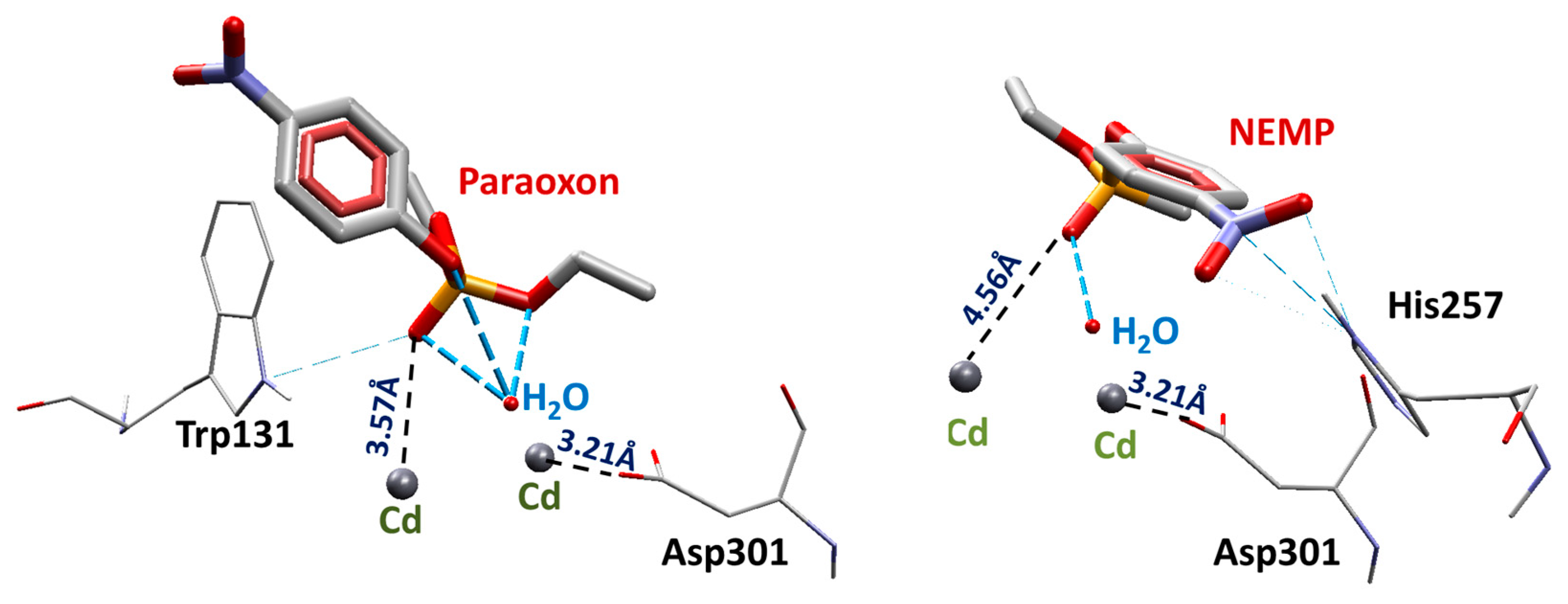
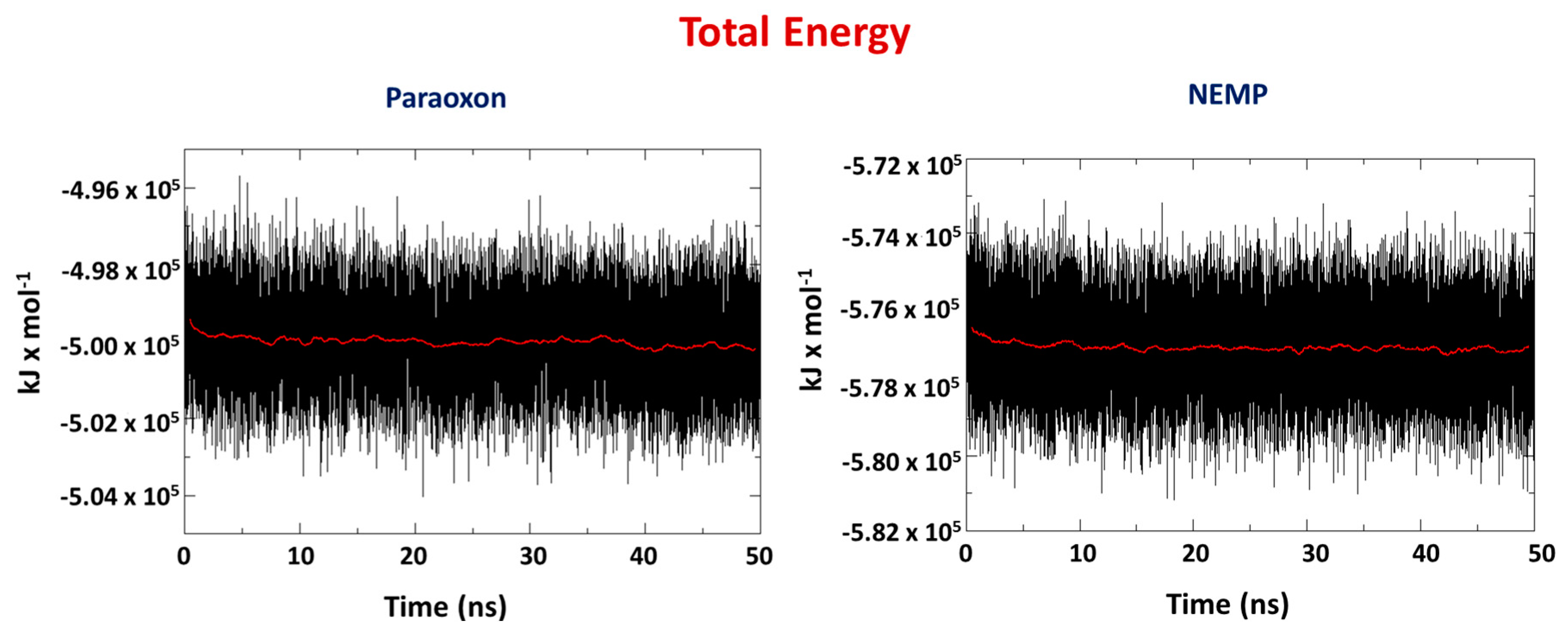
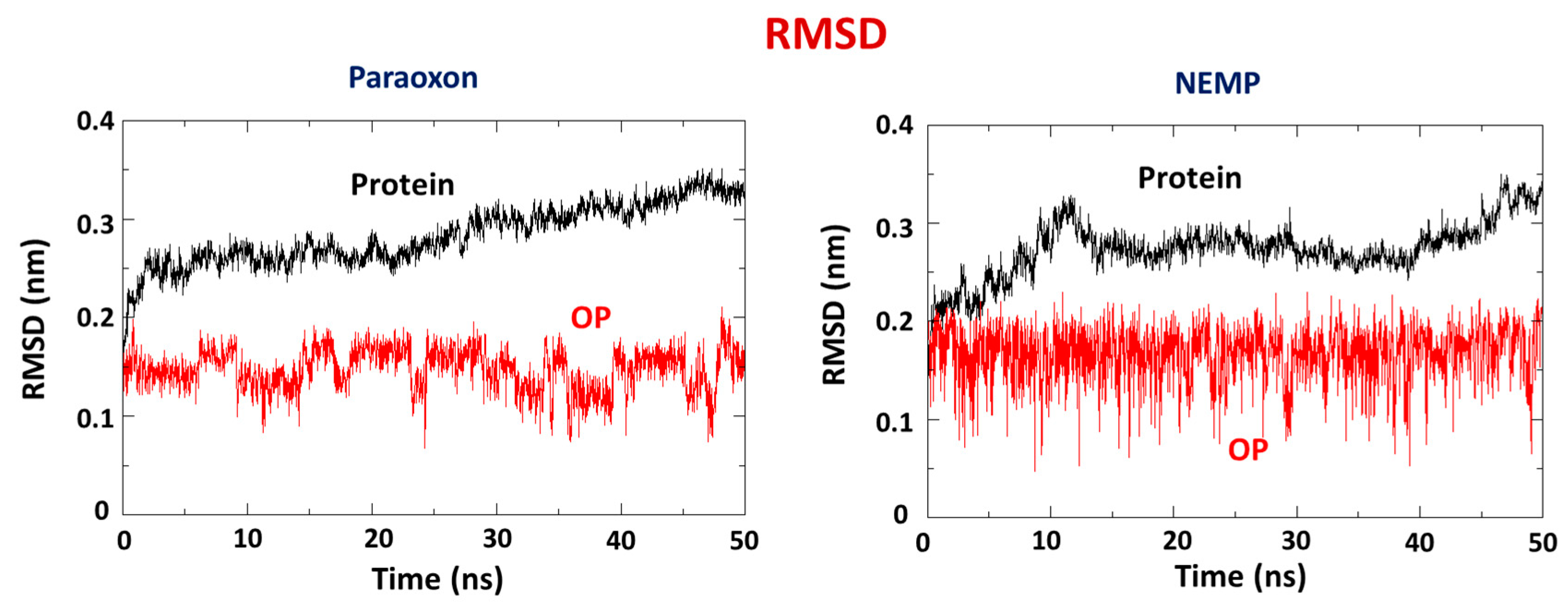
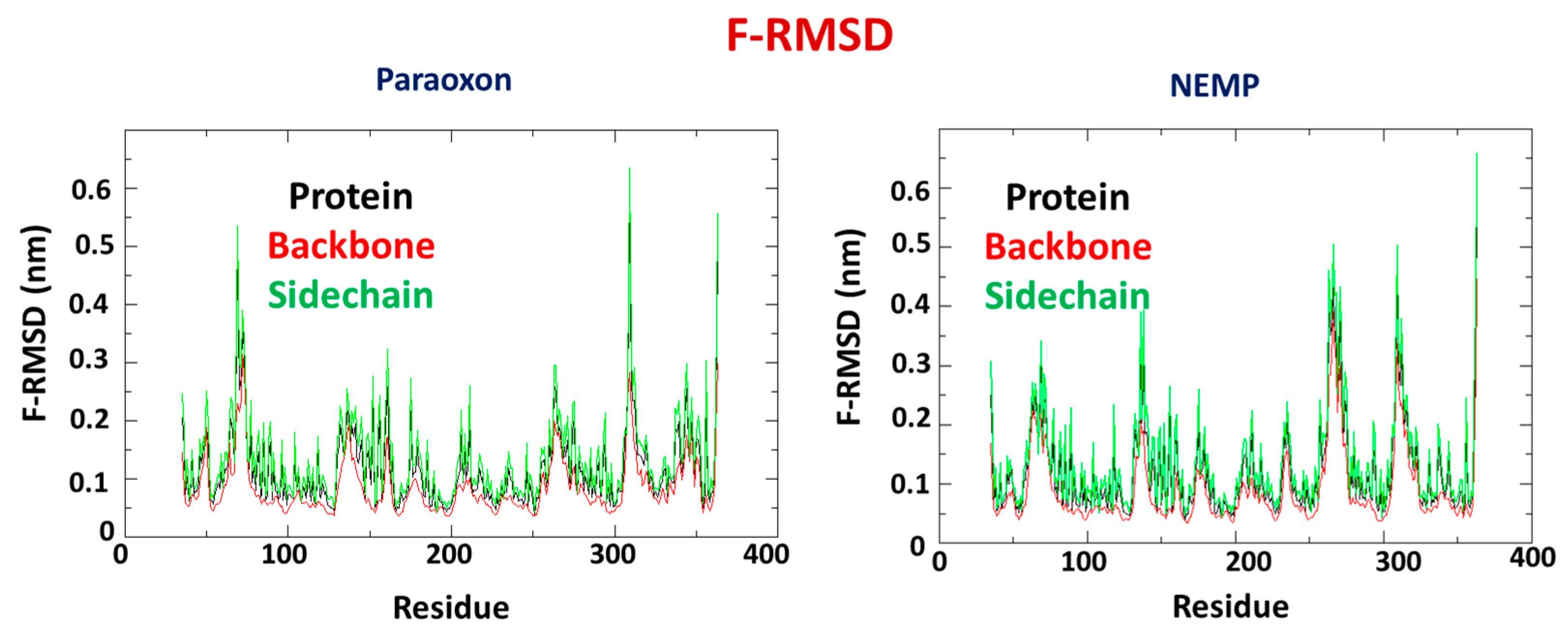

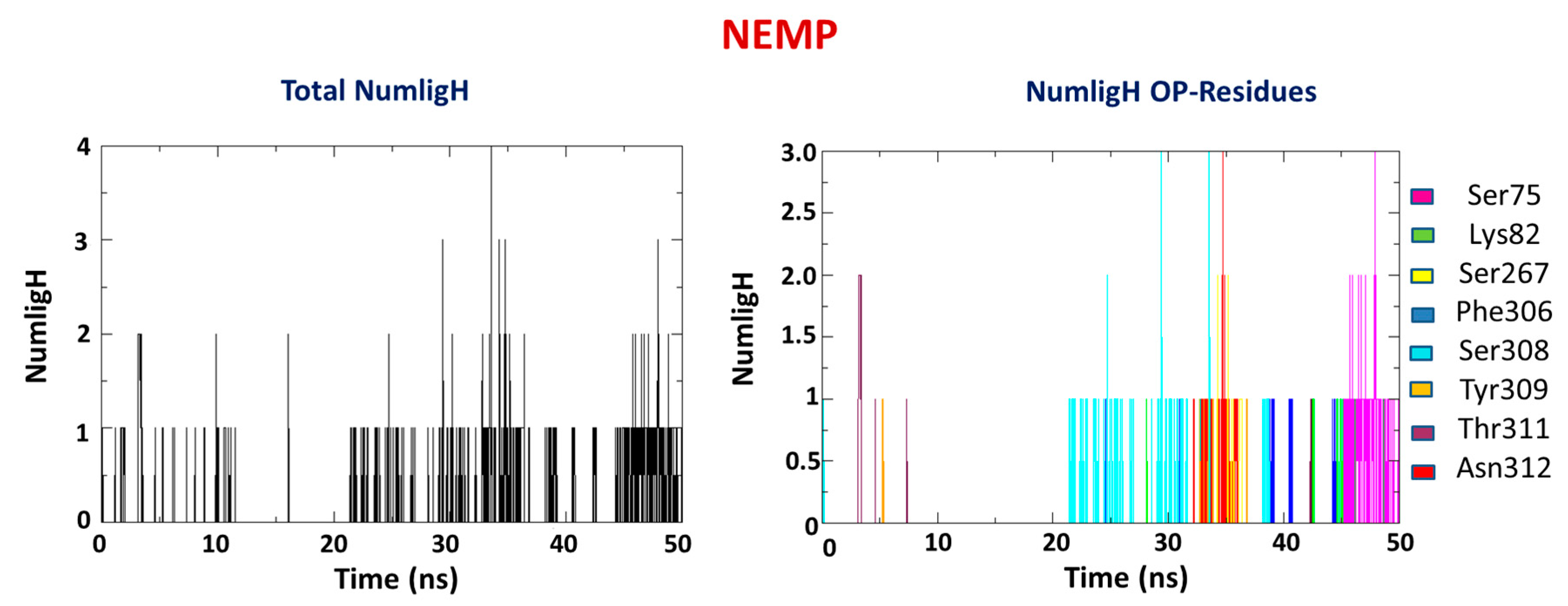
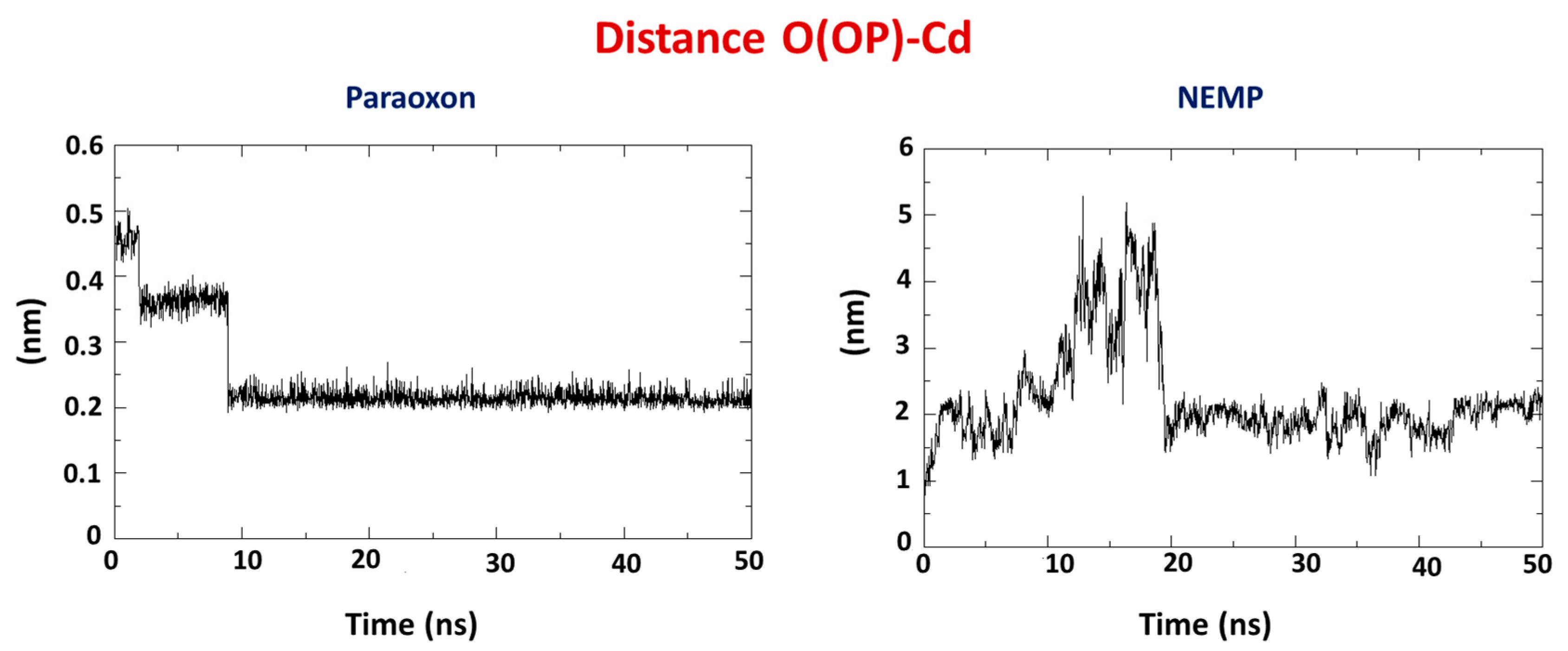

| OP | Distance O(OP)-Cd (Å) | Distance O(Asp301)-Cd | Intermolecular Energy (kcal/mol) | H-Bond Energy (kcal/mol) | H-Bond Interaction Residues |
|---|---|---|---|---|---|
| PXN | 3.57 | 3.21 | −99.33 | −0.26 | Trp131 |
| NEMP | 4.56 | 3.21 | −89.40 | −0.69 | His257 |
| OP | Average H-Bond Number | Interaction Residues |
|---|---|---|
| PXN | 1 | Asn312 |
| NEMP | 1 | Ser75 |
| Lys82 | ||
| Ser267 | ||
| Phe306 | ||
| Ser308 | ||
| Tyr309 | ||
| Thr311 | ||
| Asn312 |
| Sample | Soil with Water | Soil with PXN | Soil with NEMP |
