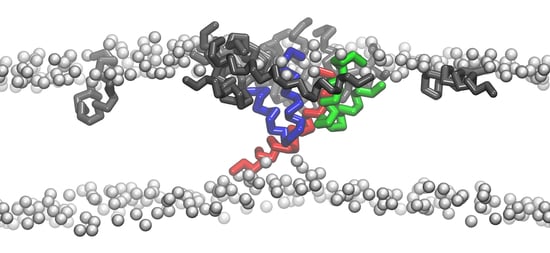
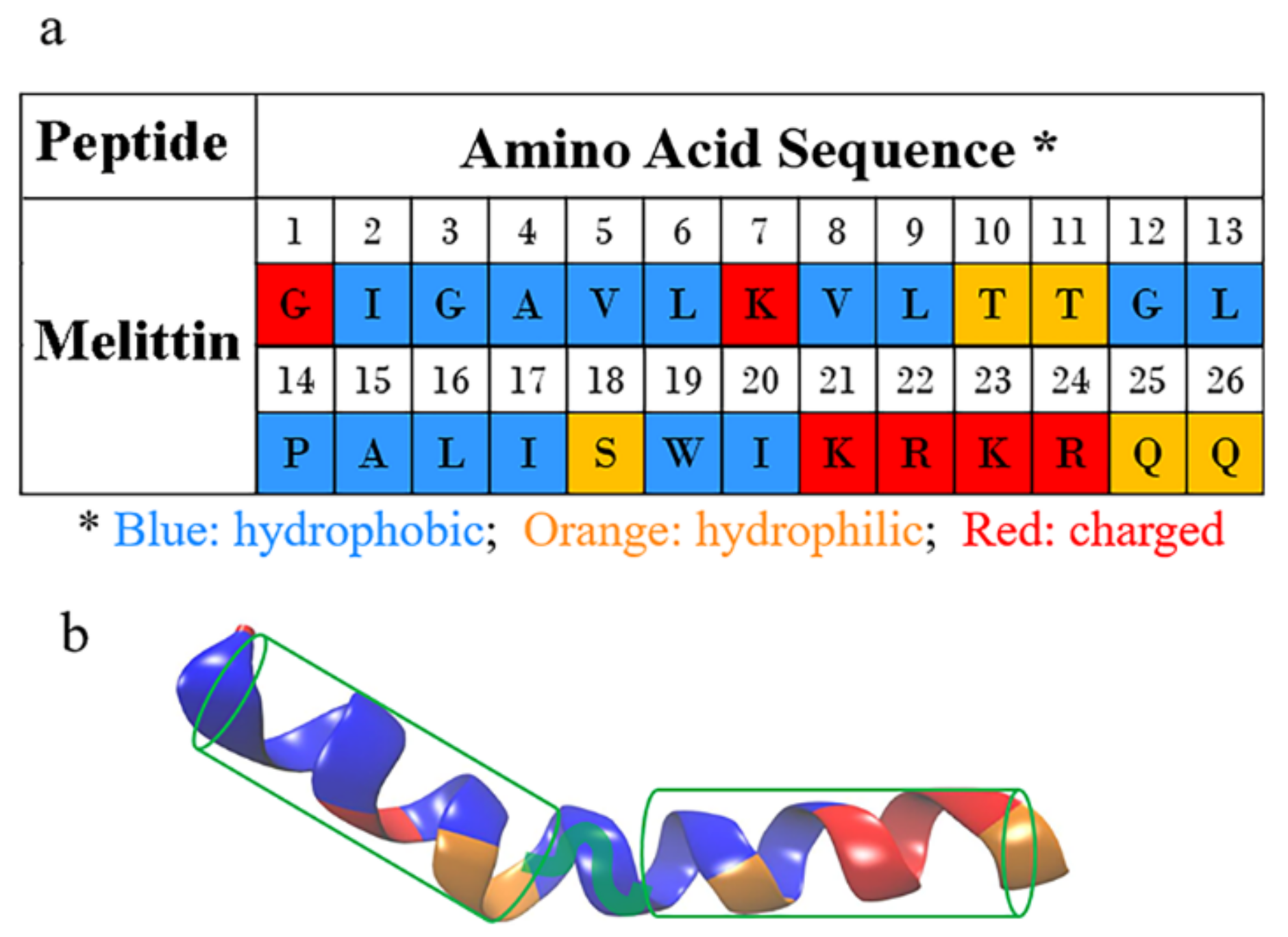
How Melittin Inserts into Cell Membrane: Conformational Changes, Inter-Peptide Cooperation, and Disturbance on the Membrane
Abstract
1. Introduction
2. Kinetic Membrane Insertion Process of Melittin: Peptide Aggregation and Deformation; Membrane Disturbance Induced by Lipid–Peptide Interaction
2.1. Simulation Models for Realization of Membrane Insertion of Melittin
2.2. Peptide-to-Lipid Ratio Dependence
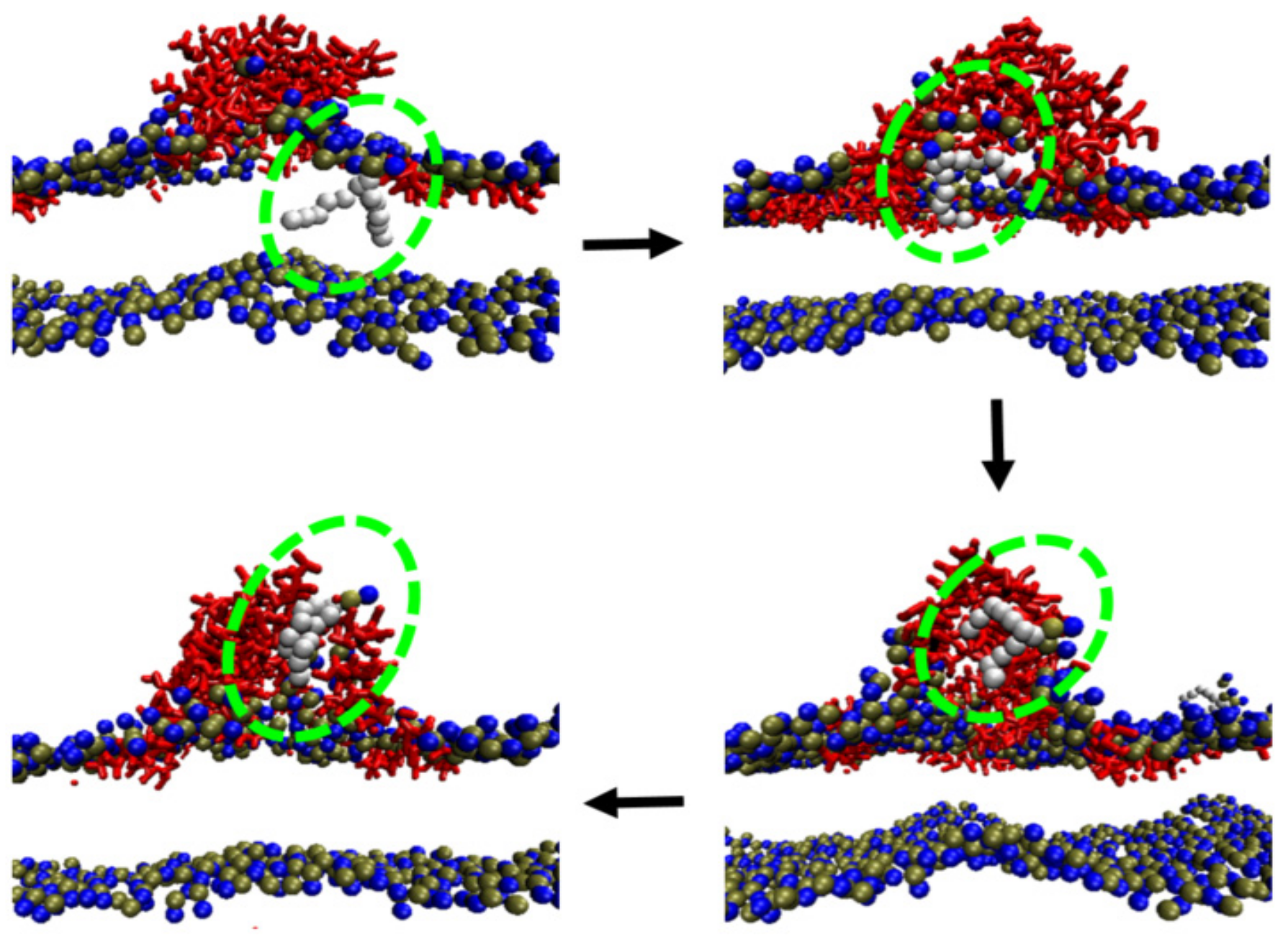
2.3. Inter-Peptide Association Based on Local Accumulation of Peptides
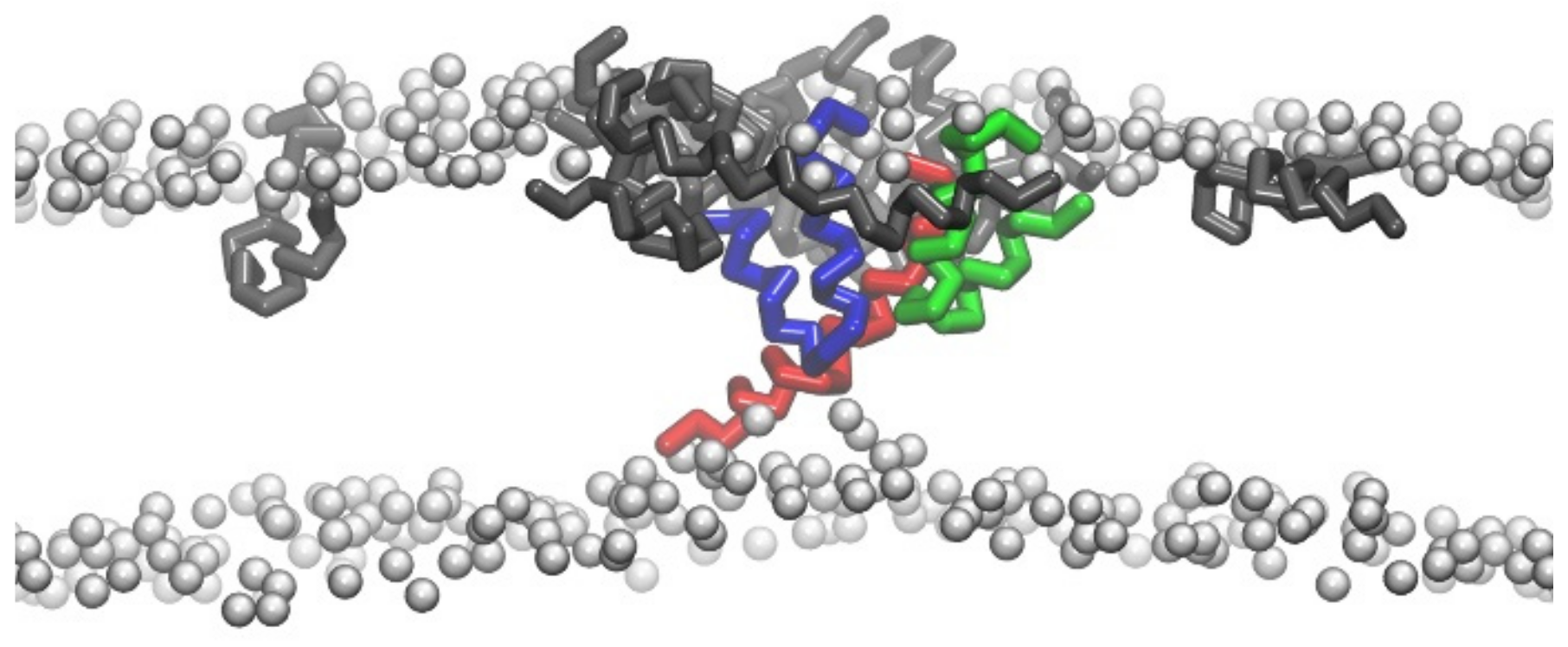
2.4. Conformational Changes of Peptide during Membrane Insertion
2.5. Structural Disturbance to Membrane during Melittin Insertion
3. Thermodynamic Analysis of Transmembrane Insertion of Melittin: Decreased Free-Energy Barrier due to Inter-Peptide Cooperation
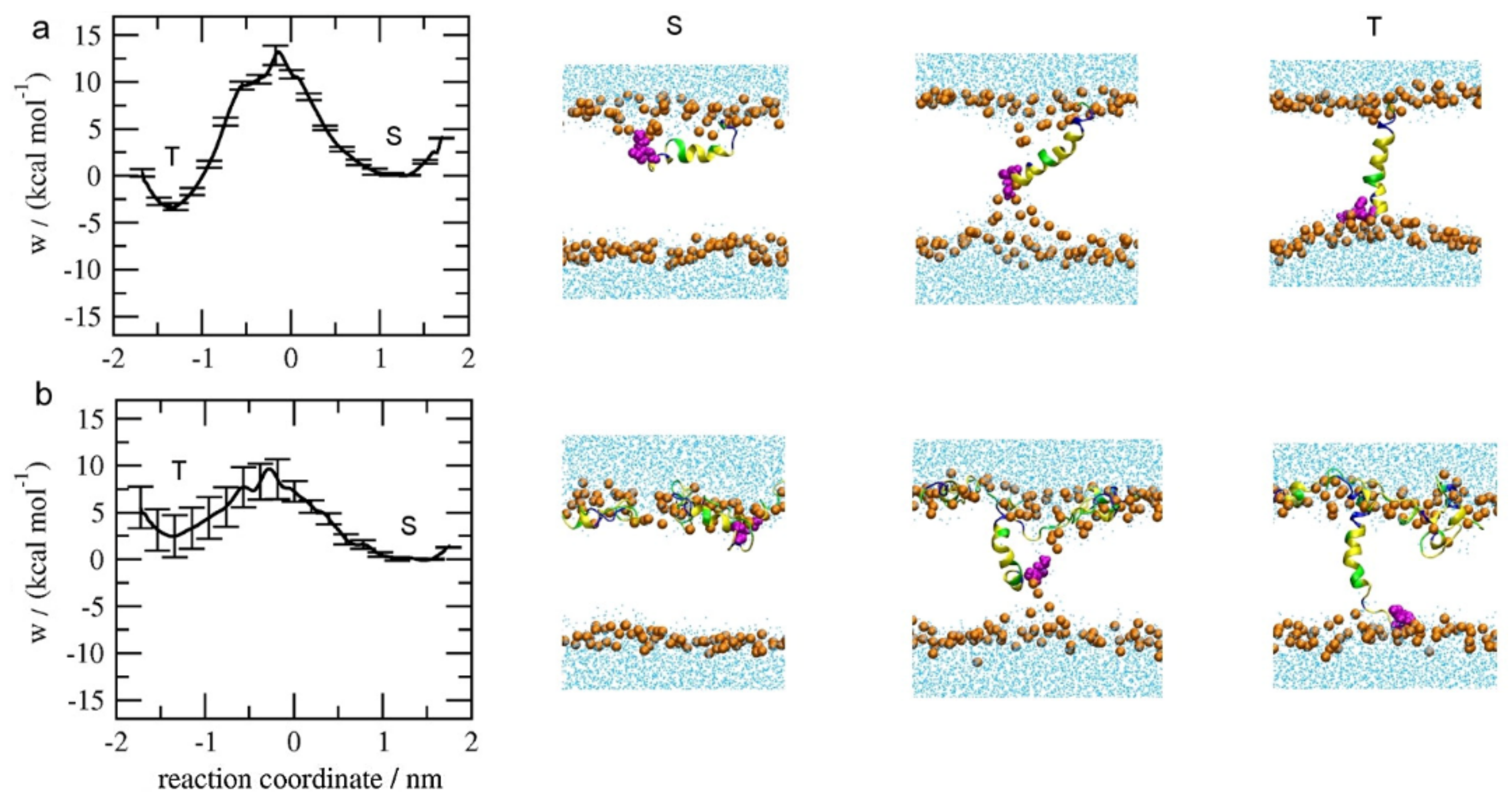
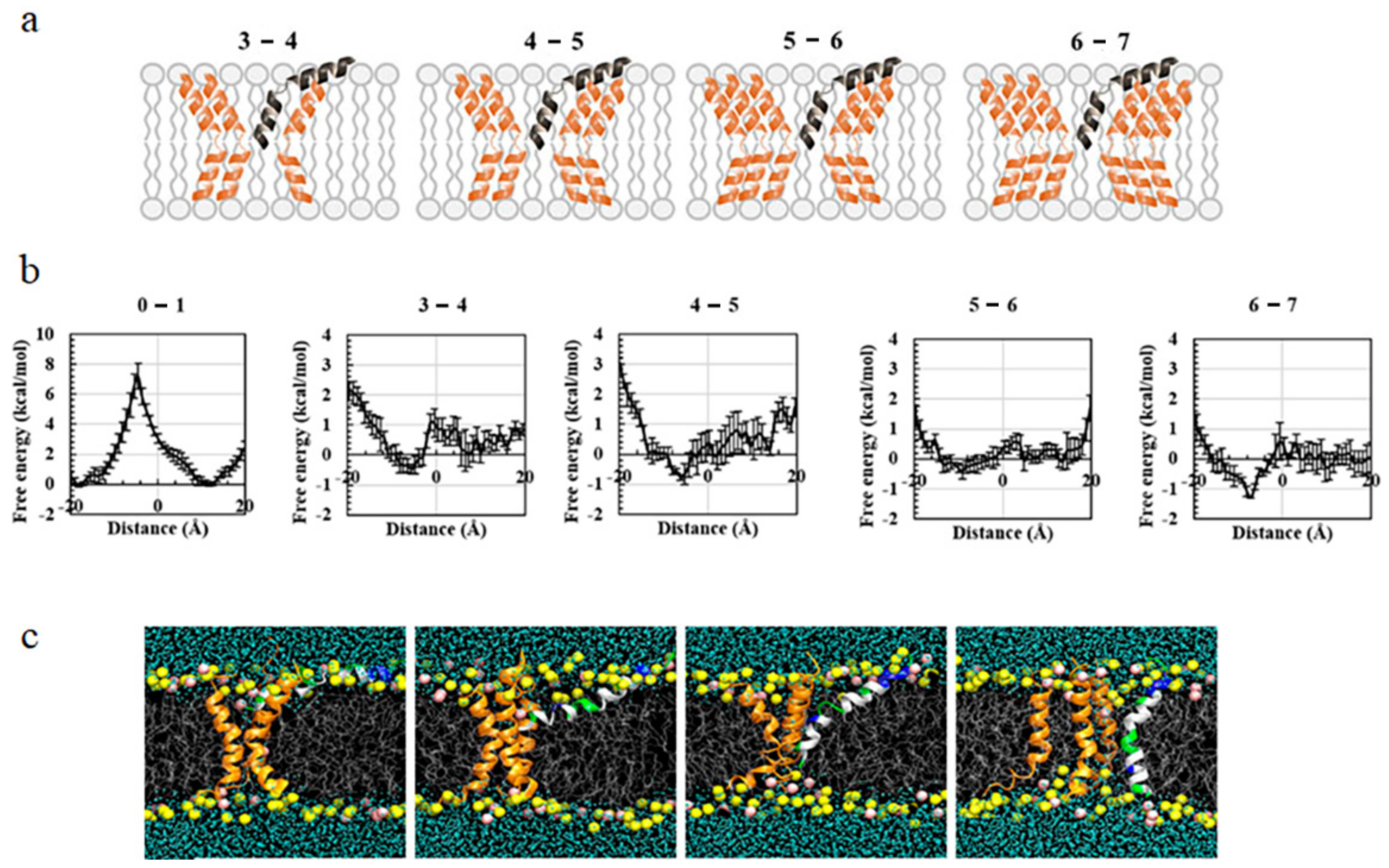
3.1. Potential of Mean Force (PMF) Distribution Demonstrating the Inter-Peptide Cooperation
3.2. Influence of Lipid State on Free-Energy Barrier during Peptide Insertion
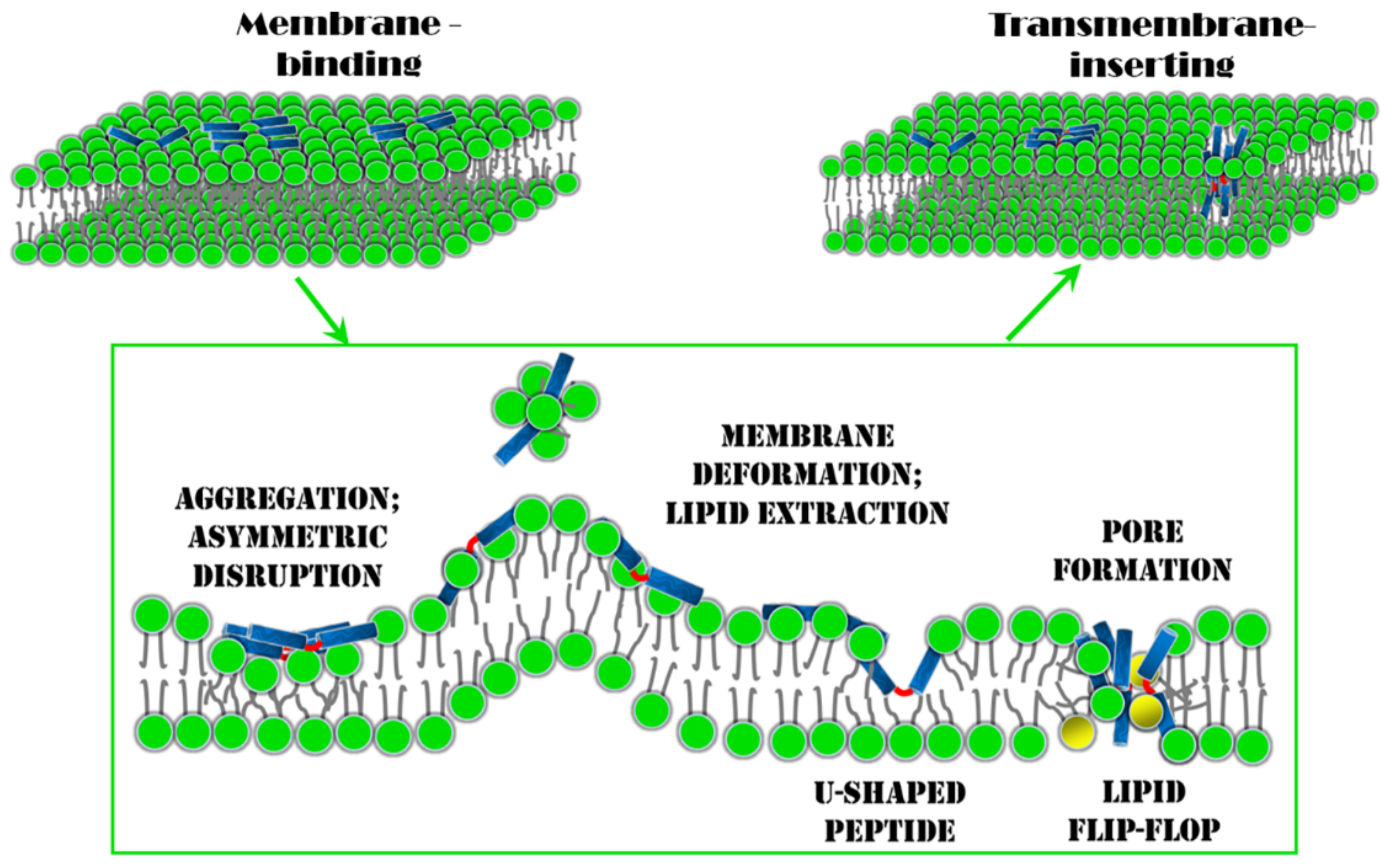
4. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Willyard, C. The drug-resistant bacteria that pose the greatest health threats. Nature 2017, 543, 15. [Google Scholar] [CrossRef]
- Li, J.; Koh, J.-J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane active antimicrobial peptides: Translating mechanistic insights to design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nation, R.L.; Turnidge, J.D.; Milne, R.W.; Coulthard, K.; Rayner, C.R.; Paterson, D.L. Colistin: The re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 2006, 6, 589–601. [Google Scholar] [CrossRef]
- Lázár, V.; Martins, A.; Spohn, R.; Daruka, L.; Grézal, G.; Fekete, G.; Számel, M.; Jangir, P.K.; Kintses, B.; Csörgő, B. Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 2018, 3, 718. [Google Scholar] [CrossRef]
- Wimley, W.C. Describing the Mechanism of Antimicrobial Peptide Action with the Interfacial Activity Model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389. [Google Scholar] [CrossRef]
- Fox, J.L. Antimicrobial peptides stage a comeback. Nat. Biotech. 2013, 31, 379. [Google Scholar] [CrossRef] [PubMed]
- Di Luca, M.; Maccari, G.; Maisetta, G.; Batoni, G. BaAMPs: The database of biofilm-active antimicrobial peptides. Biofouling 2015, 31, 193–199. [Google Scholar] [CrossRef]
- Grassi, L.; Maisetta, G.; Maccari, G.; Esin, S.; Batoni, G. Analogs of the Frog-skin Antimicrobial Peptide Temporin 1Tb Exhibit a Wider Spectrum of Activity and a Stronger Antibiofilm Potential as Compared to the Parental Peptide. Front. Chem. 2017, 5, 24. [Google Scholar] [CrossRef]
- Sancho-Vaello, E.; Zeth, K. Antimicrobial peptides: Has their time arrived? Future Microbiol. 2015, 10, 1103–1106. [Google Scholar] [CrossRef]
- Han, M.-L.; Velkov, T.; Zhu, Y.; Roberts, K.D.; Le Brun, A.P.; Chow, S.H.; Gutu, A.D.; Moskowitz, S.M.; Shen, H.-H.; Li, J. Polymyxin-Induced Lipid A Deacylation in Pseudomonas aeruginosa Perturbs Polymyxin Penetration and Confers High-Level Resistance. ACS Chem. Biol. 2018, 13, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Band, I.V.; Weiss, S.D. Mechanisms of Antimicrobial Peptide Resistance in Gram-Negative Bacteria. Antibiotics 2015, 4, 18–41. [Google Scholar] [CrossRef]
