Determination of the Structural Integrity and Stability of Polysialic Acid during Alkaline and Thermal Treatment
Abstract
:1. Introduction
2. Results
2.1. Endotoxin Removal of Biotechnologically Produced PolySia
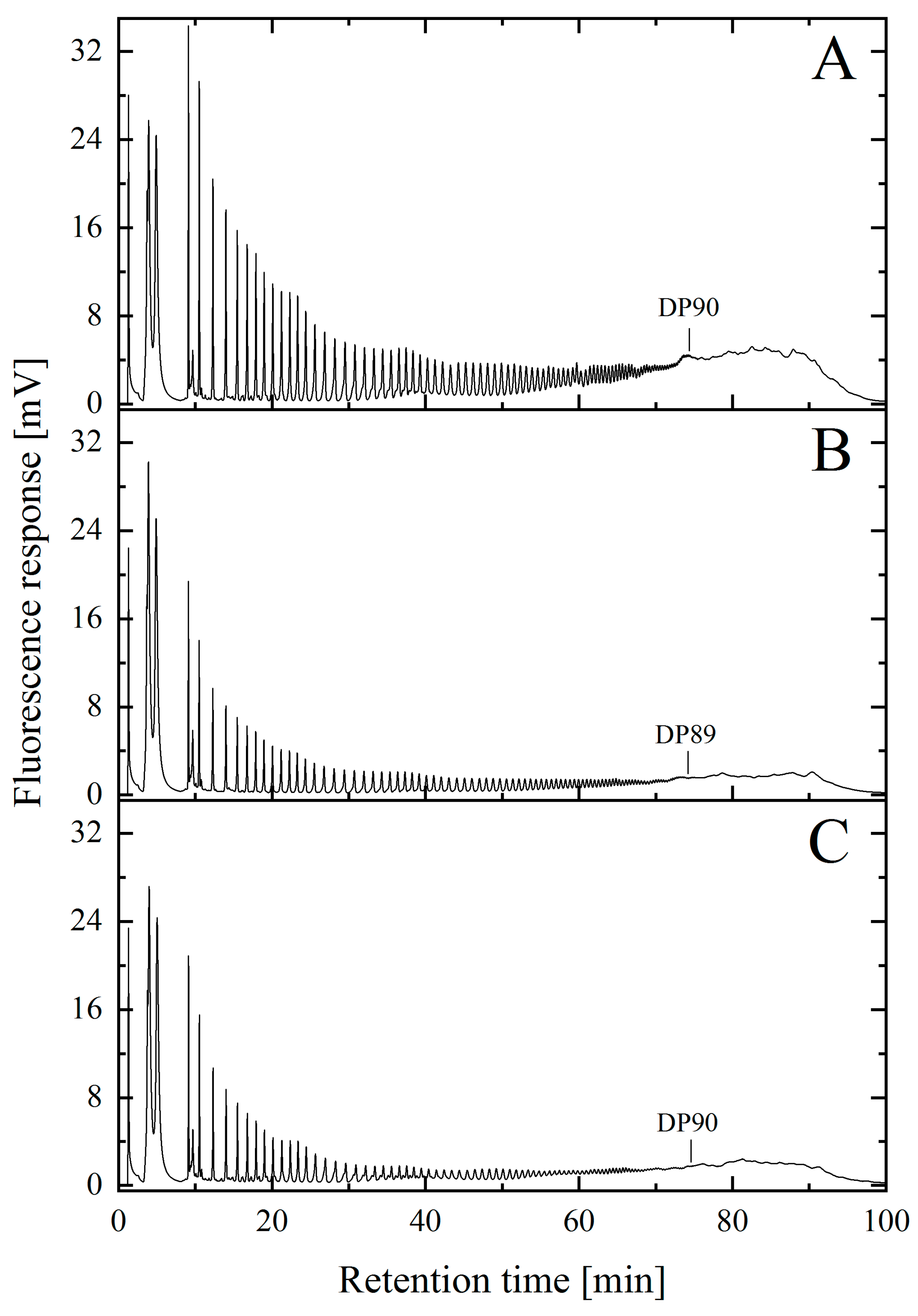
2.2. Analysis of Maximum Chain Length of NaOH-Treated and Untreated PolySia
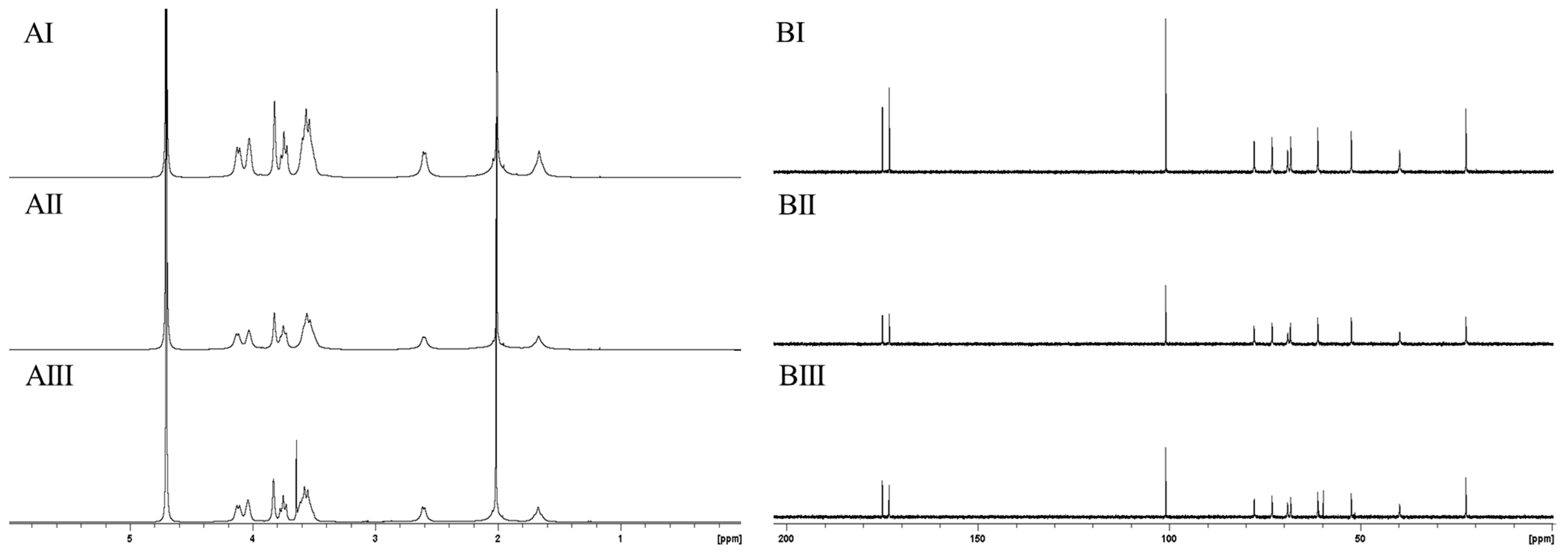
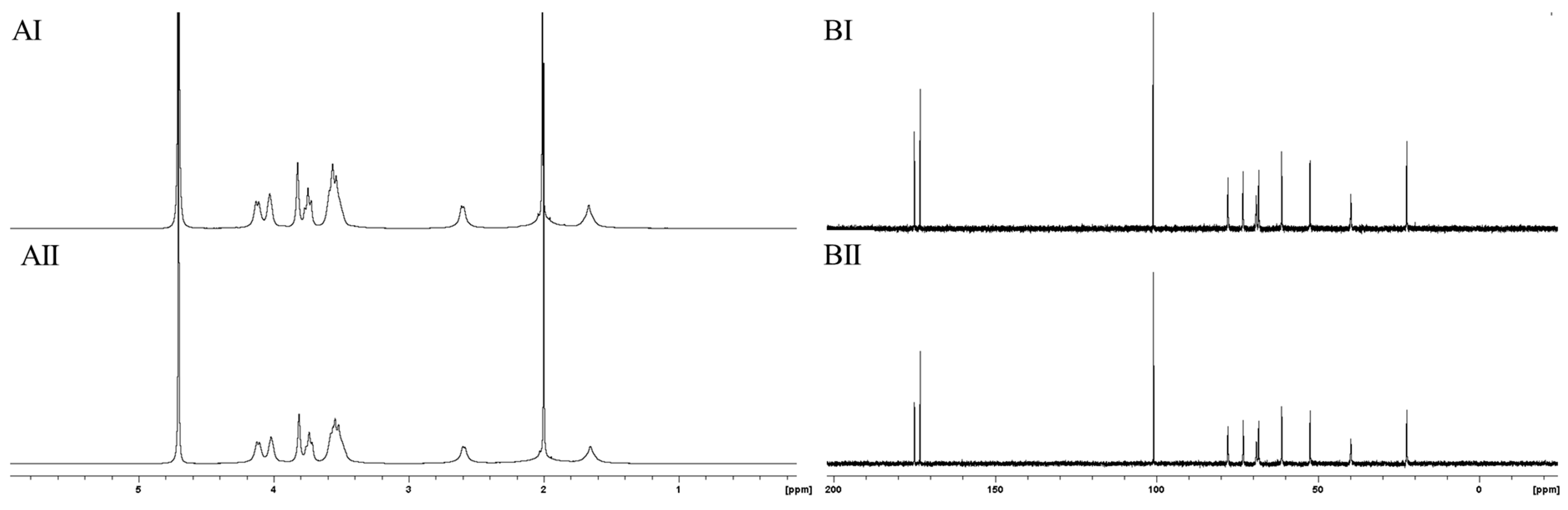
2.3. Structural Analysis of Purified PolySia Using NMR Spectroscopy
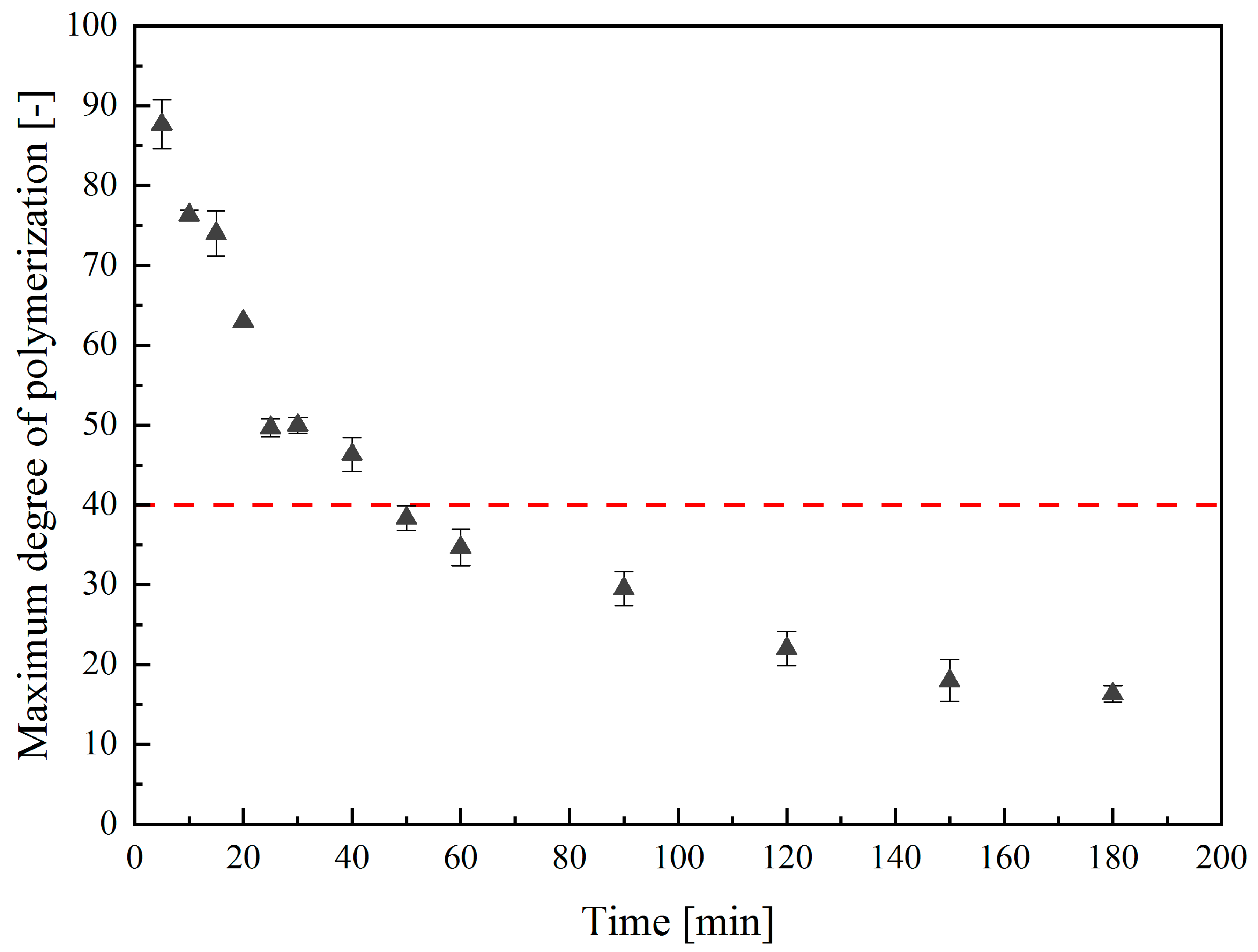
2.4. Characterization of the Thermal Hydrolysis Conditions for Low-Molecular-Weight PolySia
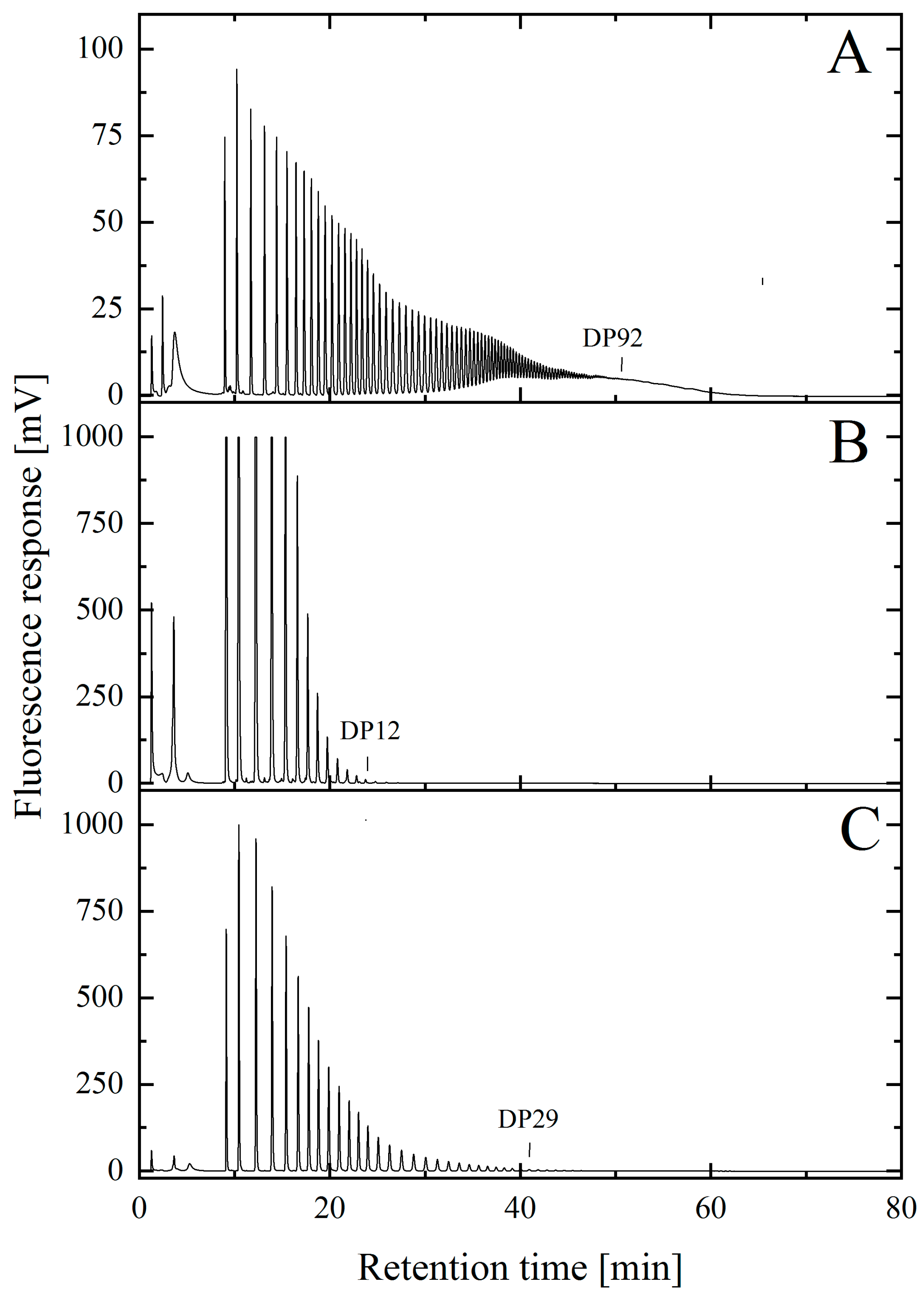
2.5. Isolation and Characterization of Low-Molecular-Weight PolySia avDP20
2.6. Analysis of the Intramolecular Stability of Thermal Hydrolyzed Low-Molecular-Weight PolySia
3. Discussion
4. Materials and Methods
4.1. Chemicals and Buffer
4.2. Bacterial Strains and Strain Maintenance
4.3. Production of PolySia in E. coli K1
4.4. Endotoxin Removal Using Combinatorial NaOH Treatment and AEX Membrane Adsorbers
4.5. Endotoxin Removal Using EndoTrap® HD Columns
4.6. Thermal Hydrolysis for the Production of Low-Molecular-Weight PolySia
4.7. Fractionation of Short Chain PolySia
4.8. Quantification of PolySia Concentrations
4.9. DMB-HPLC Analytics for Chain Length Characterization of PolySia
4.10. Structure Analysis Using 1H- and 13C-NMR Spectroscopy
4.11. Determination of Endotoxins
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Traving, C.; Schauer, R. Structure, function and metabolism of sialic acids. Cell. Mol. Life Sci. 1998, 54, 1330–1349. [Google Scholar] [CrossRef]
