Rare Acetogenins with Anti-Inflammatory Effect from the Red Alga Laurencia obtusa
Abstract
:1. Introduction
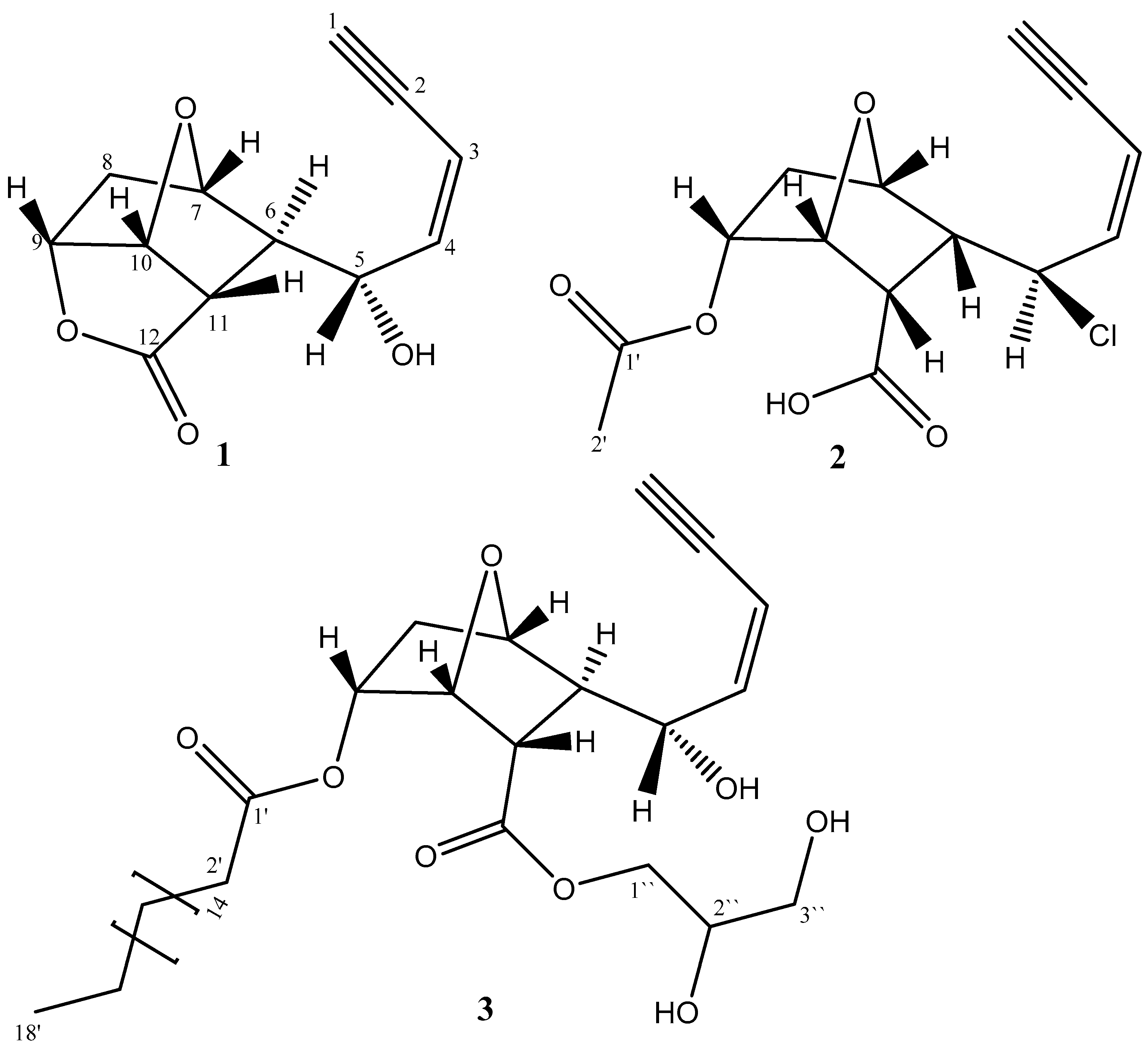
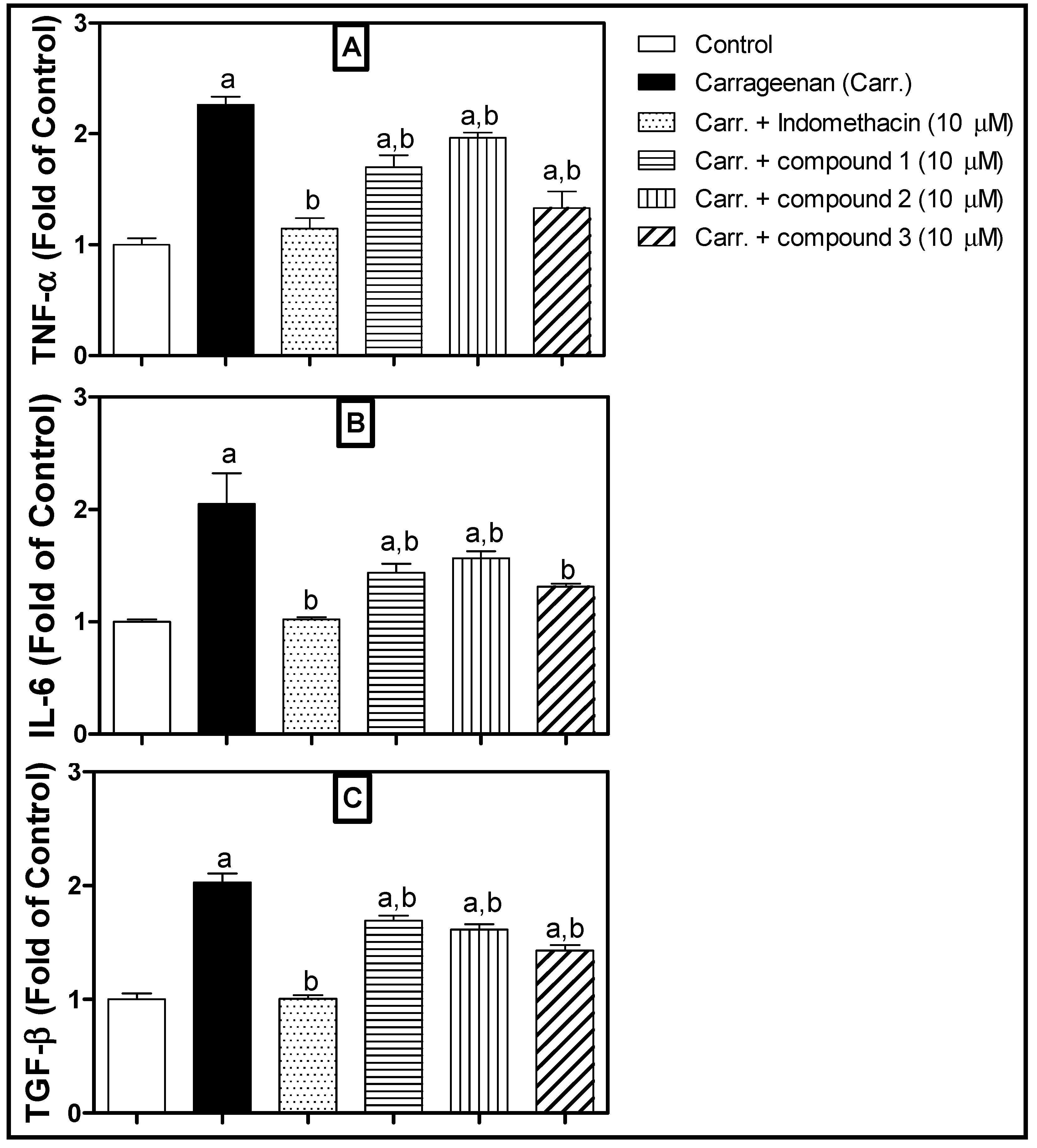
2. Results and Discussion
3. Experimental
3.1. General
3.2. Phytochemistry
3.2.1. Algael Material
3.2.2. Extraction and Isolation
3.2.3. Spectral Data
3.3. Biological Study
3.3.1. Separation of Human Lymphocytes
3.3.2. Lymphocyte Challenging
3.3.3. Assessment of Inflammatory Mediators
3.3.4. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Masudai, M.; Abe, T.; Suzuki, T.; Suzuki, M. Morphological and chemotaxonomic studies on Laurencia composita and L. okamurae (Ceramiales, Rhodophyta). Phycologia 1996, 35, 550–562. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. Algae Base. World-wide electronic publication, National University of Ireland, Galway. Available online: http://www.algaebase.org (accessed on 29 November 2015).