- Velkov, T.; Roberts, K.D.; Nation, R.L.; Thompson, P.E.; Li, J. Pharmacology of polymyxins: New insights into an ‘old’ class of antibiotics. Future Microbiol. 2013, 8, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, S.; Falanga, A.; Berisio, R.; Grieco, P.; Morelli, G.; Galdiero, M. Antimicrobial peptides as an opportunity against bacterial diseases. Curr. Med. Chem. 2015, 22, 1665–1677. [Google Scholar] [CrossRef] [PubMed]
- Faccone, D.; Veliz, O.; Corso, A.; Noguera, M.; Martínez, M.; Payes, C.; Semorile, L.; Maffía, P.C. Antimicrobial activity of de novo designed cationic peptides against multi-resistant clinical isolates. Eur. J. Med. Chem. 2014, 71, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, W.B.; Guha, S.; Wimley, W.C. Synthetic molecular evolution of hybrid cell penetrating peptides. Nat. Commun. 2018, 9, 2568. [Google Scholar] [CrossRef] [PubMed]
- Guha, S.; Ghimire, J.; Wu, E.; Wimley, W.C. Mechanistic Landscape of Membrane-Permeabilizing Peptides. Chem. Rev. 2019. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Starr, C.G.; Troendle, E.; Wiedman, G.; Wimley, W.C.; Ulmschneider, J.P.; Ulmschneider, M.B. Simulation-Guided Rational de Novo Design of a Small Pore-Forming Antimicrobial Peptide. J. Am. Chem. Soc. 2019, 141, 4839–4848. [Google Scholar] [CrossRef]
- Sahl, H.G. Optimizing Antimicrobial Host Defense Peptides. Chem. Biol. 2006, 13, 1015–1017. [Google Scholar] [CrossRef][Green Version]
- Memariani, H.; Memariani, M.; Shahidi-Dadras, M.; Nasiri, S.; Akhavan, M.M.; Moravvej, H. Melittin: From honeybees to superbugs. Appl. Microbiol. Biotechnol. 2019, 103, 3265–3276. [Google Scholar] [CrossRef]
- Habermann, E. Bee and Wasp Venoms. Science 1972, 177, 314. [Google Scholar] [CrossRef] [PubMed]
- Dorman, L.C.; Markley, L.D. Solid phase synthesis and antibacterial activity of N-terminal sequences of melittin. J. Med. Chem. 1971, 14, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Steiner, H.; Hultmark, D.; Engström, Å.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Giacometti, A.; Cirioni, O.; Kamysz, W.; D’Amato, G.; Silvestri, C.; Del Prete, M.S.; Łukasiak, J.; Scalise, G. Comparative activities of cecropin A, melittin, and cecropin A–melittin peptide CA(1–7)M(2–9)NH2 against multidrug-resistant nosocomial isolates of Acinetobacter baumannii. Peptides 2003, 24, 1315–1318. [Google Scholar] [CrossRef] [PubMed]
- Gopal, R.; Lee, J.H.; Kim, Y.G.; Kim, M.-S.; Seo, C.H.; Park, Y. Anti-Microbial, Anti-Biofilm Activities and Cell Selectivity of the NRC-16 Peptide Derived from Witch Flounder, Glyptocephalus cynoglossus. Mar. Drugs 2013, 11, 1836–1852. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.H.; Lee, D.-G.; Lee, S.H.; Ha, J.-M.; Ha, B.-J.; Kim, S.-K.; Lee, J.-H. Antibacterial activity of Ulva lactuca against methicillin-resistantStaphylococcus aureus (MRSA). Biotechnol. Bioproc. Eng. 2007, 12, 579–582. [Google Scholar] [CrossRef]
- Pandey, B.K.; Ahmad, A.; Asthana, N.; Azmi, S.; Srivastava, R.M.; Srivastava, S.; Verma, R.; Vishwakarma, A.L.; Ghosh, J.K. Cell-Selective Lysis by Novel Analogues of Melittin against Human Red Blood Cells and Escherichia coli. Biochemistry 2010, 49, 7920–7929. [Google Scholar] [CrossRef]
