Structure and Anti-Tumor Activities of Exopolysaccharides from Alternaria mali Roberts
Abstract
1. Introduction
2. Results and Discussion
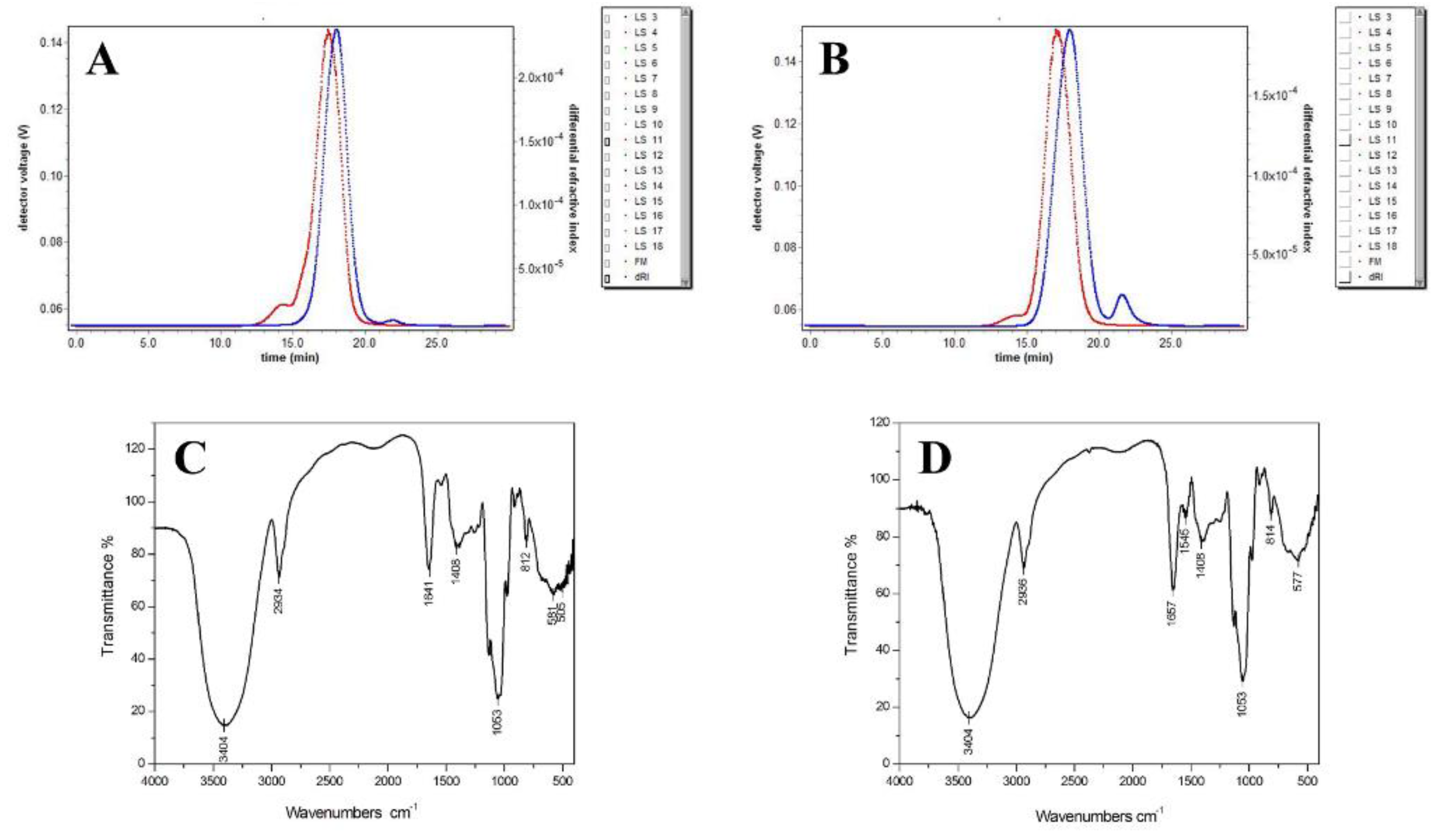
2.1. Purity and Molecular Weight of AMEP-1 and AMEP-2
2.2. Monosaccharide Composition of AMEP-1 and AMEP-2
2.3. Infrared Spectra of AMEP-1 and AMEP-2
2.4. NMR Spectra of AMEP-1 and AMEP-2
2.5. Effect of AMEP-2 on Proliferation of the Human Gastric Cancer Cell Line BGC-823
2.6. Effect of AMEP-2 on the Cell Cycle in BGC-823 Cells
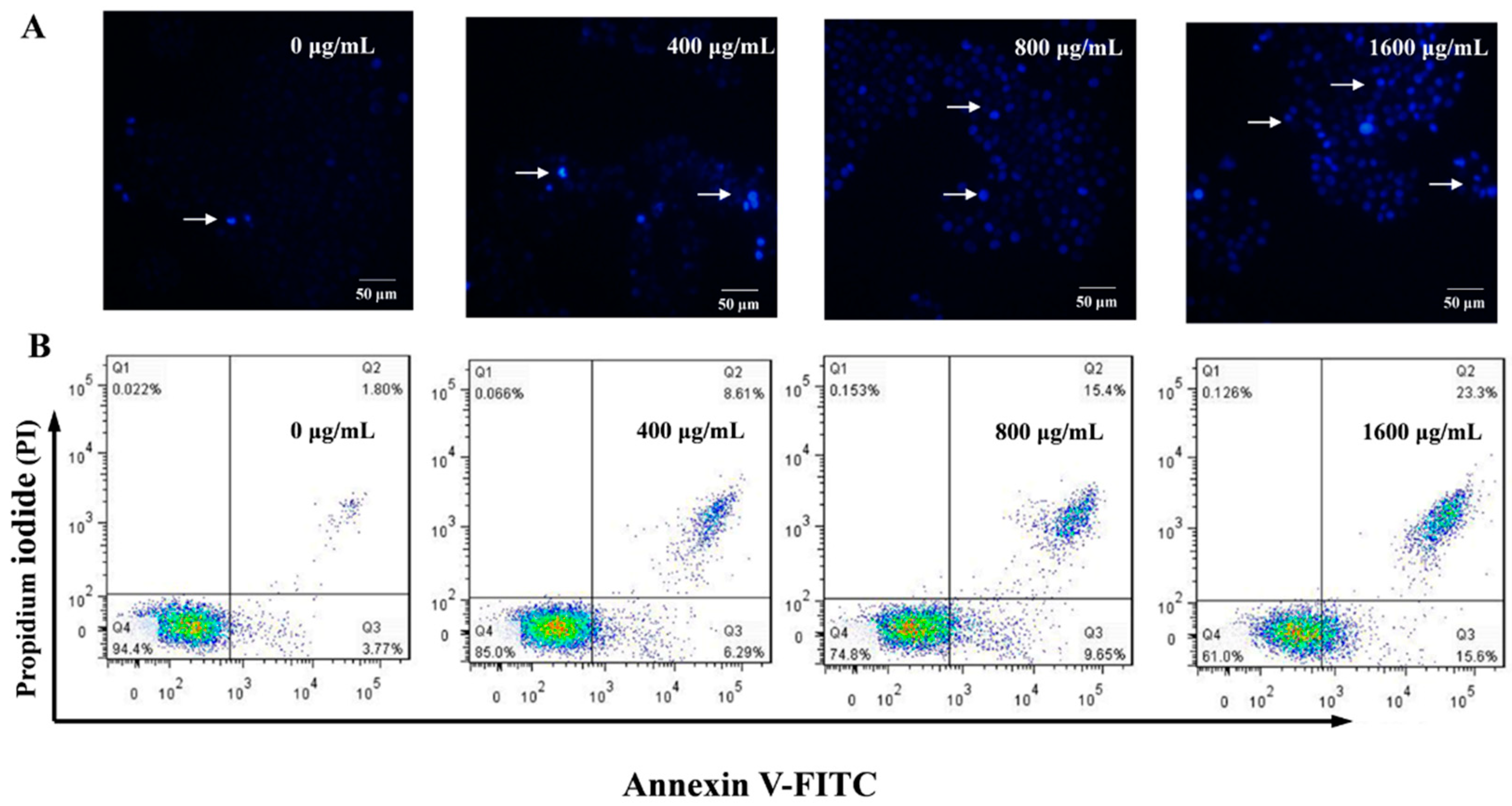
2.7. Effect of AMEP-2 on the Apoptosis of BGC-823 Cells
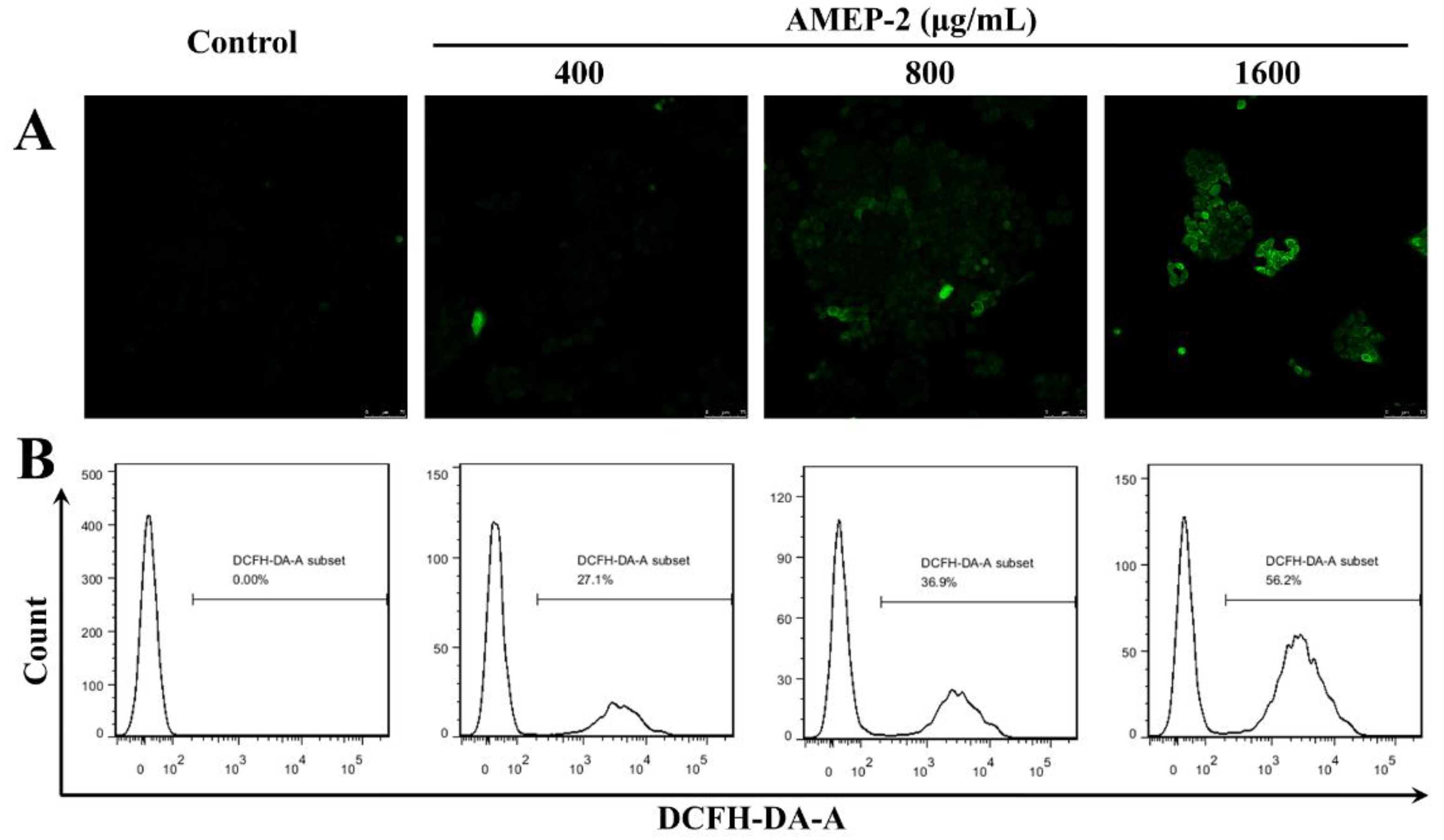
2.8. Effect of AMEP-2 on ROS Generation in BGC-823 Cells
2.9. Effect of AMEP-2 on Mitochondrial Membrane Potential in BGC-823 Cells
3. Materials and Methods
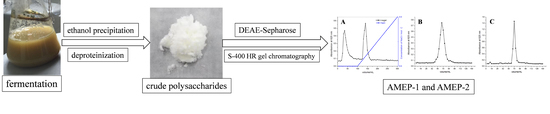
3.1. Culture and Polysaccharide Production of A. Mali Roberts
3.2. AMEP Extraction and Purification
3.3. Determination of Purity and Molecular Weight
3.4. Monosaccharide Composition Analysis
3.5. Infrared Spectrum Analysis
3.6. Nuclear Magnetic Resonance (NMR) Spectroscopy Analysis
3.7. Cell Culture
3.8. Proliferation Assay
3.9. Cell Cycle Detection
3.10. Hoechst 33258 Staining
3.11. Annexin V-FITC–PI Double Staining
3.12. Detection of ROS
3.13. Detection of Mitochondrial Membrane Potential
3.14. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gad, A.; Kydd, J.; Piel, B.; Rai, P. Targeting Cancer using Polymeric Nanoparticle mediated Combination Chemotherapy. Int. J. Nanomed. Nanosurgery 2016, 2. [Google Scholar] [CrossRef]
