Investigating the Formation of Structural Elements in Proteins Using Local Sequence-Dependent Information and a Heuristic Search Algorithm
Abstract
:1. Introduction and Related Work
2. Results and Discussion
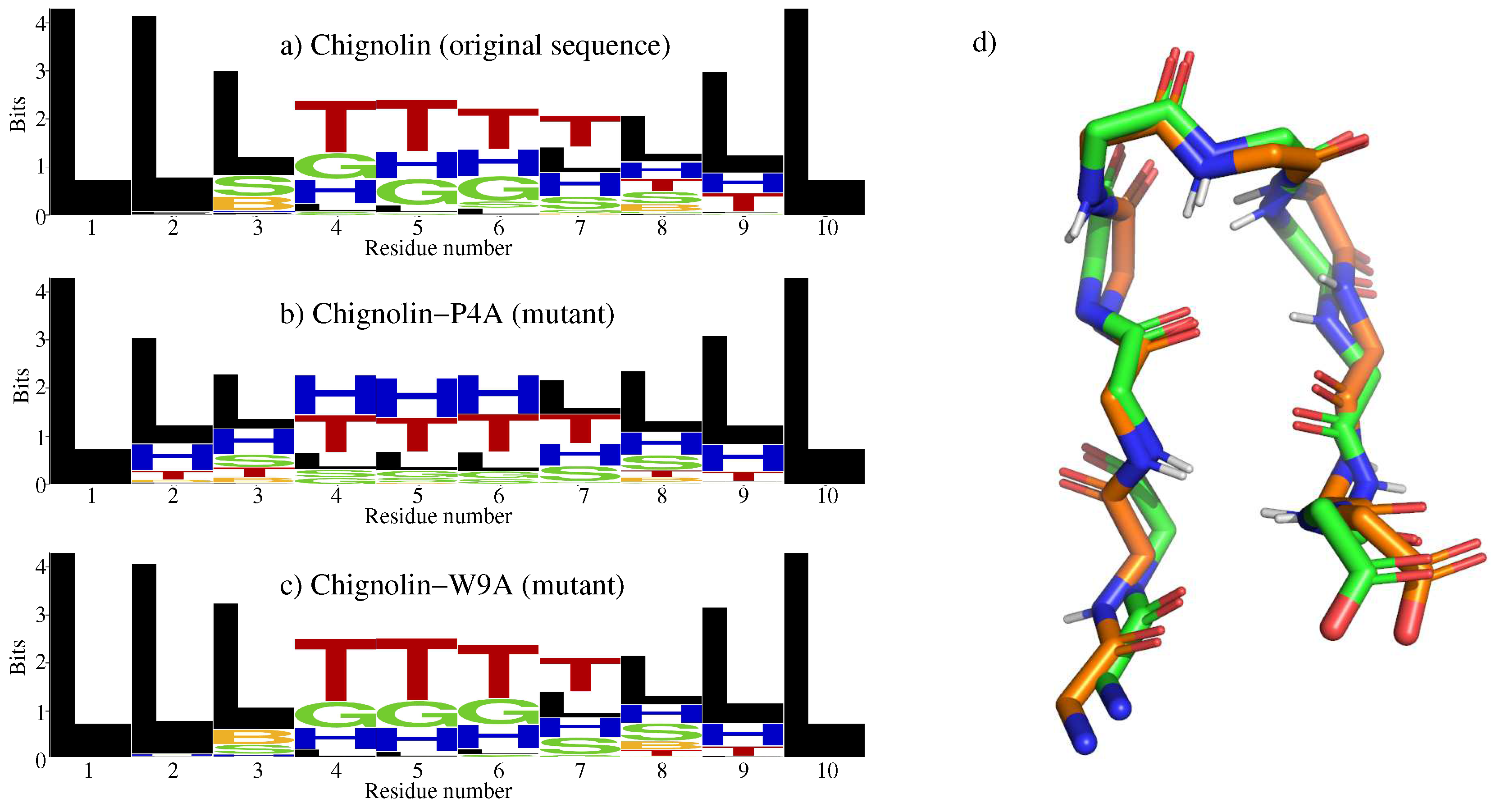
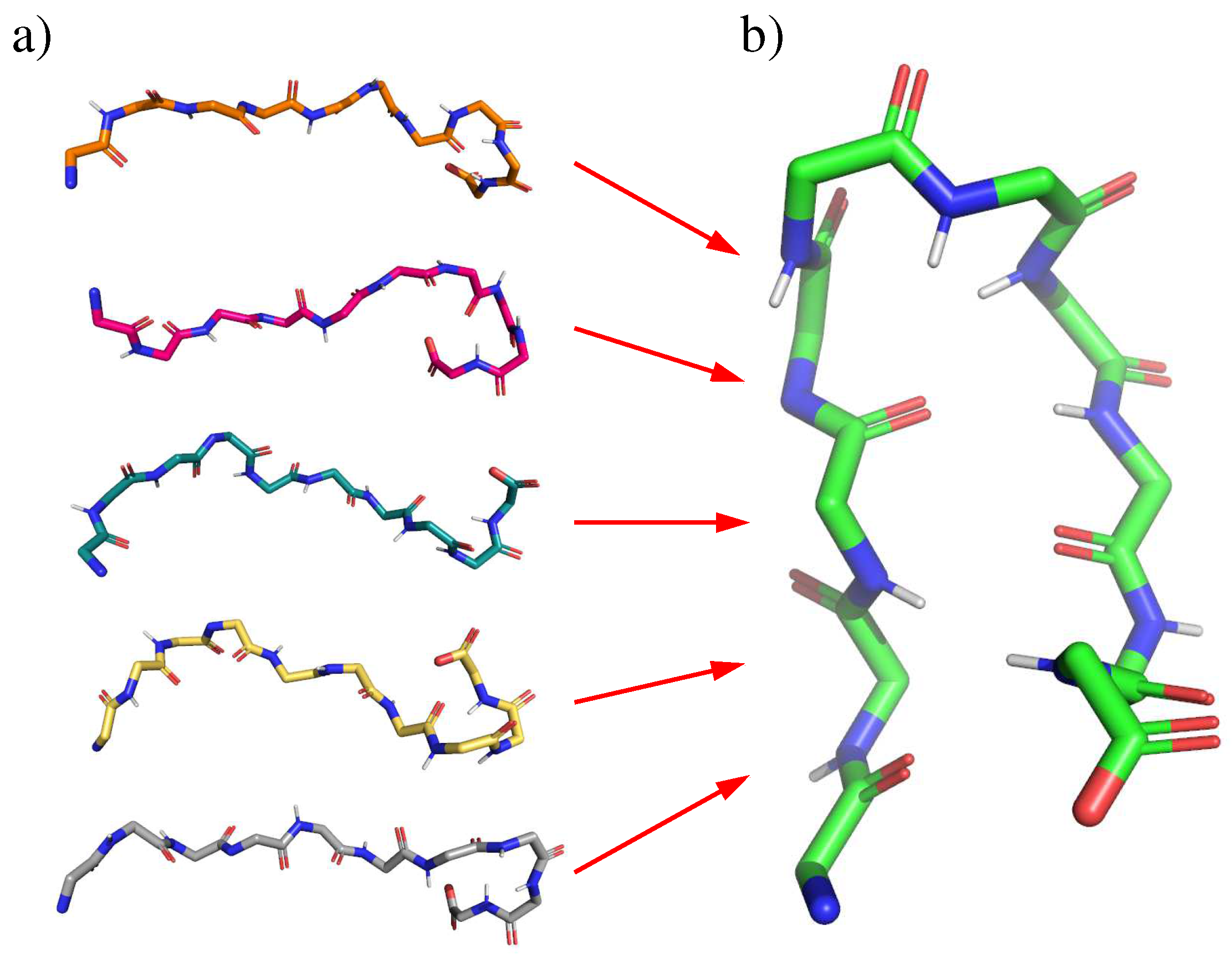
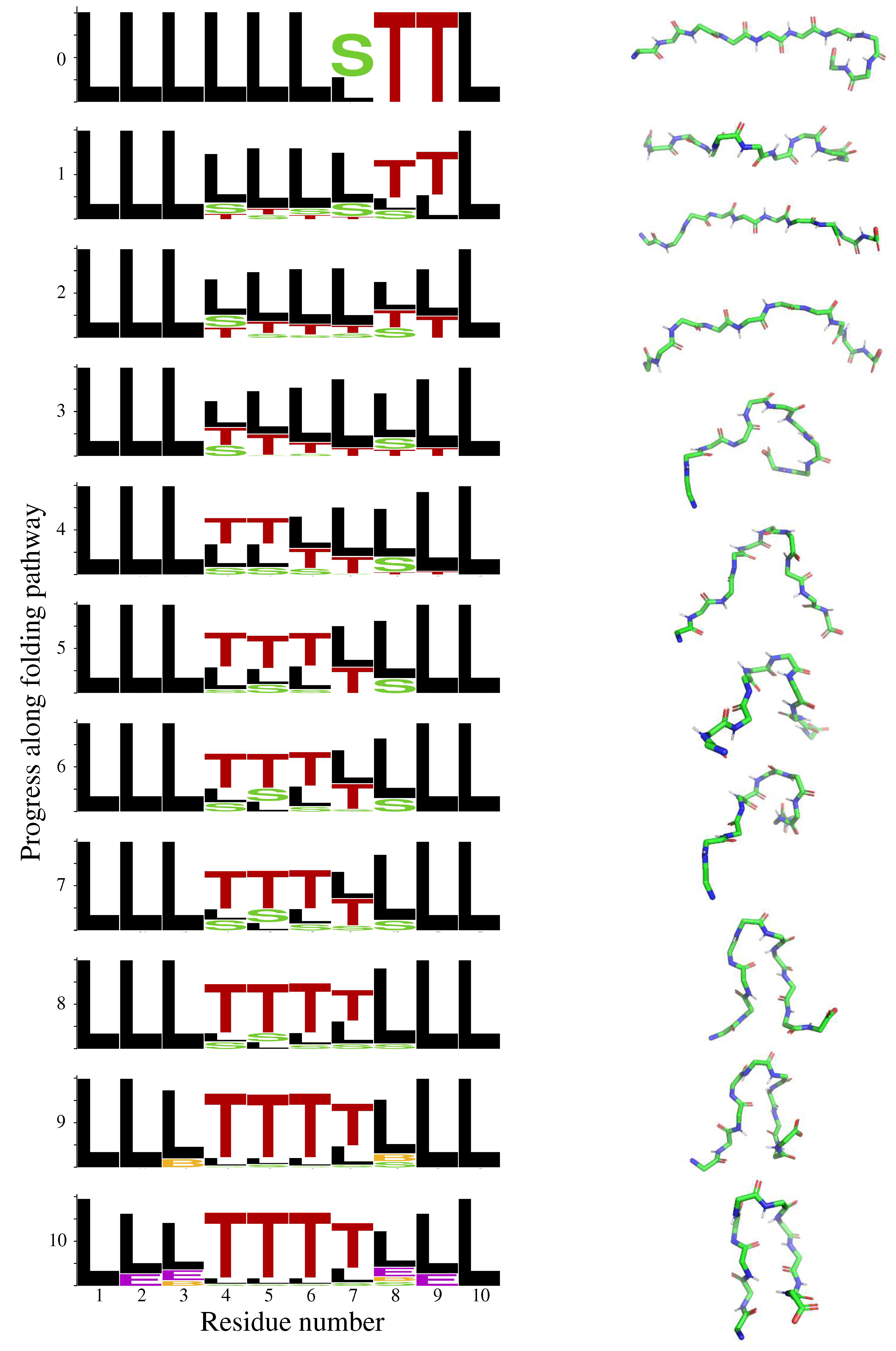
2.1. Chignolin
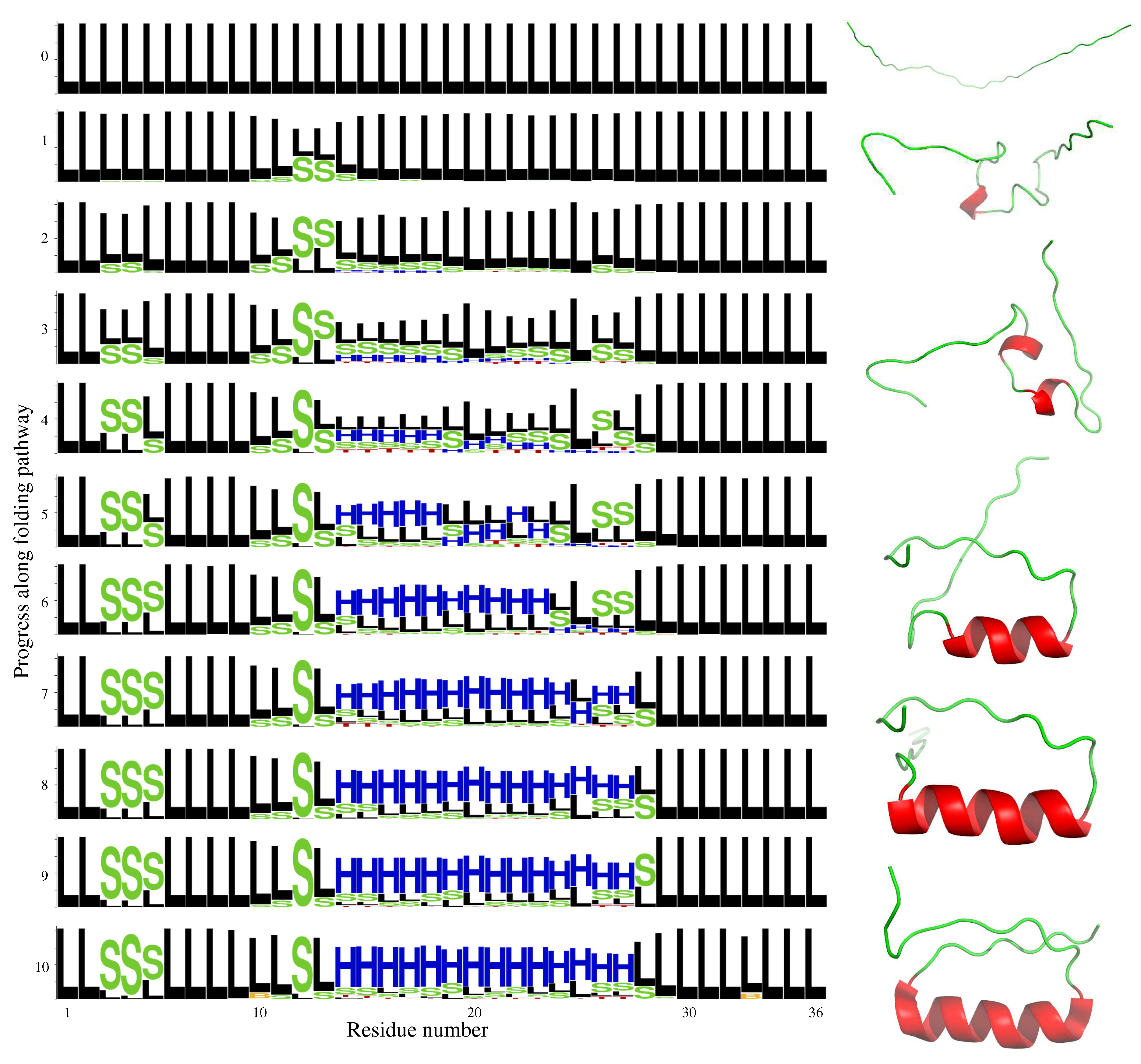
2.2. DS119
3. Materials and Methods
3.1. Structural Database
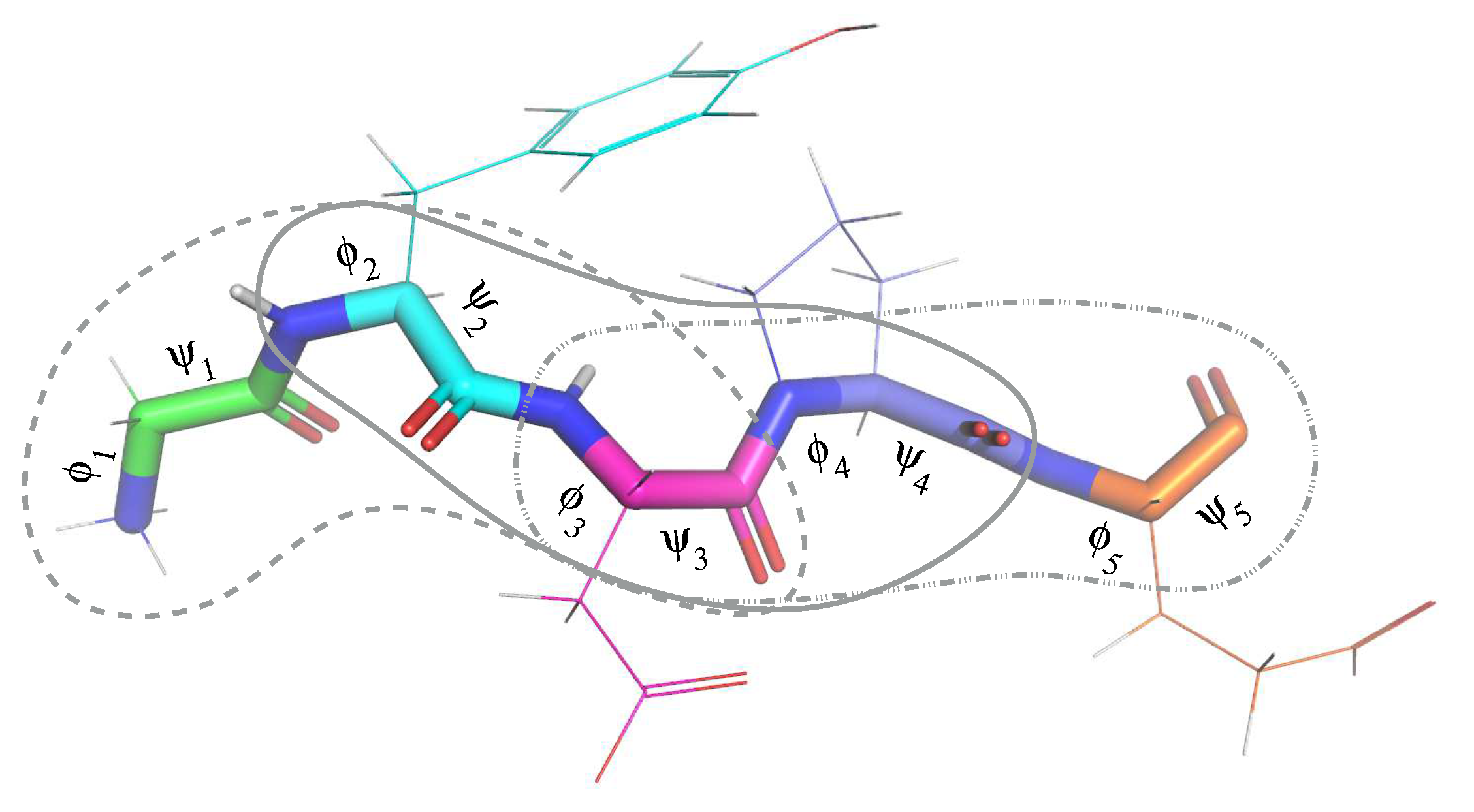
3.2. Formal Statement of the Conformation Path Finding Problem
- (i)
- the values and meet the constraints of Equation (2), and
- (ii)
- there are no collisions between the atoms of the protein in the state .
3.3. Search Algorithm
3.3.1. Heuristic Guidance Function
3.3.2. Properties of HDFS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Vendruscolo, M.; Zurdo, J.; Macphee, C.; M Dobson, C. Protein folding and misfolding: A paradigm of self-assembly and regulation in complex biological systems. Philos. Trans. Ser. A Math. Phys. Eng. Sci. 2003, 361, 1205–1222. [Google Scholar] [CrossRef]
- Valastyan, J.S.; Lindquist, S. Mechanisms of protein-folding diseases at a glance. Dis. Models Mech. 2014, 7, 9–14. [Google Scholar] [CrossRef] [Green Version]
- Knowles, T.; Vendruscolo, M.; Dobson, C. The amyloid state and its association with protein misfolding diseases. Structure 2014, 15, 384–396. [Google Scholar] [CrossRef]
- Baldwin, R.L. Protein folding: Matching speed and stability. Nature 1994, 369, 183–184. [Google Scholar] [CrossRef]