- Moerman, L.; Bosteels, S.; Noppe, W.; Willems, J.; Clynen, E.; Schoofs, L.; Thevissen, K.; Tytgat, J.; Van Eldere, J.; van der Walt, J.; et al. Antibacterial and antifungal properties of α-helical, cationic peptides in the venom of scorpions from southern Africa. Eur. J. Biochem. 2002, 269, 4799–4810. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Harroun, T.A.; Weiss, T.M.; Ding, L.; Huang, H.W. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 2001, 81, 1475–1485. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Weissman, L.; Eisenberg, D. The structure of melittin in the form I crystals and its implication for melittin’s lytic and surface activities. Biophys. J. 1982, 37, 353–361. [Google Scholar] [CrossRef]
- Andersson, M.; Ulmschneider, J.P.; Ulmschneider, M.B.; White, S.H. Conformational states of melittin at a bilayer interface. Biophys. J. 2013, 104, L12–L14. [Google Scholar] [CrossRef]
- Lee, M.-T.; Sun, T.-L.; Hung, W.-C.; Huang, H.W. Process of inducing pores in membranes by melittin. Proc. Natl. Acad. Sci. USA 2013, 110, 14243–14248. [Google Scholar] [CrossRef]
- Dempsey, C.E. The actions of melittin on membranes. Biochim. Biophys. Acta. 1990, 1031, 143–161. [Google Scholar] [CrossRef]
- Sansom, M.S. The biophysics of peptide models of ion channels. Prog. Biophys. Mol. Biol. 1991, 55, 139–235. [Google Scholar] [CrossRef]
- Wu, Y.; Ma, L.; Cheley, S.; Bayley, H.; Cui, Q.; Chapman, E.R. Permeation of Styryl Dyes through Nanometer-Scale Pores in Membranes. Biochemistry 2011, 50, 7493–7502. [Google Scholar] [CrossRef]
- Huang, H.W. Action of Antimicrobial Peptides: Two-State Model. Biochemistry 2000, 39, 8347–8352. [Google Scholar] [CrossRef]
- Huang, H.W.; Chen, F.-Y.; Lee, M.-T. Molecular mechanism of Peptide-induced pores in membranes. Phys. Rev. Lett. 2004, 92, 198304. [Google Scholar] [CrossRef]
- Chen, F.-Y.; Lee, M.-T.; Huang, H.W. Evidence for membrane thinning effect as the mechanism for peptide-induced pore formation. Biophys. J. 2003, 84, 3751–3758. [Google Scholar] [CrossRef]
- Huang, H.-W. Molecular mechanism of antimicrobial peptides: The origin of cooperativity. Biochim. Biophys. Acta (BBA)—Biomembr. 2006, 1758, 1292–1302. [Google Scholar] [CrossRef]
- Sengupta, D.; Leontiadou, H.; Mark, A.E.; Marrink, S.-J. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim. Biophys. Acta 2008, 1778, 2308–2317. [Google Scholar] [CrossRef]
- Xiao, S.; Lu, X.; Gou, L.; Li, J.; Ma, Y.; Liu, J.; Yang, K.; Yuan, B. Graphene oxide as antibacterial sensitizer: Mechanically disturbed cell membrane for enhanced poration efficiency of melittin. Carbon 2019, 149, 248–256. [Google Scholar] [CrossRef]
- Naito, A.; Matsumori, N.; Ramamoorthy, A. Dynamic membrane interactions of antibacterial and antifungal biomolecules, and amyloid peptides, revealed by solid-state NMR spectroscopy. Biochim. Biophys. Acta. Gen. Subj. 2018, 1862, 307–323. [Google Scholar] [CrossRef]
- Sani, M.-A.; Separovic, F. How membrane-active peptides get into lipid membranes. Acc. Chem. Res. 2016, 49, 1130–1138. [Google Scholar] [CrossRef]
- Afonin, S.; Glaser, R.W.; Sachse, C.; Salgado, J.; Wadhwani, P.; Ulrich, A.S. 19F NMR screening of unrelated antimicrobial peptides shows that membrane interactions are largely governed by lipids. Biochim. Biophys. Acta 2014, 1838, 2260–2268. [Google Scholar] [CrossRef]
- Tremouilhac, P.; Strandberg, E.; Wadhwani, P.; Ulrich, A.S. Conditions affecting the re-alignment of the antimicrobial peptide PGLa in membranes as monitored by solid state 2 H-NMR. Biochim. Biophys. Acta 2006, 1758, 1330–1342. [Google Scholar] [CrossRef] [PubMed]