- Colley, K.J.; Kitajima, K.; Sato, C. Polysialic acid: Biosynthesis, novel functions and applications. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 498–532. [Google Scholar] [CrossRef]
- Johnson, J.R. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 1991, 4, 80–128. [Google Scholar] [CrossRef]
- Janas, T.; Janas, T. Membrane oligo- and polysialic acids. Biochim. Biophys. Acta Biomembr. 2011, 1808, 2923–2932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenman, A.; Manuel Liaci, A.; Liu, Y.; Frängsmyr, L.; Frank, M.; Blaum, B.S.; Chai, W.; Podgorski, I.I.; Harrach, B.; Benko, M.; et al. Polysialic acid is a cellular receptor for human adenovirus 52. Proc. Natl. Acad. Sci. USA 2018, 115, E4264–E42736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlstetter, M.; Kopatz, J.; Aslanidis, A.; Shahraz, A.; Caramoy, A.; Linnartz-Gerlach, B.; Lin, Y.; Lückoff, A.; Fauser, S.; Düker, K.; et al. Polysialic acid blocks mononuclear phagocyte reactivity, inhibits complement activation, and protects from vascular damage in the retina. EMBO Mol. Med. 2017, 9, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Mühlenhoff, M.; Eckhardt, M.; Gerardy-Schahn, R. {P}olysialic acid: three-dimensional structure, biosynthesis and function. Curr. Opin. Struct. Biol. 1998, 8, 558–564. [Google Scholar] [CrossRef]
- Almagro-Moreno, S.; Boyd, E.F. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol. Biol. 2009, 9, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Elkashef, S.M.; Loadman, P.M.; Patterson, L.H.; Falconer, R.A. Recent advances in the analysis of polysialic acid from complex biological systems. Carbohydr. Polym. 2019, 224, 115145. [Google Scholar] [CrossRef]