- Bahirwani, R.; Reddy, K.R. Drug-induced liver injury due to cancer chemotherapeutic agents. Semin. Liver Dis. 2014, 34, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, M.M.; Lavi, O.; Hall, M.D.; Gillet, J.P. Toward a Better Understanding of the Complexity of Cancer Drug Resistance. Annu. Rev. Pharmacol. Toxicol. 2016, 56, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Luo, P.; Liu, H. A Potential Adjuvant Agent of Chemotherapy: Sepia Ink Polysaccharides. Mar. Drugs 2018, 16, 485. [Google Scholar]
- Wang, G.; Zhu, L.; Yu, B.; Chen, K.; Liu, B.; Liu, J.; Qin, G.; Liu, C.; Liu, H.; Chen, K. Exopolysaccharide from Trichoderma pseudokoningii induces macrophage activation. Carbohydr. Polym. 2016, 149, 112–120. [Google Scholar] [CrossRef]
- Wang, G.; Liu, C.; Liu, J.; Liu, B.; Li, P.; Qin, G.; Xu, Y.; Chen, K.; Liu, H.; Chen, K. Exopolysaccharide from Trichoderma pseudokoningii induces the apoptosis of MCF-7 cells through an intrinsic mitochondrial pathway. Carbohydr. Polym. 2016, 136, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Sun, Q.; Liu, J.; Zhang, X.; Song, G.; Wang, G.; Zhang, P.; Chen, K. Polysaccharide from Rhizopus nigricans inhibits the invasion and metastasis of colorectal cancer. Biomed. Pharmacother. 2018, 103, 738–745. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tian, Y.; Shao, J.; Shu, X.; Jia, J.; Ren, X.; Guan, Y. Macrophage immunomodulatory activity of the polysaccharide isolated from Collybia radicata mushroom. Int. J. Biol. Macromol. 2018, 108, 300–306. [Google Scholar] [CrossRef]
- Chen, Y.; Yao, F.; Ming, K.; Wang, D.; Hu, Y.; Liu, J. Polysaccharides from Traditional Chinese Medicines: Extraction, Purification, Modification, and Biological Activity. Molecules 2016, 21, 1705. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Jiao, Y.; Yang, X.; Yuan, L.; Guo, J.; Zhao, Y. Antioxidant and antitumor effects of polysaccharides from the fungus Pleurotus abalonus. Chem. Biol. Interact. 2015, 237, 166–174. [Google Scholar] [CrossRef]