- Strandberg, E.; Horn, D.; Reißer, S.; Zerweck, J.; Wadhwani, P.; Ulrich, A.S. 2H-NMR and MD simulations reveal membrane-bound conformation of magainin 2 and its synergy with PGLa. Biophys. J. 2016, 111, 2149–2161. [Google Scholar] [CrossRef]
- Norisada, K.; Javkhlantugs, N.; Mishima, D.; Kawamura, I.; Saitô, H.; Ueda, K.; Naito, A. Dynamic Structure and Orientation of Melittin Bound to Acidic Lipid Bilayers, As Revealed by Solid-State NMR and Molecular Dynamics Simulation. J. Phys. Chem. B 2017, 121, 1802–1811. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, C.H.; Hu, D.; Ulmschneider, M.B.; Ulmschneider, J.P. Spontaneous formation of structurally diverse membrane channel architectures from a single antimicrobial peptide. Nat. Commun. 2016, 7, 13535. [Google Scholar] [CrossRef]
- Bürck, J.; Wadhwani, P.; Fanghänel, S.; Ulrich, A.S. Oriented Circular Dichroism: A Method to Characterize Membrane-Active Peptides in Oriented Lipid Bilayers. Acc. Chem. Res. 2016, 49, 184–192. [Google Scholar] [CrossRef]
- Miles, A.J.; Wallace, B.A. Circular dichroism spectroscopy of membrane proteins. Chem. Soc. Rev. 2016, 45, 4859–4872. [Google Scholar] [CrossRef]
- Lee, M.W.; Han, M.; Bossa, G.V.; Snell, C.; Song, Z.; Tang, H.; Yin, L.; Cheng, J.; May, S.; Luijten, E.; et al. Interactions between Membranes and “Metaphilic” Polypeptide Architectures with Diverse Side-Chain Populations. ACS Nano 2017, 11, 2858–2871. [Google Scholar] [CrossRef]
- Lee, M.W.; Lee, E.Y.; Lai, G.H.; Kennedy, N.W.; Posey, A.E.; Xian, W.; Ferguson, A.L.; Hill, R.B.; Wong, G.C.L. Molecular Motor Dnm1 Synergistically Induces Membrane Curvature To Facilitate Mitochondrial Fission. ACS Cent. Sci. 2017, 3, 1156–1167. [Google Scholar] [CrossRef]
- Schmidt, N.W.; Mishra, A.; Lai, G.H.; Davis, M.; Sanders, L.K.; Tran, D.; Garcia, A.; Tai, K.P.; McCray, P.B.; Ouellette, A.J.; et al. Criterion for Amino Acid Composition of Defensins and Antimicrobial Peptides Based on Geometry of Membrane Destabilization. J. Am. Chem. Soc. 2011, 133, 6720–6727. [Google Scholar] [CrossRef]
- Irudayam, S.J.; Berkowitz, M.L. Binding and reorientation of melittin in a POPC bilayer: Computer simulations. Biochim. Biophys. Acta 2012, 1818, 2975–2981. [Google Scholar] [CrossRef][Green Version]
- Gumbart, J.C.; Ulmschneider, M.B.; Hazel, A.; White, S.H.; Ulmschneider, J.P. Computed Free Energies of Peptide Insertion into Bilayers are Independent of Computational Method. J. Membr. Biol. 2018, 251, 345–356. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, T.; Wei, D.; Strandberg, E.; Ulrich, A.S.; Ulmschneider, J.P. How reliable are molecular dynamics simulations of membrane active antimicrobial peptides? Biochim. Biophys. Acta 2014, 1838, 2280–2288. [Google Scholar] [CrossRef]
- Lee, E.H.; Hsin, J.; Sotomayor, M.; Comellas, G.; Schulten, K. Discovery through the computational microscope. Structure 2009, 17, 1295–1306. [Google Scholar] [CrossRef]
- Bereau, T.; Deserno, M. Enhanced Sampling of Coarse-Grained Transmembrane-Peptide Structure Formation from Hydrogen-Bond Replica Exchange. J. Membr. Biol. 2015, 248, 395–405. [Google Scholar] [CrossRef]
- Dong, X.; Qiao, Q.; Qian, Z.; Wei, G. Recent computational studies of membrane interaction and disruption of human islet amyloid polypeptide: Monomers, oligomers and protofibrils. Biochim. Biophys. Acta 2018, 1860, 1826–1839. [Google Scholar] [CrossRef] [PubMed]