- Rutishauser, U. Polysialic acid at the cell surface: Biophysics in service of cell interactions and tissue plasticity. J. Cell. Biochem. 1998, 70, 304–312. [Google Scholar] [CrossRef]
- Shahraz, A.; Kopatz, J.; Mathy, R.; Kappler, J.; Winter, D.; Kapoor, S.; Schütza, V.; Scheper, T.; Gieselmann, V.; Neumann, H. Anti-inflammatory activity of low molecular weight polysialic acid on human macrophages. Sci. Rep. 2015, 5, 16800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schütza, V. Anti-Angiogenic Effects of Polysialic Acids with an Average Degree of Polymerization of 20. Master’s Thesis, University of Bonn, Bonn, Germany, 2016. [Google Scholar]
- Lin, B.-X.; Qiao, Y.; Shi, B.; Tao, Y. Polysialic acid biosynthesis and production in Escherichia coli: Current state and perspectives. Appl. Microbiol. Biotechnol. 2016, 100, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhan, X.; Liu, L.; Xia, X. Bioproduction, purification, and application of polysialic acid. Appl. Environ. Microbiol. 2018, 102, 9403–9409. [Google Scholar] [CrossRef]
- De Vries, I.; Busse, C.; Kopatz, J.; Neumann, H.; Beutel, S.; Scheper, T. Polysialic acid production using Escherichia coli K1 in a disposable bag reactor. Eng. Life Sci. 2017, 17, 723–731. [Google Scholar] [CrossRef] [Green Version]