|---|---|---|---|
| 0 days | |||
| 10 days | |||
| 20 days | |||
| 30 days | |||
| 40 days |
| Parameter | Criteria | NEMP | PXN |
|---|---|---|---|
| Selectivity | Not find any interference in the analysis of soil blank. | No interference at retention time 17.4 ± 1. | No interference at retention time 18.3 ± 1 |
| Linearity | Linear regression; R2 values above 0.9 was considered satisfactory. Variation range: 0 to 40 µmol/kg. Points: 0, 8, 16, 24, 32, 40 µmol/kg. | y = 157,640.29 x − 413,553.34 R2 = 0.92 (satisfactory) | y = 847,053.87 x − 1592,824.70 R2 = 0.94 (satisfactory) |
| Precision | Repeatability measured by relative standard deviation; mean value below 20% was considered satisfactory. Levels: 8, 24, 40 µmol/kg. | 15.81 ± 5.57% (high, satisfactory) | 16.28 ± 5.61% (high, satisfactory) |
| Detection limit | Method based on analytical curve parameters; ; s = standard deviation of linear coeficiente of equation and S = angular coeficient. | 0.86 µmol/kg of soil | 0.62 µmol/kg of soil |
| Quantification limit | . | 2.90 µmol/kg of soil | 2.07 µmol/kg of soil |
| Accuracy | ||||||
|---|---|---|---|---|---|---|
| OP | Concentration (µmol/kg) | Mean Value + SD | Criteria | Aceptance | ||
| 8 | 24 | 40 | ||||
| Recovery for NEMP | 42.37% | 46.45% | 53.14% | 47.32 ± 5.44% | Recovery test; mean value for all variation range above or near to 50% was considered satisfactory | Satisfactory |
| Recovery for PXN | 41.30% | 59.59% | 69.95% | 56.95 ± 14.50% | Satisfactory | |
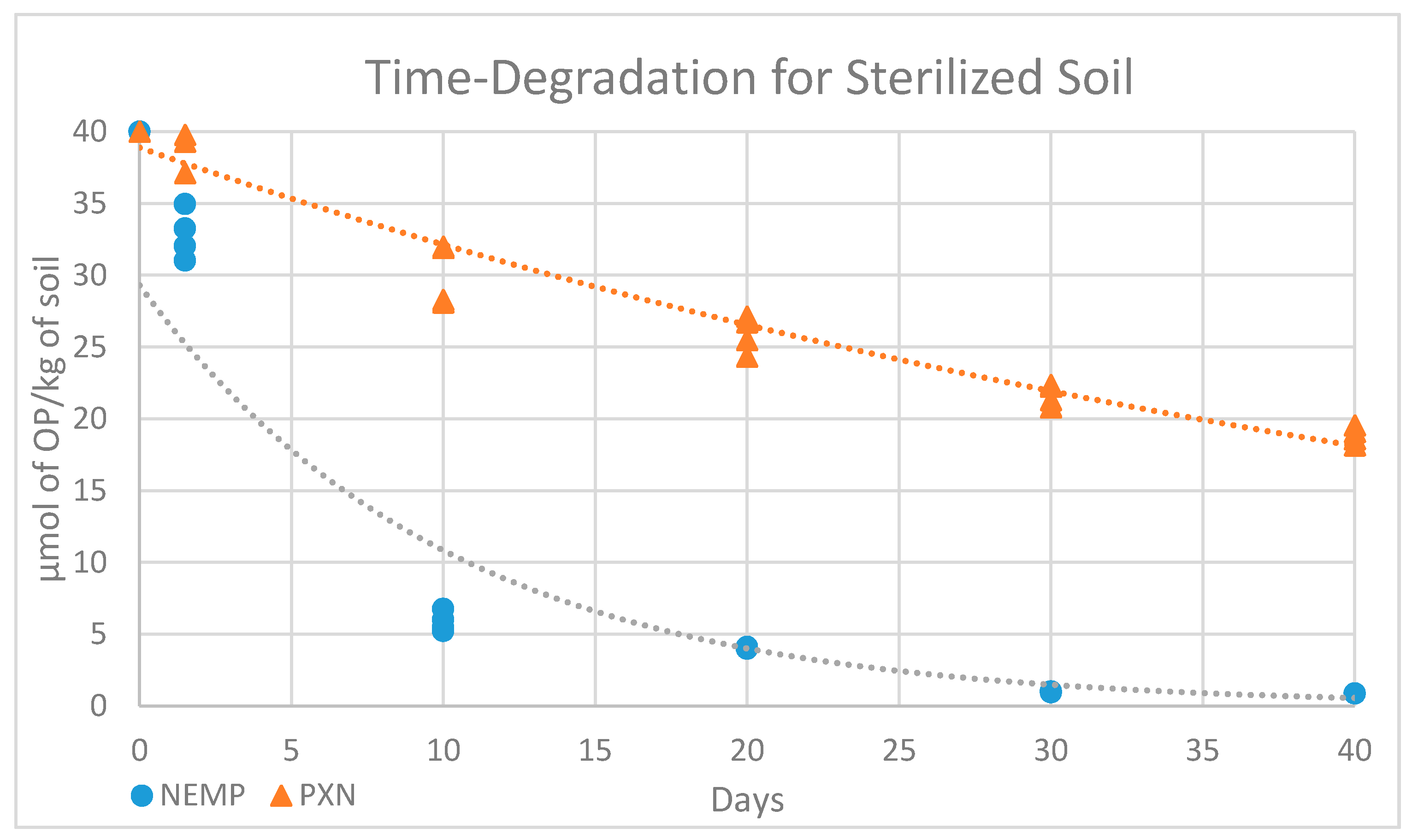