- Yen, H.-Y.; Hoi, K.K.; Liko, I.; Hedger, G.; Horrell, M.R.; Song, W.; Wu, D.; Heine, P.; Warne, T.; Lee, Y.; et al. PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature 2018, 559, 423–427. [Google Scholar] [CrossRef]
- Caffalette, C.A.; Corey, R.A.; Sansom, M.S.P.; Stansfeld, P.J.; Zimmer, J. A lipid gating mechanism for the channel-forming O antigen ABC transporter. Nat. Commun. 2019, 10, 824. [Google Scholar] [CrossRef]
- Ingólfsson, H.I.; Arnarez, C.; Periole, X.; Marrink, S.J. Computational ‘microscopy’ of cellular membranes. J. Cell Sci. 2016, 129, 257. [Google Scholar] [CrossRef]
- Cheng, X.; Smith, J.C. Biological Membrane Organization and Cellular Signaling. Chem. Rev. 2019. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.J.; Lakshminarayanan, R.; Beuerman, R.W.; Li, J.; Verma, C.S. Conformational Transitions of Melittin between Aqueous and Lipid Phases: Comparison of Simulations with Experiments. J. Phys. Chem. B 2018, 122, 8698–8705. [Google Scholar] [CrossRef] [PubMed]
- Avci, F.G.; Akbulut, B.S.; Ozkirimli, E. Membrane Active Peptides and Their Biophysical Characterization. Biomolecules 2018, 8, 77. [Google Scholar] [CrossRef]
- Sun, D.; Forsman, J.; Woodward, C.E. Molecular Simulations of Melittin-Induced Membrane Pores. J. Phys. Chem. B 2017, 121, 10209–10214. [Google Scholar] [CrossRef]
- Leveritt, J.M.; Pino-Angeles, A.; Lazaridis, T. The Structure of a Melittin-Stabilized Pore. Biophys. J. 2015, 108, 2424–2426. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, M. Chapter One—A Molecular Look at Membranes. In Current Topics in Membranes; Bennett, V., Ed.; Academic Press: Cambridge, MA, USA, 2016; Volume 77, pp. 1–25. ISBN 1063-5823. [Google Scholar]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef]
- Monticelli, L.; Kandasamy, S.K.; Periole, X.; Larson, R.G.; Tieleman, D.P.; Marrink, S.-J. The MARTINI coarse-grained force field: Extension to proteins. J. Chem. Theory Comp. 2008, 4, 819–834. [Google Scholar] [CrossRef]
- De Jong, D.H.; Singh, G.; Bennett, W.D.; Arnarez, C.; Wassenaar, T.A.; Schäfer, L.V.; Periole, X.; Tieleman, D.P.; Marrink, S.J. Improved parameters for the martini coarse-grained protein force field. J. Chem. Theory Comp. 2012, 9, 687–697. [Google Scholar] [CrossRef]
- Pannuzzo, M.; McDargh, Z.A.; Deserno, M. The role of scaffold reshaping and disassembly in dynamin driven membrane fission. eLife 2018, 7, e39441. [Google Scholar] [CrossRef]
- Lelimousin, M.; Limongelli, V.; Sansom, M.S. Conformational changes in the epidermal growth factor receptor: Role of the transmembrane domain investigated by coarse-grained metadynamics free energy calculations. J. Am. Chem. Soc. 2016, 138, 10611–10622. [Google Scholar] [CrossRef]
- Marrink, S.J.; Corradi, V.; Souza, P.C.T.; Ingólfsson, H.I.; Tieleman, D.P.; Sansom, M.S.P. Computational Modeling of Realistic Cell Membranes. Chem. Rev. 2019. [Google Scholar] [CrossRef] [PubMed]
- Santo, K.P.; Irudayam, S.J.; Berkowitz, M.L. Melittin creates transient pores in a lipid bilayer: Results from computer simulations. J. Phys. Chem. B 2013, 117, 5031–5042. [Google Scholar] [CrossRef] [PubMed]
- Santo, K.P.; Berkowitz, M.L. Difference between Magainin-2 and Melittin Assemblies in Phosphatidylcholine Bilayers: Results from Coarse-Grained Simulations. J. Phys. Chem. B 2012, 116, 3021–3030. [Google Scholar] [CrossRef] [PubMed]
- Han, E.; Lee, H. Synergistic effects of magainin 2 and PGLa on their heterodimer formation, aggregation, and insertion into the bilayer. RSC Adv. 2015, 5, 2047–2055. [Google Scholar] [CrossRef]