- Wolynes, P.; Onuchic, J.; Thirumalai, D. Navigating the folding routes. Science 1995, 267, 1619–1620. [Google Scholar] [CrossRef]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef]
- Rose, G.D.; Fleming, P.J.; Banavar, J.R.; Maritan, A. A backbone-based theory of protein folding. Proc. Natl. Acad. Sci. USA 2006, 103, 16623–16633. [Google Scholar] [CrossRef] [Green Version]
- Lindorff-Larsen, K.; Piana, S.; Dror, R.O.; Shaw, D.E. How fast-folding proteins fold. Science 2011, 334, 517–520. [Google Scholar] [CrossRef]
- Best, R.B. Atomistic molecular simulations of protein folding. Curr. Opin. Struct. Biol. 2012, 22, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Pancsa, R.; Fuxreiter, M. Interactions via intrinsically disordered regions: What kind of motifs? IUBMB Life 2012, 64, 513–520. [Google Scholar] [CrossRef] [Green Version]
- Tompa, P.; Davey, N.E.; Gibson, T.J.; Babu, M.M. A Million Peptide Motifs for the Molecular Biologist. Mol. Cell 2014, 55, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P.; Schad, E.; Tantos, A.; Kalmar, L. Intrinsically disordered proteins: Emerging interaction specialists. Curr. Opin. Struct. Biol. 2015, 35, 49–59. [Google Scholar] [CrossRef]
- Dunbrack, R. Rotamer libraries in the 21st century. Curr. Opin. Struct. Biol. 2002, 12, 431–440. [Google Scholar] [CrossRef]
- Smith, L.J.; Bolin, K.A.; Schwalbe, H.; MacArthur, M.W.; Thornton, J.M.; Dobson, C.M. Analysis of main chain torsion angles in proteins: Prediction of NMR coupling constants for native and random coil conformations. J. Mol. Biol. 1996, 255, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.K.; Colubri, A.; Freed, K.F.; Sosnick, T.R. Statistical coil model of the unfolded state: Resolving the reconciliation problem. Proc. Natl. Acad. Sci. USA 2005, 102, 13099–13104. [Google Scholar] [CrossRef]