- De Vries, I.; Schreiber, S.; Boßmann, D.; Hellmann, Z.; Kopatz, J.; Neumann, H.; Beutel, S. Single-use membrane adsorbers for endotoxin removal and purification of endogenous polysialic acid from Escherichia coli K1. Biotechnol. Rep. 2018, 17, 110–116. [Google Scholar] [CrossRef]
- Boßmann, D.; Bartling, B.; de Vries, I.; Winkler, J.; Neumann, H.; Lammers, F.; Beutel, S.; Scheper, T. Charged aerosol detector HPLC as a characterization and quantification application of biopharmaceutically relevant polysialic acid from E. coli K1. J. Chromatogr. A 2019, 20, 1–10. [Google Scholar] [CrossRef]
- Ma, R.; Fan, D.D.; Xue, W.J.; Xing, J.Y.; Zhu, C.H.; Ma, X.X. Endotoxin removal during the purification process of human-like collagen. Sep. Sci. Technol. 2010, 45, 2400–2405. [Google Scholar] [CrossRef]
- Ongkudon, C.M.; Chew, J.H.; Liu, B.; Danquah, M.K. Chromatographic Removal of Endotoxins: A Bioprocess Engineer’ s Chromatographic Removal of Endotoxins: A Bioprocess Engineer’ s Perspective. ISRN Chromatogr. 2012, 2012, 649746. [Google Scholar]
- Lee, S.Y.; Choi, J.I.; Han, K.; Song, J.Y. Removal of endotoxin during purification of poly(3-hydroxybutyrate) from gram-negative bacteria. Appl. Environ. Microbiol. 1999, 65, 2762–2764. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; She, Z.; Huang, Z.; Li, J.; Luo, X.; Deng, Y. Application of sialic acid/polysialic acid in the drug delivery systems. Asian J. Pharm. Sci. 2014, 9, 75–81. [Google Scholar] [CrossRef] [Green Version]