- Woo, S.Y.; Lee, H. Aggregation and insertion of melittin and its analogue MelP5 into lipid bilayers at different concentrations: Effects on pore size, bilayer thickness and dynamics. Phys. Chem. Chem. Phys. 2017, 19, 7195–7203. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K.; Wang, Y.; Zhao, T.; Ulmschneider, J.P. Insights from micro-second atomistic simulations of melittin in thin lipid bilayers. J. Membrane. Biol. 2015, 248, 497–503. [Google Scholar] [CrossRef]
- Therrien, A.; Fournier, A.; Lafleur, M. Role of the Cationic C-Terminal Segment of Melittin on Membrane Fragmentation. J. Phys. Chem. B 2016, 120, 3993–4002. [Google Scholar] [CrossRef]
- Illya, G.; Deserno, M. Coarse-Grained Simulation Studies of Peptide-Induced Pore Formation. Biophys. J. 2008, 95, 4163–4173. [Google Scholar] [CrossRef]
- Chen, C.H.; Wiedman, G.; Khan, A.; Ulmschneider, M.B. Absorption and folding of melittin onto lipid bilayer membranes via unbiased atomic detail microsecond molecular dynamics simulation. Biochim. Biophys. Acta 2014, 1838, 2243–2249. [Google Scholar] [CrossRef]
- Sun, D.; Forsman, J.; Woodward, C.E. Multistep molecular dynamics simulations identify the highly cooperative activity of melittin in recognizing and stabilizing membrane pores. Langmuir 2015, 31, 9388–9401. [Google Scholar] [CrossRef]
- Goliaei, A.; Santo, K.P.; Berkowitz, M.L. Local Pressure Changes in Lipid Bilayers Due to Adsorption of Melittin and Magainin-h2 Antimicrobial Peptides: Results from Computer Simulations. J. Phys. Chem. B 2014, 118, 12673–12679. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, S.; Li, J.; Yuan, B.; Yang, K.; Ma, Y. Molecular details on the intermediate states of melittin action on a cell membrane. Biochim. Biophys. Acta 2018, 1860, 2234–2241. [Google Scholar] [CrossRef]
- Lu, N.-Y.; Yang, K.; Li, J.-L.; Yuan, B.; Ma, Y.-Q. Vesicle deposition and subsequent membrane–melittin interactions on different substrates: A QCM-D experiment. Biochim. Biophys. Acta 2013, 1828, 1918–1925. [Google Scholar] [CrossRef]
- Dufourc, E.J.; Smith, I.C.P.; Dufourcq, J. Molecular details of melittin-induced lysis of phospholipid membranes as revealed by deuterium and phosphorus NMR. Biochemistry 1986, 25, 6448–6455. [Google Scholar] [CrossRef]
- Therrien, A.; Lafleur, M. Melittin-induced lipid extraction modulated by the methylation level of phosphatidylcholine headgroups. Biophys. J. 2016, 110, 400–410. [Google Scholar] [CrossRef]
- Irudayam, S.J.; Pobandt, T.; Berkowitz, M.L. Free energy barrier for melittin reorientation from a membrane-bound state to a transmembrane state. J. Phys. Chem. B 2013, 117, 13457–13463. [Google Scholar] [CrossRef][Green Version]
- Lu, N.; Yang, K.; Yuan, B.; Ma, Y. Molecular Response and Cooperative Behavior during the Interactions of Melittin with a Membrane: Dissipative Quartz Crystal Microbalance Experiments and Simulations. J. Phys. Chem. B 2012, 116, 9432–9438. [Google Scholar] [CrossRef]
- Lyu, Y.; Xiang, N.; Zhu, X.; Narsimhan, G. Potential of mean force for insertion of antimicrobial peptide melittin into a pore in mixed DOPC/DOPG lipid bilayer by molecular dynamics simulation. J. Chem. Phys. 2017, 146, 155101. [Google Scholar] [CrossRef]