- Bernadó, P.; Blanchard, L.; Timmins, P.; Marion, D.; Ruigrok, R.W.H.; Blackledge, M. A structural model for unfolded proteins from residual dipolar couplings and small-angle X-ray scattering. Proc. Natl. Acad. Sci. USA 2005, 102, 17002–17007. [Google Scholar] [CrossRef]
- Kolodny, R.; Koehl, P.; Guibas, L.; Levitt, M. Small libraries of protein fragments model native protein structures accurately. J. Mol. Biol. 2002, 323, 297–307. [Google Scholar] [CrossRef]
- Rohl, C.A.; Strauss, C.E.; Misura, K.M.; Baker, D. Protein structure prediction using Rosetta. In Numerical Computer Methods, Part D; Academic Press: Cambridge, UK, 2004; Volume 383, pp. 66–93. [Google Scholar]
- Baeten, L.; Reumers, J.; Tur, V.; Stricher, F.; Lenaerts, T.; Serrano, L.; Rousseau, F.; Schymkowitz, J. Reconstruction of protein backbones from the BriX collection of canonical protein fragments. PLOS Comput. Biol. 2008, 4, e1000083. [Google Scholar] [CrossRef]
- Maupetit, J.; Derreumaux, P.; Tufféry, P. A fast method for large-scale De Novo peptide and miniprotein structure prediction. J. Comput. Chem. 2010, 31, 726–738. [Google Scholar]
- Molloy, K.; Shehu, A. A general, adaptive, roadmap-based algorithm for protein motion computation. IEEE Trans. Nano Biosci. 2016, 15, 158–165. [Google Scholar] [CrossRef]