- Rode, B.; Endres, C.; Ran, C.; Stahl, F.; Beutel, S.; Kasper, C.; Galuska, S.; Geyer, R.; Mühlenhoff, M.; Gerardy-Schahn, R.; et al. Large-scale production and homogenous purification of long chain polysialic acids from E. coli K1. J. Biotechnol. 2008, 135, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Bice, I.; Celik, H.; Wolff, C.; Beutel, S.; Zahid, M.; Hitzmann, B.; Rinas, U.; Kasper, C.; Gerardy-Schahn, R.; Scheper, T. Downstream processing of high chain length polysialic acid using membrane adsorbers and clay minerals for application in tissue engineering. Eng. Life Sci. 2013, 13, 140–148. [Google Scholar] [CrossRef] [Green Version]
- Xiao, R.; Grinstaff, M.W. Chemical synthesis of polysaccharides and polysaccharide mimetics. Prog. Polym. Sci. 2017, 74, 78–116. [Google Scholar] [CrossRef]
- Si, A.; Misra, A.K. Expedient Synthesis of the Pentasaccharide Repeating Unit of the Polysaccharide O-Antigen of Escherichia coli O11. ChemistryOpen 2016, 5, 47–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohrs, T.E.; Troy, F.A. Structure and Biosynthesis of Surface Polymers Containing Polysialic Acid in Escherichia coli. J. Biol. Chem. 1980, 255, 2332–2342. [Google Scholar]
- Cheng, M.; Wang, K.; Inoue, S.; Inoue, Y.; Khoo, K.; Wu, S. Controlled Acid Hydrolysis of Colominic Acid under Microwave Irradiation. Anal. Biochem. 1999, 293, 287–293. [Google Scholar] [CrossRef]
- Davies, L.R.L.; Pearce, O.M.T.; Tessier, M.B.; Assar, S.; Smutova, V.; Pajunen, M.; Sumida, M.; Sato, C.; Kitajima, K.; Finne, J.; et al. Metabolism of vertebrate amino sugars with N-glycolyl groups: Resistance of α2-8-linked N-glycolylneuraminic acid to enzymatic cleavage. J. Biol. Chem. 2012, 287, 28917–28931. [Google Scholar] [CrossRef] [Green Version]