- Li, Y.; Qian, Z.; Ma, L.; Hu, S.; Nong, D.; Xu, C.; Ye, F.; Lu, Y.; Wei, G.; Li, M. Single-molecule visualization of dynamic transitions of pore-forming peptides among multiple transmembrane positions. Nat. Commun. 2016, 7, 12906. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Sinha, S.K.; Patel, S. Investigating Hydrophilic Pores in Model Lipid Bilayers Using Molecular Simulations: Correlating Bilayer Properties with Pore-Formation Thermodynamics. Langmuir 2015, 31, 6615–6631. [Google Scholar] [CrossRef] [PubMed]
- McNulty, R.; Ulmschneider, J.P.; Luecke, H.; Ulmschneider, M.B. Mechanisms of molecular transport through the urea channel of Helicobacter pylori. Nat. Commun. 2013, 4, 2900. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Kim, S.Y.; Pittman, A.E.; King, G.M.; Wimley, W.C.; Hristova, K. Potent Macromolecule-Sized Poration of Lipid Bilayers by the Macrolittins, A Synthetically Evolved Family of Pore-Forming Peptides. J. Am. Chem. Soc. 2018, 140, 6441–6447. [Google Scholar] [CrossRef]
- Wiedman, G.; Kim, S.Y.; Zapata-Mercado, E.; Wimley, W.C.; Hristova, K. pH-triggered, macromolecule-sized poration of lipid bilayers by synthetically evolved peptides. J. Am. Chem. Soc. 2017, 139, 937–945. [Google Scholar] [CrossRef]
- Krauson, A.J.; He, J.; Wimley, W.C. Gain-of-function analogues of the pore-forming peptide melittin selected by orthogonal high-throughput screening. J. Am. Chem. Soc. 2012, 134, 12732–12741. [Google Scholar] [CrossRef] [PubMed]
- Wiedman, G.; Fuselier, T.; He, J.; Searson, P.C.; Hristova, K.; Wimley, W.C. Highly efficient macromolecule-sized poration of lipid bilayers by a synthetically evolved peptide. J. Am. Chem. Soc. 2014, 136, 4724–4731. [Google Scholar] [CrossRef]
- Krauson, A.J.; Hall, O.M.; Fuselier, T.; Starr, C.G.; Kauffman, W.B.; Wimley, W.C. Conformational fine-tuning of pore-forming peptide potency and selectivity. J. Am. Chem. Soc. 2015, 137, 16144–16152. [Google Scholar] [CrossRef]
- Cherkasov, A.; Hilpert, K.; Jenssen, H.; Fjell, C.D.; Waldbrook, M.; Mullaly, S.C.; Volkmer, R.; Hancock, R.E.W. Use of Artificial Intelligence in the Design of Small Peptide Antibiotics Effective against a Broad Spectrum of Highly Antibiotic-Resistant Superbugs. ACS Chem. Biol. 2009, 4, 65–74. [Google Scholar] [CrossRef]
- Lata, S.; Sharma, B.; Raghava, G. Analysis and prediction of antibacterial peptides. BMC Bioinformatics 2007, 8, 263. [Google Scholar] [CrossRef]
- Mee, R.P.; Auton, T.R.; Morgan, P.J. Design of active analogues of a 15-residue peptide using D-optimal design, QSAR and a combinatorial search algorithm. J. Pept. Res. 1997, 49, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Fulan, B.M.; Wong, G.C.; Ferguson, A.L. Mapping membrane activity in undiscovered peptide sequence space using machine learning. Proc. Natl Acad. Sci. USA 2016, 113, 13588–13593. [Google Scholar] [CrossRef]
- Lee, M.W.; Lee, E.Y.; Ferguson, A.L.; Wong, G.C.L. Machine learning antimicrobial peptide sequences: Some surprising variations on the theme of amphiphilic assembly. Curr. Opin. Colloid Interface Sci. 2018, 38, 204–213. [Google Scholar] [CrossRef]
- Lee, E.Y.; Wong, G.C.L.; Ferguson, A.L. Machine learning-enabled discovery and design of membrane-active peptides. Bioorg. Med. Chem. 2018, 26, 2708–2718. [Google Scholar] [CrossRef]