- Huang, J.R.; Ozenne, V.; Jensen, M.R.; Blackledge, M. Direct prediction of NMR residual dipolar couplings from the primary sequence of unfolded proteins. Angew. Chem. Int. Ed. 2013, 52, 687–690. [Google Scholar] [CrossRef] [PubMed]
- Estaña, A.; Sibille, N.; Delaforge, E.; Vaisset, M.; Cortés, J.; Bernadó, P. Realistic ensemble models of intrinsically disordered proteins using a structure-encoding coil database. Structure 2019, 27, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Levinthal, C. How to fold graciously. Mössbauer Spectroscopy in Biological Systems Proceedings. In Proceedings of a meeting held at Allerton House, Chicago, IL, USA, 17–18 March 1969; pp. 22–24. [Google Scholar]
- Rooman, M.; Dehouck, Y.; Kwasigroch, J.; Biot, C.; Gilis, D. What is paradoxical about Levinthal paradox? J. Biomol. Struct. Dyn. 2003, 20, 327–329. [Google Scholar] [CrossRef]
- Al-Bluwi, I.; Siméon, T.; Cortés, J. Motion planning algorithms for molecular simulations: A survey. Comput. Sci. Rev. 2012, 6, 125–143. [Google Scholar] [CrossRef]
- Gipson, B.; Hsu, D.; Kavraki, L.; Latombe, J.C. Computational models of protein kinematics and dynamics: Beyond simulation. Ann. Rev. Anal. Chem. 2012, 5, 273–291. [Google Scholar] [CrossRef] [PubMed]
- Shehu, A.; Plaku, E. A survey of computational treatments of biomolecules by robotics-inspired methods modeling equilibrium structure and dynamic. J. Artif. Intell. Res. 2016, 57, 509–572. [Google Scholar] [CrossRef]
- Ghallab, M.; Nau, D.; Traverso, P. Automated Planning: Theory and Practice; The Morgan Kaufmann Series in Artificial Intelligence; Elsevier: Amsterdam, The Netherlands, 2004. [Google Scholar]