- Suzuki, M.; Takahashi, Y.; Nakano, S.; Abe, T.; Masuda, M.; Ohnishi, T.; Noya, Y.; Seki, K.-I. An experimental approach to study the biosynthesis of brominated metabolites by the red algal genus Laurencia. Phytochemistry 2009, 70, 1410–1415. [Google Scholar] [CrossRef] [PubMed]
- Kurata, K.; Taniguchi, K.; Agatsuma, Y.; Suzuki, M. Diterpenoid feeding-deterrents from Laurencia saitoi. Phytochemistry 1998, 47, 363–369. [Google Scholar] [CrossRef]
- Paul, V.J.; Ritson-Williams, R.; Sharp, K. Marine chemical ecology in benthic environments. Nat. Prod. Rep. 2011, 28, 345–387. [Google Scholar] [CrossRef] [PubMed]
- Davyt, D.; Fernandez, R.; Suescun, L.; Mombru, A.W.; Saldana, J.; Dominguez, L.; Coll, J.; Fujii, M.T.; Manta, E. New sesquiterpene derivatives from the red alga Laurencia scoparia. Isolation, structure determination, and anthelmintic activity. J. Nat. Prod. 2001, 64, 1552–1555. [Google Scholar] [CrossRef] [PubMed]
- Bawakid, N.O.; Alarif, W.M.; Alburae, N.A.; Alorfi, H.S.; Al-Footy, K.O.; Al-Lihaibi, S.S. Isolaurenidificin and Bromlaurenidificin, two new C15-acetogenins from the red alga Laurencia obtusa. Molecules 2017, 22, 807. [Google Scholar] [CrossRef] [PubMed]
- Bawakid, N.O.; Alarif, W.M.; Elhefnawy, M.E.; Ismael, A.; Al-Footy, K.O.; Al-Lihaibi, S.S. Bio-active maneonenes and isomaneonenes from Laurencia obtusa. Phytochemistry 2017, 143, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Alarif, W.M.; Al-Lihaibi, S.S.; Ayyad, S.-E.; Abdel-Rhman, M.H.; Badria, F.A. Laurene-type sesquiterpenes from the Red Sea red alga Laurencia obtusa as potential antitumor-antimicrobial agents. Eur. J. Med. Chem. 2012, 55, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Alarif, W.M.; Al-Footy, K.O.; Zubair, M.S.; Ghandourah, M.H.; Basaif, S.A.; Al-Lihaibi, S.S.; Ayyad, S.-E.N.; Badria, F.A. The role of new eudesmane-type sesquiterpenoid and known eudesmane derivatives from the red alga Laurencia obtusa as potential antifungal–antitumour agents. Nat. Prod. Res. 2015, 30, 1150–1155. [Google Scholar] [CrossRef] [PubMed]
- Angawi, R.F.; Alarif, W.M.; Hamza, R.I.; Badria, F.A.; Ayyad, S.-E.N. New cytotoxic laurene, cuparene and laurokamurene type-sesquiterpenes from the red alga Laurencia obtusa. Helv. Chem. Acta 2014, 97, 1388–1395. [Google Scholar] [CrossRef]
- Ayyad, S.-E.N.; Al-Footy, K.O.; Alarif, W.M.; Sobahi, T.R.; Basaif, S.A.; Makki, M.S.; Asiri, A.M.; Al Halwani, A.Y.; Badria, A.F.; Badria, F.A. Bioactive C15 Acetogenins from red alga Laurencia obtusa. Chem. Pharm. Bull. 2011, 59, 1294–1298. [Google Scholar] [CrossRef] [PubMed]
- Vairappan, C.S.; Suzuki, M.; Abe, T.; Masuda, M. Halogenated metabolites with antibacterial activity from the Okinawan Laurencia species. Phytochemistry 2001, 58, 517–523. [Google Scholar] [CrossRef]
- Bovey, F.A. Nuclear Magnetic Resonance Spectroscopy; Academic Press: New York, NY, USA, 1969. [Google Scholar]
- Waraszkiewicz, S.M.; Sun, H.H.; Erickson, K.L.; Finer, J.; Clardy, J. C15 halogenated compounds from the Hawaiian marine alga Laurencia nidifica. Maneonenenes and isomaneonenens. J. Org. Chem. 1978, 43, 3194–3204. [Google Scholar] [CrossRef]
- Gaver, R.C.; Sweeley, C.C. Methods for methanolysis of sphingolipids and direct determination of long-chain bases by gas chromatography. J. Am. Oil Chem. Soc. 1965, 42, 294–298. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
 ) and HMBC (
) and HMBC (  ) correlations of 1.