- Lifely, M.R.; Gilbert, A.S.; Moreno, C. Rate, Mechanism, and Immunochemical studies of Lactonisation in Serogroup B and Polysaccharides of Neisseria meningitidis. Carbohydr. Res. 1984, 134, 229–243. [Google Scholar] [CrossRef]
- Cheng, M.C.; Lin, C.H.; Lin, H.J.H.J.; Yu, Y.P.; Wu, S.H. Hydrolysis, lactonization, and identification of α(2⟶8 /α(2⟶9) alternatively linked tri-, tetra-, and polysialic acids. Glycobiology 2004, 14, 147–155. [Google Scholar] [CrossRef]
- Zhang, Y.; Lee, Y.C. Acid-catalyzed lactonization of α2,8-linked oligo/polysialic acids studied by high performance anion-exchange chromatography. J. Biol. Chem. 1999, 274, 6183–6189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aminoff, D. Methods for the quantitative estimation of N-acetylneuraminic acid and their application to hydrolysates of sialomucoids. Biochem. J. 1961, 81, 384–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, L. The Thiobarbituric Acid Assay of Sialic Acids*. J. Biol. Chem. 1959, 234, 1971–1975. [Google Scholar] [PubMed]
- Inoue, S.; Inoue, Y. Developmental Profile of Neural Cell Adhesion Molecule Glycoforms with a Varying Degree of Polymerization of Polysialic Acid Chains. J. Biol. Chem. 2001, 276, 31863–31870. [Google Scholar] [CrossRef] [Green Version]
- Egan, W.; Liu, T.Y.; Dorow, D.; Cohen, J.S.; Robbins, J.D.; Gotschlich, E.C.; Robbins, J.B. Structural Studies on the Sialic Acid Polysaccharide Antigen of Escherichia coli Strain Bos-12. Biochemistry 1977, 16, 3687–3692. [Google Scholar] [CrossRef]