- Richter, S.; Westphal, M. The LAMA planner: Guiding cost-based anytime planning with landmarks. J. Artif. Intell. Res. 2010, 39, 127–177. [Google Scholar] [CrossRef]
- Wales, D. Energy Landscapes: Applications to Clusters, Biomolecules and Glasses; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Honda, S.; Yamasaki, K.; Sawada, Y.; Morii, H. 10 residue folded peptide designed by segment statistics. Structure 2004, 12, 1507–1518. [Google Scholar] [CrossRef]
- Liang, H.; Chen, H.; Fan, K.; Wei, P.; Guo, X.; Jin, C.; Zeng, C.; Tang, C.; Lai, L. De novo design of a βαβ Motif. Angew. Chem. Int. Ed. 2009, 48, 3301–3303. [Google Scholar] [CrossRef]
- Enemark, S.; Kurniawan, N.A.; Rajagopalan, R. β-Hairpin forms by rolling up from C-terminal: Topological guidance of early folding dynamics. Sci. Rep. 2012, 2, 649. [Google Scholar] [CrossRef]
- Qi, Y.; Huang, Y.; Liang, H.; Liu, Z.; Lai, L. Folding simulations of a de novo designed protein with a βαβ fold. Biophys. J. 2010, 98, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Rapaport, D.C. The Art of Molecular Dynamics Simulation; Academic Press: Cambridge, MA, USA, 2007. [Google Scholar]
- Snow, C.; Zagrovic, B.; Pande, V. The Trp-cage: Folding kinetics and unfolded state topology via molecular dynamics simulations. J. Am. Chem. Soc. 2003, 124, 14548–14549. [Google Scholar] [CrossRef]
- Satoh, D.; Shimizu, K.; Nakamura, S.; Terada, T. Folding free-energy landscape of a 10-residue mini-protein, chignolin. FEBS Lett. 2006, 580, 3422–3426. [Google Scholar] [CrossRef] [Green Version]