- Ryu, D.S.; Baek, G.O.; Kim, E.Y.; Kim, K.H.; Lee, D.S. Effects of polysaccharides derived from Orostachys japonicus on induction of cell cycle arrest and apoptotic cell death in human colon cancer cells. BMB Rep. 2010, 43, 750–755. [Google Scholar] [CrossRef]
- Almeida, E.A.; Facchi, S.P.; Martins, A.F.; Nocchi, S.; Schuquel, I.T.; Nakamura, C.V.; Rubira, A.F.; Muniz, E.C. Synthesis and characterization of pectin derivative with antitumor property against Caco-2 colon cancer cells. Carbohydr. Polym. 2015, 115, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Chien, R.C.; Yen, M.T.; Tseng, Y.H.; Mau, J.L. Chemical characteristics and anti-proliferation activities of Ganoderma tsugae polysaccharides. Carbohydr. Polym. 2015, 128, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Cheng, Y.; Wang, F.; Sun, L.; Liu, C.; Chen, G.; Li, Y.; Ward, S.G.; Qu, X. Inhibition activity of sulfated polysaccharide of Sepiella maindroni ink on matrix metalloproteinase (MMP)-2. Biomed. Pharmacother. 2008, 62, 297–302. [Google Scholar] [CrossRef]
- Wang, X.; Ding, J.; Feng, Y.; Weng, L.; Zhao, G.; Xiang, J.; Zhang, M.; Xing, D. Targeting of growth factors in the treatment of hepatocellular carcinoma: The potentials of polysaccharides. Oncol. Lett. 2017, 13, 1509–1517. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Yang, W.; Zhang, L.; Cao, G. Anti-hepatoma activity in mice of a polysaccharide from the rhizome of Anemone raddeana. Int. J. Biol. Macromol. 2012, 50, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Meng, M.; Ni, H. Chemosensitizing Effect of Astragalus Polysaccharides on Nasopharyngeal Carcinoma Cells by Inducing Apoptosis and Modulating Expression of Bax/Bcl-2 Ratio and Caspases. Med. Sci. Monit.: Int. Med. J. Exp. Clin. Res. 2017, 23, 462–469. [Google Scholar] [CrossRef]