- Liu, J.L.; Zhan, X.B.; Wu, J.R.; Lin, C.C.; Yu, D.F. An efficient and large-scale preparation process for polysialic acid by Escherichia coli CCTCC M208088. Biochem. Eng. J. 2010, 53, 97–103. [Google Scholar] [CrossRef]
- Sonnenburg, J.L.; van Halbeek, H.; Varki, A. Characterization of the Acid Stability of Glycosidically Linked Neuraminic Acid. J. Biol. Chem. 2002, 277, 17502–17510. [Google Scholar] [CrossRef] [Green Version]
- Gottschalk, A. The Chemistry and Biology of Sialic Acids and Related Substances. AIBS Bull. 1961, 11, 30. [Google Scholar]
- Gee, A.P.; Sumstad, D.; Stanson, J.; Watson, P.; Proctor, J.; Kadidlo, D.; Koch, E.; Sprague, J.; Wood, D.; Styers, D.; et al. A multicenter comparison study between the Endosafe® PTSTM rapid-release testing system and traditional methods for detecting endotoxin in cell-therapy products. Cytotherapy 2008, 10, 427–435. [Google Scholar] [CrossRef] [Green Version]
- Inoue, S.; Lin, S.G.; Lee, Y.C.; Inoue, Y. An ultrasensitive chemical method for polysialic acid analysis. Glycobiology 2001, 11, 759–767. [Google Scholar] [CrossRef] [Green Version]
- Jakobsson, E.; Jokilammi, A.; Aalto, J.; Ollikka, P.; Lehtonen, J.V.; Hirvonen, H.; Finne, J. Identification of amino acid residues at the active site of endosialidase that dissociate the polysialic acid binding and cleaving activities in Escherichia coli K1 bacteriophages. Biochem. J. 2007, 472, 465–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frosch, M.; Roberts, I.; Görgen, I.; Metzger, S.; Boulnois, G.J.; Bitter-Suermann, D. Serotyping and genotyping of encapsulated Escherichia coli K1 sepsis isolates with a monoclonal IgG anti K1 antibody and K1 gene probes. Microb. Pathog. 1987, 2, 319–326. [Google Scholar] [CrossRef]