- The PyMOL Molecular Graphics System, Version 2.0; Schrödinger, LLC: New York, NY, USA, 2018.
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: a sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Kührová, P.; De Simone, A.; Otyepka, M.; Best, R.B. Force-field dependence of Chignolin folding and misfolding: Comparison with experiment and redesign. Biophys. J. 2012, 102, 1897–1906. [Google Scholar] [CrossRef]
- Fox, N.K.; Brenner, S.E.; Chandonia, J.M. SCOPe: Structural classification of proteins–extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 2014, 42, D304–D309. [Google Scholar] [CrossRef]
- Scott, R.A.; Scheraga, H.A. Conformational analysis of macromolecules. III. Helical structures of polyglycine and poly-l-alanine. J. Chem. Phys. 1966, 45, 2091–2101. [Google Scholar] [CrossRef]
- Levitt, M. A simplified representation of protein conformations for rapid simulation of protein folding. J. Mol. Biol. 1976, 104, 59–107. [Google Scholar] [CrossRef] [Green Version]
- Ruiz de Angulo, V.; Cortés, J.; Porta, J.M. Rigid-CLL: Avoiding constant-distance computations in cell linked-lists algorithms. J. Comput. Chem. 2012, 33, 294–300. [Google Scholar] [CrossRef]
- Bondi, A. Van der Waals Volumes and Radii. J. Phys. Chem. 1964, 68, 441–451. [Google Scholar] [CrossRef]
- Devaurs, D.; Molloy, K.; Vaisset, M.; Shehu, A.; Siméon, T.; Cortés, J. Characterizing energy landscapes of peptides using a combination of stochastic algorithms. IEEE Trans. NanoBiosci. 2015, 14, 545–552. [Google Scholar] [CrossRef] [PubMed]


| Tripeptide Sequence | No. of Conformations |
|---|---|
| Gly-Tyr-Asp | 994 |