- Mao, G.H.; Ren, Y.; Feng, W.W.; Li, Q.; Wu, H.Y.; Jin, D.; Zhao, T.; Xu, C.Q.; Yang, L.Q.; Wu, X.Y. Antitumor and immunomodulatory activity of a water-soluble polysaccharide from Grifola frondosa. Carbohydr. Polym. 2015, 134, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Gokce, E.C.; Kahveci, R.; Atanur, O.M.; Gurer, B.; Aksoy, N.; Gokce, A.; Sargon, M.F.; Cemil, B.; Erdogan, B.; Kahveci, O. Neuroprotective effects of Ganoderma lucidum polysaccharides against traumatic spinal cord injury in rats. Injury 2015, 46, 2146–2155. [Google Scholar] [CrossRef]
- Zhu, Z.Y.; Chen, C.J.; Sun, H.Q.; Chen, L.J. Structural characterisation and ACE-inhibitory activities of polysaccharide from Gastrodia elata Blume. Nat. Prod. Res. 2018, 2, 1–6. [Google Scholar] [CrossRef]
- Xu, Y.; Li, J.; Ju, J.; Shen, B.; Chen, G.; Qian, W.; Zhu, L.; Lu, J.; Liu, C.; Qin, G.; et al. Exopolysaccharide from Trichoderma pseudokoningii promotes maturation of murine dendritic cells. Int. J. Biol. Macromol. 2016, 92, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.M.; Duan, W.Q.; Liu, Y.; Cen, Y. Anti-inflammatory effect of the polysaccharides of golden needle mushroom in burned rats. Int. J. Biol. Macromol. 2010, 46, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Peng, Q.; Wang, M. Anti-colon-cancer effects of polysaccharides: A mini-review of the mechanisms. Int. J. Biol. Macromol. 2018, 114, 1127–1133. [Google Scholar] [CrossRef]
- Jin, M.; Huang, Q.; Zhao, K.; Shang, P. Biological activities and potential health benefit effects of polysaccharides isolated from Lycium barbarum L. Int. J. Biol. Macromol. 2013, 54, 16–23. [Google Scholar] [CrossRef]
- Raafat, E.M.; Gamal-Eldeen, A.M.; El-Hussieny, E.A.; Ahmed, E.F.; Eissa, A.A. Polysaccharide extracts of the brown alga Sargassum asperifolium possess in vitro cancer chemopreventive properties. Nat. Prod. Res. 2014, 28, 2304–2311. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Song, L.; Wang, H.; Liu, S.; Yu, H.; Wang, X.; Li, R.; Liu, T.; Li, P. Partial Characterization, the Immune Modulation and Anticancer Activities of Sulfated Polysaccharides from Filamentous Microalgae Tribonema sp. Molecules 2019, 24, 322. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Hou, D.; Lu, J.; Zhu, L.; Zhang, P.; Zhou, N.; Chen, K. Anti-tumor activity of exopolysaccharide from Rhizopus nigricans Ehrenb on S180 tumor-bearing mice. Bioorg. Med. Chem. Lett. 2016, 26, 2098–2104. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zhang, P.; Huang, T.; Yu, W.; Lin, J.; Li, P.; Chen, K. Polysaccharides from Rhizopus nigricans mycelia induced apoptosis and G2/M arrest in BGC-823 cells. Carbohydr. Polym. 2013, 97, 800–808. [Google Scholar] [CrossRef]
- Lu, Y.; Xu, L.; Cong, Y.; Song, G.; Han, J.; Wang, G.; Zhang, P.; Chen, K. Structural characteristics and anticancer/antioxidant activities of a novel polysaccharide from Trichoderma kanganensis. Carbohydr. Polym. 2019, 205, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Feng, J.; Zhang, J.S.; Lin, C.C.; Wang, W.H.; Chen, H.G. Structural Characteristics of the Novel Polysaccharide FVPA1 from Winter Culinary-Medicinal Mushroom, Flammulina velutipes (Agaricomycetes), Capable of Enhancing Natural Killer Cell Activity against K562 Tumor Cells. Int. J. Med. Mushrooms 2017, 19, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Zhang, J.; Gao, F.; Zhou, J.; Xiang, Z.; Zhou, C.; Wan, L.; Chen, J. Structure features and in vitro hypoglycemic activities of polysaccharides from different species of Maidong. Carbohydr. Polym. 2017, 173, 215–222. [Google Scholar] [CrossRef]
- Vinogradov, E.; Aubry, A.; Logan, S.M. Structural characterization of wall and lipidated polysaccharides from Clostridium perfringens ATCC 13124. Carbohydr. Res. 2017, 448, 88–94. [Google Scholar] [CrossRef]