- Beutel, S. B2032/82 K1 (DSM-Nr. 107164), DSMZ-Stammhinterlegung Escherichia coli B2032/82 Serotype K1. 2018. Available online: https://www.dsmz.de/catalogues/details/culture/DSM-107164.html?tx_dsmzresources_pi5%5BreturnPid%5D=304 (accessed on 31 December 2019).
- Rodríguez-Aparicio, L.B.; Reglero, A.; Ortiz, A.I.; Luengo, J.M. Effect of physical and chemical conditions on the production of colominic acid by E. coli in defined medium. Appl. Microbiol. Biotechnol. 1988, 27, 474–483. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartling, B.; Rehfeld, J.S.; Boßmann, D.; de Vries, I.; Fohrer, J.; Lammers, F.; Scheper, T.; Beutel, S. Determination of the Structural Integrity and Stability of Polysialic Acid during Alkaline and Thermal Treatment. Molecules 2020, 25, 165. https://doi.org/10.3390/molecules25010165
Bartling B, Rehfeld JS, Boßmann D, de Vries I, Fohrer J, Lammers F, Scheper T, Beutel S. Determination of the Structural Integrity and Stability of Polysialic Acid during Alkaline and Thermal Treatment. Molecules. 2020; 25(1):165. https://doi.org/10.3390/molecules25010165
Chicago/Turabian StyleBartling, Bastian, Johanna S. Rehfeld, Daniel Boßmann, Ingo de Vries, Jörg Fohrer, Frank Lammers, Thomas Scheper, and Sascha Beutel. 2020. "Determination of the Structural Integrity and Stability of Polysialic Acid during Alkaline and Thermal Treatment" Molecules 25, no. 1: 165. https://doi.org/10.3390/molecules25010165