| Tyr-Asp-Pro | 710 |
| Asp-Pro-Glu | 1541 |
| Pro-Glu-Thr | 1030 |
| Glu-Thr-Gly | 1446 |
| Thr-Gly-Thr | 1779 |
| Gly-Thr-Trp | 545 |
| Thr-Trp-Gly | 240 |
| Chignolin (Original Sequence) | ||||
|---|---|---|---|---|
| C-ter T→folded | C-ter T→misfolded | extended→folded | extended→misfolded | |
| CPU time (s) | 11.1 | 8.7 | 5.2 | 3.5 |
| # states | 5416.4 | 2587.7 | 2800.1 | 849.5 |
| # backtracks | 234.6 | 136.6 | 124.6 | 39.2 |
| Path length (# steps) | 133.8 | 54.5 | 106.3 | 48.7 |
| Path distance (rad) | 8.8 | 5.1 | 6.0 | 7.0 |
| Path density | 31.9 | 5.5 | 23.3 | 4.5 |
| Chignolin-W9A (Mutant) | ||||
| C-ter T→folded | C-ter T→misfolded | extended→folded | extended→misfolded | |
| CPU time (s) | 12.2 | 8.8 | 5.6 | 5.1 |
| # states | 4943.6 | 2567.8 | 2317.0 | 2946.0 |
| # backtracks | 219.6 | 139.0 | 101.3 | 126.3 |
| Path length (# steps) | 140.3 | 51.3 | 103.0 | 125.7 |
| Path distance (rad) | 8.2 | 9.0 | 5.8 | 8.2 |
| Path density | 31.2 | 4.6 | 23.4 | 23.8 |
| DS119: Extended→Folded | |
|---|---|
| CPU time (s) | 25.2 |
| # states | 70558.2 |
| # backtracks | 8210.4 |
| Path length (# steps) | 158.2 |
| Path distance (rad) | 11.3 |
| Path density | 124.4 |
| Tripeptide Sequence | No. Conformations | Tripeptide Sequence | No. Conformations |
|---|---|---|---|
| Gly-Ser-Gly | 3727 | Lys-Lys-Leu | 2286 |
| Ser-Gly-Gln | 1118 | Lys-Leu-Lys | 1996 |
| Gly-Gln-Val | 1294 | Leu-Lys-Glu | 3100 |
| Gln-Val-Arg | 607 | Leu-Glu-Glu | 1631 |
| Val-Arg-Thr | 970 | Glu-Glu-Ala | 2591 |
| Arg-Thr-Ile | 757 | Glu-Ala-Lys | 1514 |
| Thr-Ile-Trp | 181 | Ala-Lys-Lys | 1714 |