| 0 | 1 Day | 10 Days | 20 Days | 30 Days | 40 Days | |
|---|---|---|---|---|---|---|
| NEMP | 40 | 32.82 ± 1.8 | 5.86 ± 0.73 | 4.04 ± 0.05 | 0.98 ± 0.03 | 0.86 |
| PXN | 40 | 39.4 ± 1.90 | 30.06 ± 2.31 | 25.91 ± 1.35 | 21.66 ± 0.80 | 18.83 ± 0.65 |
| 0 | 1 Day | 10 Days | 20 Days | 30 Days | 40 Days | |
|---|---|---|---|---|---|---|
| NEMP | 40 | 6.02 ± 0.34 | 1.44 ± 0.07 | 0.89 ± 0.01 | 0.86 | 0.86 |
| PXN | 40 | 13.45 ± 1.77 | 6.82 ± 0.90 | 0.85 ± 0.13 | 0.62 | 0.62 |
| NEMP | PXN | |||
|---|---|---|---|---|
| Equation | R2 Value | Equation | R2 Value | |
| Natural soil | 0.89 | 0.94 | ||
| Sterilized soil | 0.93 | 0.96 | ||
| NEMP t1/2-Value | PXN t1/2-Value | ||
|---|---|---|---|
| Natural Soil | Sterilized Soil | Natural Soil | Sterilized Soil |
| 5.49 | 6.97 | 5.62 | 36.29 |