- Liu, X.; Hao, J.; He, X.; Wang, S.; Cao, S.; Qin, L.; Mao, W. A rhamnan-type sulfated polysaccharide with novel structure from Monostroma angicava Kjellm (Chlorophyta) and its bioactivity. Carbohydr. Polym. 2017, 173, 732–748. [Google Scholar] [CrossRef]
- Sevag, M.G. The Presence of a Type-and Species-Specific Conjugated Polysaccharide in Type I Pneumococcus. Science 1938, 87, 304–305. [Google Scholar] [CrossRef]
- Huang, K.; Li, Y.; Tao, S.; Wei, G.; Huang, Y.; Chen, D.; Wu, C. Purification, Characterization and Biological Activity of Polysaccharides from Dendrobium officinale. Molecules 2016, 21, 701. [Google Scholar] [CrossRef]
- Yu, X.H.; Liu, Y.; Wu, X.L.; Liu, L.Z.; Fu, W.; Song, D.D. Isolation, purification, characterization and immunostimulatory activity of polysaccharides derived from American ginseng. Carbohydr. Polym. 2017, 156, 9–18. [Google Scholar] [CrossRef]
- Hu, H.; Liang, H.; Wu, Y. Isolation, purification and structural characterization of polysaccharide from Acanthopanax brachypus. Carbohydr. Polym. 2015, 127, 94–100. [Google Scholar] [CrossRef]
- Luo, Q.L.; Tang, Z.H.; Zhang, X.F.; Zhong, Y.H.; Yao, S.Z.; Wang, L.S.; Lin, C.W.; Luo, X. Chemical properties and antioxidant activity of a water-soluble polysaccharide from Dendrobium officinale. Int. J. Biol. Macromol. 2016, 89, 219–227. [Google Scholar] [CrossRef]
- Zhang, Z.P.; Shen, C.C.; Gao, F.L.; Wei, H.; Ren, D.F.; Lu, J. Isolation, Purification and Structural Characterization of Two Novel Water-Soluble Polysaccharides from Anredera cordifolia. Molecules 2017, 22, 1276. [Google Scholar] [CrossRef]
Sample Availability: Not available. |





| Group | Polydispersity | Molar Mass Moments (g/mol) | Root Mean Square Radius Moments (nm) | |||||
|---|---|---|---|---|---|---|---|---|
| Mw/Mn | Mz/Mn | Mn | Mw | Mz | Rn | Rw | Rz | |