| Ile-Trp-Val | 180 | Lys-Lys-Ala | 1629 |
| Trp-Val-Gly | 279 | Lys-Ala-Asn | 1009 |
| Val-Gly-Gly | 2443 | Ala-Asn-Ile | 1010 |
| Gly-Gly-Thr | 2510 | Asn-Ile-Arg | 647 |
| Gly-Thr-Pro | 1428 | Ile-Arg-Val | 998 |
| Thr-Pro-Glu | 1738 | Arg-Val-Thr | 1351 |
| Pro-Glu-Glu | 1752 | Val-Thr-Phe | 888 |
| Glu-Glu-Leu | 3433 | Thr-Phe-Trp | 151 |
| Glu-Leu-Lys | 2378 | Phe-Trp-Gly | 192 |
| Leu-Lys-Lys | 2528 | Trp-Gly-Asp | 257 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Estaña, A.; Ghallab, M.; Bernadó, P.; Cortés, J. Investigating the Formation of Structural Elements in Proteins Using Local Sequence-Dependent Information and a Heuristic Search Algorithm. Molecules 2019, 24, 1150. https://doi.org/10.3390/molecules24061150
Estaña A, Ghallab M, Bernadó P, Cortés J. Investigating the Formation of Structural Elements in Proteins Using Local Sequence-Dependent Information and a Heuristic Search Algorithm. Molecules. 2019; 24(6):1150. https://doi.org/10.3390/molecules24061150
Chicago/Turabian StyleEstaña, Alejandro, Malik Ghallab, Pau Bernadó, and Juan Cortés. 2019. "Investigating the Formation of Structural Elements in Proteins Using Local Sequence-Dependent Information and a Heuristic Search Algorithm" Molecules 24, no. 6: 1150. https://doi.org/10.3390/molecules24061150
APA StyleEstaña, A., Ghallab, M., Bernadó, P., & Cortés, J. (2019). Investigating the Formation of Structural Elements in Proteins Using Local Sequence-Dependent Information and a Heuristic Search Algorithm. Molecules, 24(6), 1150. https://doi.org/10.3390/molecules24061150