) correlations of 1.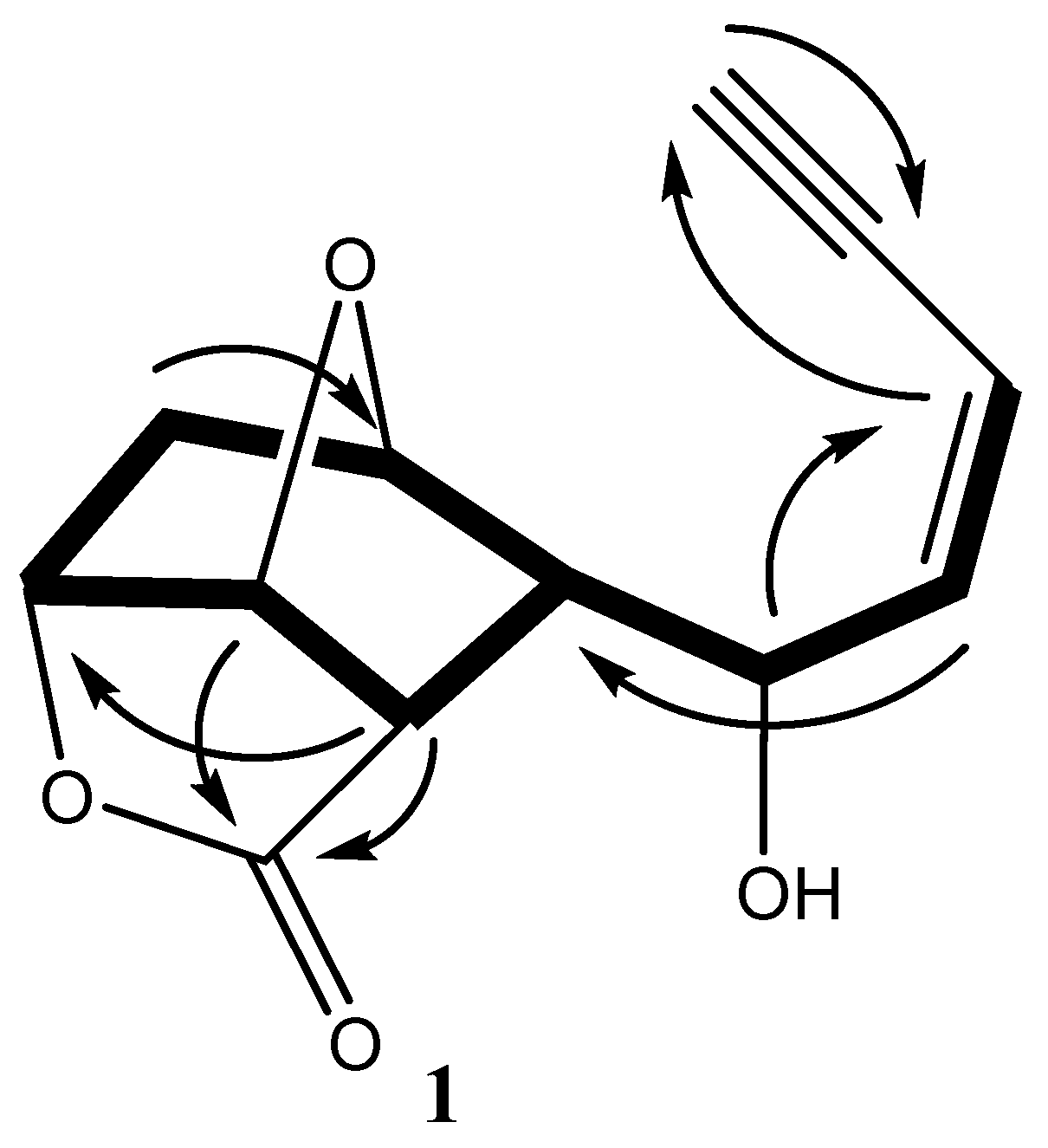

| No. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | |
| 1 | 3.20 dd (2.6, 0.9) | 84.1 | 3.28 dd (2.6, 0.9) | 85.4 | 3.21 dd (2.6, 0.9) | 83.2 |
| 2 | - | 79.1 | - | 78.1 | - | 79.1 |
| 3 | 5.64 ddd (11.1, 2.6, 1.7) | 110.5 | 5.75 dd (11.1, 2.6) | 113.2 | 5.65 dd (11.1, 2.6) | 110.6 |
| 4 | 6.01 ddd (11.1, 10.2, 0.9) | 143.0 | 5.98 ddd (11.1, 10.2) | 139.9 | 6.01 ddd (11.1, 10.2, 0.9) | 143.0 |
| 5 | 4.73 dd, (10.2, 10.2) | 67.8 | 4.76 dd (11.1, 10.2) | 57.1 | 4.74 dd (10.2, 10.2) | 67.8 |
| 6 | 2.59 dddd (10.2, 10.2, 4.3, 1.7) | 53.3 | 2.42 dd (11.1, 1.7) | 55.9 | 2.60 dddd (10,2, 10.2, 4.3,1.7) | 53.5 |
| 7 | 4.37 dd (5.1, 4.3) | 78.3 | 4.98 d (5.1) | 78.8 | 4.37 dd (5.1, 4.3) | 78.3 |
| 8a 8b | 2.25 d (14.5) 1.93 dddd (14.5, 7.7, 5.1, 1.7) | 35.1 | 2.11 m 1.80 d (14.5) | 38.3 | 2.26 d (14.5) 1.93 dddd (14.5, 7.7, 5.1, 1.7) | 35.1 |
| 9 | 4.90 dd (7.7, 5.1) | 79.9 | 4.86 dd (7.7, 5.1) | 78.8 | 4.91 dd (7.7, 5.1) | 79.9 |
| 10 | 5.37 dd (5.1, 5.1) | 83.2 | 5.31 dd (5.1, 5.1) | 81.6 | 5.37 dd (5.1, 5.1) | 84.1 |
| 11 | 2.97 dd (10.2, 5.1) | 43.0 | 2.46 brd (5.1) | 43.6 | 2.97 dd (10.2, 5.1) | 43.0 |
| 12 | 177.7 | 175.9 | 174.4 | |||
| 1′ | 174.9 | 177.7 | ||||
| 2′ | 2.09 s | 20.3 | 2.35 t (7.7) | 35.1 | ||