| AMEP-1 | 1.492 | 2.858 | 2.588 × 104 | 3.861 × 104 | 7.395 × 104 | 15.2 | 12.5 | 11.6 |
| AMEP-2 | 1.849 | 3.47 | 2.536 × 104 | 4.689 × 104 | 8.798 × 104 | 35.5 | 30.2 | 22.7 |
| Group | Concentration μg·mL−1 | Inhibitory Rate of Cell Growth | ||
|---|---|---|---|---|
| 24 h | 48 h | 72 h | ||
| Control | 0 | 0 ± 6.32 | 0 ± 5.55 | 0 ± 6.68 |
| AMEP-2 | 200 | 5.12 ± 2.82 | 6.69 ± 4.12 | 8.5 ± 2.89 |
| 400 | 8.65 ± 2.15 * | 7.82 ± 5.54 | 10.86 ± 3.92 * | |
| 600 | 9.63 ± 1.53 ** | 11.84 ± 6.61 | 24.25 ± 1.3 * | |
| 800 | 13.37 ± 3.52 ** | 20.48 ± 4.1 ** | 27.37 ± 5.13 * | |
| 1200 | 14.48 ± 5.98 ** | 26.84 ± 6.33 ** | 38.42 ± 3.67 ** | |
| 1600 | 19.6 ± 1.65 ** | 34.34 ± 4.59 ** | 43.18 ± 2.62 ** | |
| 2000 | 25.05 ± 2.74 ** | 35.86 ± 2.14 ** | 46.84 ± 2.5 ** | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, P.; Fang, F.; Shao, T.; Li, P.; Hu, W.; Zhou, Y.; Wang, G.; Han, J.; Chen, K. Structure and Anti-Tumor Activities of Exopolysaccharides from Alternaria mali Roberts. Molecules 2019, 24, 1345. https://doi.org/10.3390/molecules24071345
Yuan P, Fang F, Shao T, Li P, Hu W, Zhou Y, Wang G, Han J, Chen K. Structure and Anti-Tumor Activities of Exopolysaccharides from Alternaria mali Roberts. Molecules. 2019; 24(7):1345. https://doi.org/10.3390/molecules24071345
Chicago/Turabian StyleYuan, Pingchuan, Fang Fang, Taili Shao, Ping Li, Wei Hu, Yuyan Zhou, Guodong Wang, Jun Han, and Kaoshan Chen. 2019. "Structure and Anti-Tumor Activities of Exopolysaccharides from Alternaria mali Roberts" Molecules 24, no. 7: 1345. https://doi.org/10.3390/molecules24071345
APA StyleYuan, P., Fang, F., Shao, T., Li, P., Hu, W., Zhou, Y., Wang, G., Han, J., & Chen, K. (2019). Structure and Anti-Tumor Activities of Exopolysaccharides from Alternaria mali Roberts. Molecules, 24(7), 1345. https://doi.org/10.3390/molecules24071345