| pH (In Water) | P | K | Ca | Mg | Al | H + Al | Na |
|---|---|---|---|---|---|---|---|
| 5.9 (moderate) | 69 mg/dm3 | 72 mg/dm3 | 2.8 cmolc/dm3 (high) | 0.8 cmolc/dm3 | 0.0 cmolc/dm3 | 2.2 cmolc/dm3 | 0.1 cmolc/dm3 |
| N | C | Electric Conditivity | Cation Exchange Capacity | Organic Matter | Granulometry | Class/Texture | ||
|---|---|---|---|---|---|---|---|---|
| 0.29% | 1.99% | 0.21 dS/cm | Effective | 3.9 cmolc/dm3 | 34.3 g/dm3 (high) | Sand | 778 g/kg | Sandy |
| pH 7 | 6.1 cmolc/dm3 | Silt | 156 g/kg | |||||
| Sum of bases | 3.9 cmolc/dm3 | Clay | 66 g/kg | |||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardozo, M.; de Almeida, J.S.F.D.; Cavalcante, S.F.d.A.; Salgado, J.R.S.; Gonçalves, A.S.; França, T.C.C.; Kuca, K.; Bizzo, H.R. Biodegradation of Organophosphorus Compounds Predicted by Enzymatic Process Using Molecular Modelling and Observed in Soil Samples Through Analytical Techniques and Microbiological Analysis: A Comparison. Molecules 2020, 25, 58. https://doi.org/10.3390/molecules25010058
Cardozo M, de Almeida JSFD, Cavalcante SFdA, Salgado JRS, Gonçalves AS, França TCC, Kuca K, Bizzo HR. Biodegradation of Organophosphorus Compounds Predicted by Enzymatic Process Using Molecular Modelling and Observed in Soil Samples Through Analytical Techniques and Microbiological Analysis: A Comparison. Molecules. 2020; 25(1):58. https://doi.org/10.3390/molecules25010058
Chicago/Turabian StyleCardozo, Monique, Joyce S. F. D. de Almeida, Samir F. de A. Cavalcante, Jacqueline R. S. Salgado, Arlan S. Gonçalves, Tanos C. C. França, Kamil Kuca, and Humberto R. Bizzo. 2020. "Biodegradation of Organophosphorus Compounds Predicted by Enzymatic Process Using Molecular Modelling and Observed in Soil Samples Through Analytical Techniques and Microbiological Analysis: A Comparison" Molecules 25, no. 1: 58. https://doi.org/10.3390/molecules25010058
APA StyleCardozo, M., de Almeida, J. S. F. D., Cavalcante, S. F. d. A., Salgado, J. R. S., Gonçalves, A. S., França, T. C. C., Kuca, K., & Bizzo, H. R. (2020). Biodegradation of Organophosphorus Compounds Predicted by Enzymatic Process Using Molecular Modelling and Observed in Soil Samples Through Analytical Techniques and Microbiological Analysis: A Comparison. Molecules, 25(1), 58. https://doi.org/10.3390/molecules25010058