| 3′-16′ | 1.64–1.62 m | 24.9 | ||||
| 17′ | 1.30–1.24 m | 29.7 | ||||
| 18′ | 0.88 t (6.8) | 14.1 | ||||
| 1″ | 4.20 dd (11.9, 4.3) 4.15 dd (11.9, 6.0) | 65.2 | ||||
| 2″ | 3.94–3.92 m | 70.3 | ||||
| 3″ | 3.70 dd (11.9, 4.3) 3.60 dd (11.1, 6.0) | 63.3 | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alarif, W.M.; Al-Lihaibi, S.S.; Bawakid, N.O.; Abdel-Lateff, A.; Al-malky, H.S. Rare Acetogenins with Anti-Inflammatory Effect from the Red Alga Laurencia obtusa. Molecules 2019, 24, 476. https://doi.org/10.3390/molecules24030476
Alarif WM, Al-Lihaibi SS, Bawakid NO, Abdel-Lateff A, Al-malky HS. Rare Acetogenins with Anti-Inflammatory Effect from the Red Alga Laurencia obtusa. Molecules. 2019; 24(3):476. https://doi.org/10.3390/molecules24030476
Chicago/Turabian StyleAlarif, Walied Mohamed, Sultan Semran Al-Lihaibi, Nahed Obaid Bawakid, Ahmed Abdel-Lateff, and Hamdan Salem Al-malky. 2019. "Rare Acetogenins with Anti-Inflammatory Effect from the Red Alga Laurencia obtusa" Molecules 24, no. 3: 476. https://doi.org/10.3390/molecules24030476
APA StyleAlarif, W. M., Al-Lihaibi, S. S., Bawakid, N. O., Abdel-Lateff, A., & Al-malky, H. S. (2019). Rare Acetogenins with Anti-Inflammatory Effect from the Red Alga Laurencia obtusa. Molecules, 24(3), 476. https://doi.org/10.3390/molecules24030476