- Mei, L.; Lu, Z.; Zhang, W.; Wu, Z.; Zhang, X.; Wang, Y.; Luo, Y.; Li, C.; Jia, Y. Bioconjugated nanoparticles for attachment and penetration into pathogenic bacteria. Biomaterials 2013, 34, 10328–10337. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; Yang, J.; Jia, H.-R.; Li, Y.-H.; Chen, Z.; Wu, F.-G. Quaternized Silicon Nanoparticles with Polarity-Sensitive Fluorescence for Selectively Imaging and Killing Gram-Positive Bacteria. Adv. Funct. Mater. 2016, 26, 5958–5970. [Google Scholar] [CrossRef]
- Pillai, P.P.; Kowalczyk, B.; Kandere-Grzybowska, K.; Borkowska, M.; Grzybowski, B.A. Engineering Gram Selectivity of Mixed-Charge Gold Nanoparticles by Tuning the Balance of Surface Charges. Angew. Chem. Int. Ed. 2016, 55, 8610–8614. [Google Scholar] [CrossRef]
- Li, J.; Chen, Z.; Zhou, M.; Jing, J.; Li, W.; Wang, Y.; Wu, L.; Wang, L.; Wang, Y.; Lee, M. Polyoxometalate-Driven Self-Assembly of Short Peptides into Multivalent Nanofibers with Enhanced Antibacterial Activity. Angew. Chem. Int. Ed. 2016, 55, 2592–2595. [Google Scholar] [CrossRef]
- Liu, L.; Xu, K.; Wang, H.; Jeremy Tan, P.K.; Fan, W.; Venkatraman, S.S.; Li, L.; Yang, Y.-Y. Self-assembled cationic peptide nanoparticles as an efficient antimicrobial agent. Nat. Nanotech. 2009, 4, 457. [Google Scholar] [CrossRef]
- Lam, S.J.; O’Brien-Simpson, N.M.; Pantarat, N.; Sulistio, A.; Wong, E.H.; Chen, Y.-Y.; Lenzo, J.C.; Holden, J.A.; Blencowe, A.; Reynolds, E.C. Combating multidrug-resistant Gram-negative bacteria with structurally nanoengineered antimicrobial peptide polymers. Nat. Microbiol. 2016, 1, 16162. [Google Scholar] [CrossRef]
- Ding, H.-M.; Tian, W.; Ma, Y.-Q. Designing nanoparticle translocation through membranes by computer simulations. ACS Nano 2012, 6, 1230–1238. [Google Scholar] [CrossRef]
- Tian, W.; Ma, Y. Theoretical and computational studies of dendrimers as delivery vectors. Chem. Soc. Rev. 2013, 42, 705–727. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Ma, Y. Theoretical and computational investigations of nanoparticle-biomembrane interactions in cellular delivery. Small 2015, 11, 1055–1071. [Google Scholar] [CrossRef]
- Ji, Q.-J.; Yuan, B.; Lu, X.-M.; Yang, K.; Ma, Y.-Q. Controlling the nanoscale rotational behaviors of nanoparticles on the cell membranes: A computational model. Small 2016, 12, 1140–1146. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Liu, J.; Gou, L.; Li, J.; Yuan, B.; Yang, K.; Ma, Y. Designing Melittin-Graphene Hybrid Complexes for Enhanced Antibacterial Activity. Adv. Healthc. Mater. 2019, 0, 1801521. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, J.; Lu, X.; Deng, Z.; Xiao, S.; Yuan, B.; Yang, K. How Melittin Inserts into Cell Membrane: Conformational Changes, Inter-Peptide Cooperation, and Disturbance on the Membrane. Molecules 2019, 24, 1775. https://doi.org/10.3390/molecules24091775
Hong J, Lu X, Deng Z, Xiao S, Yuan B, Yang K. How Melittin Inserts into Cell Membrane: Conformational Changes, Inter-Peptide Cooperation, and Disturbance on the Membrane. Molecules. 2019; 24(9):1775. https://doi.org/10.3390/molecules24091775
Chicago/Turabian StyleHong, Jiajia, Xuemei Lu, Zhixiong Deng, Shufeng Xiao, Bing Yuan, and Kai Yang. 2019. "How Melittin Inserts into Cell Membrane: Conformational Changes, Inter-Peptide Cooperation, and Disturbance on the Membrane" Molecules 24, no. 9: 1775. https://doi.org/10.3390/molecules24091775
APA StyleHong, J., Lu, X., Deng, Z., Xiao, S., Yuan, B., & Yang, K. (2019). How Melittin Inserts into Cell Membrane: Conformational Changes, Inter-Peptide Cooperation, and Disturbance on the Membrane. Molecules, 24(9), 1775. https://doi.org/10.3390/molecules24091775