PpNAC187 Enhances Lignin Synthesis in ‘Whangkeumbae’ Pear (Pyrus pyrifolia) ‘Hard-End’ Fruit
Abstract
1. Introduction
2. Results
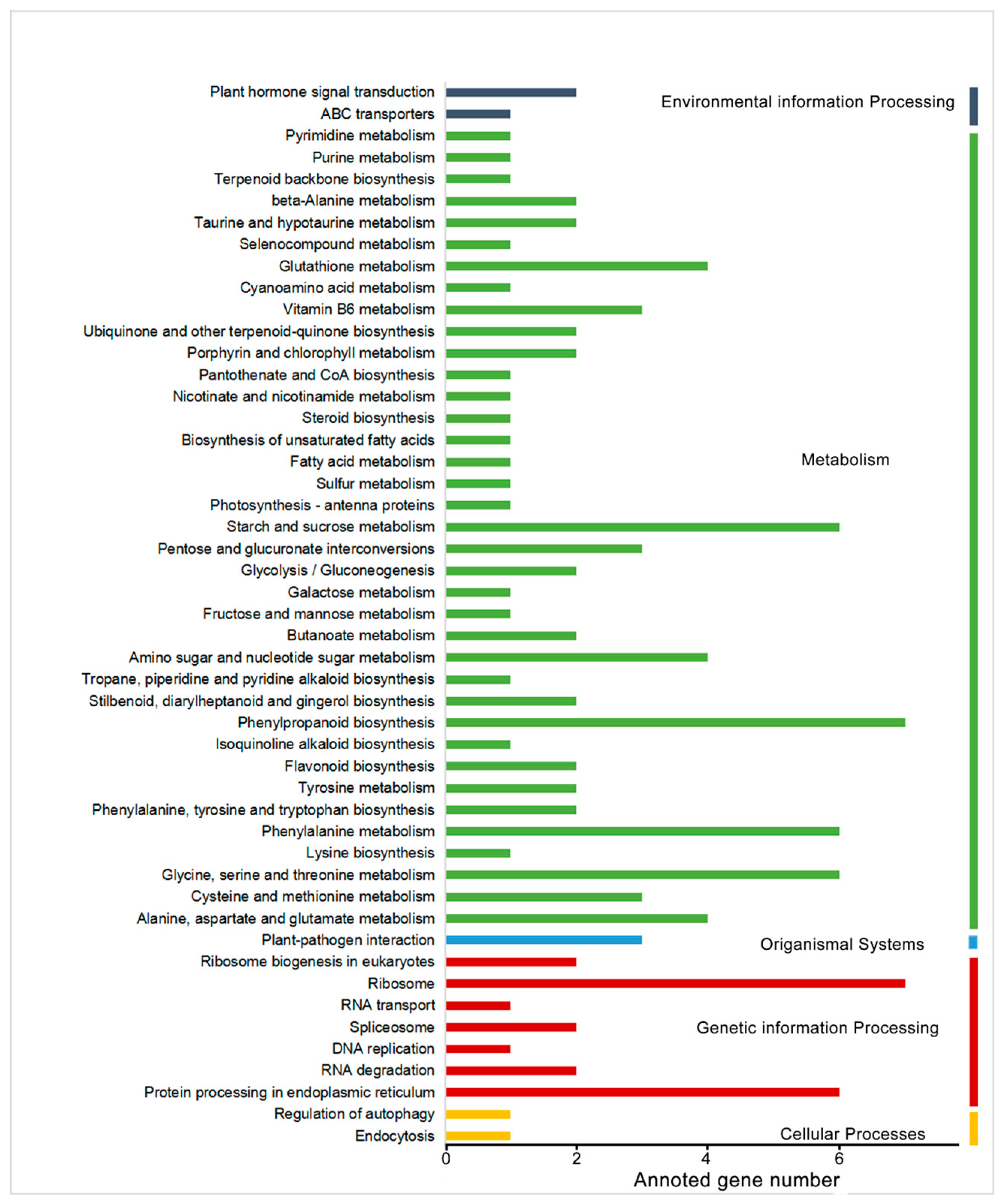
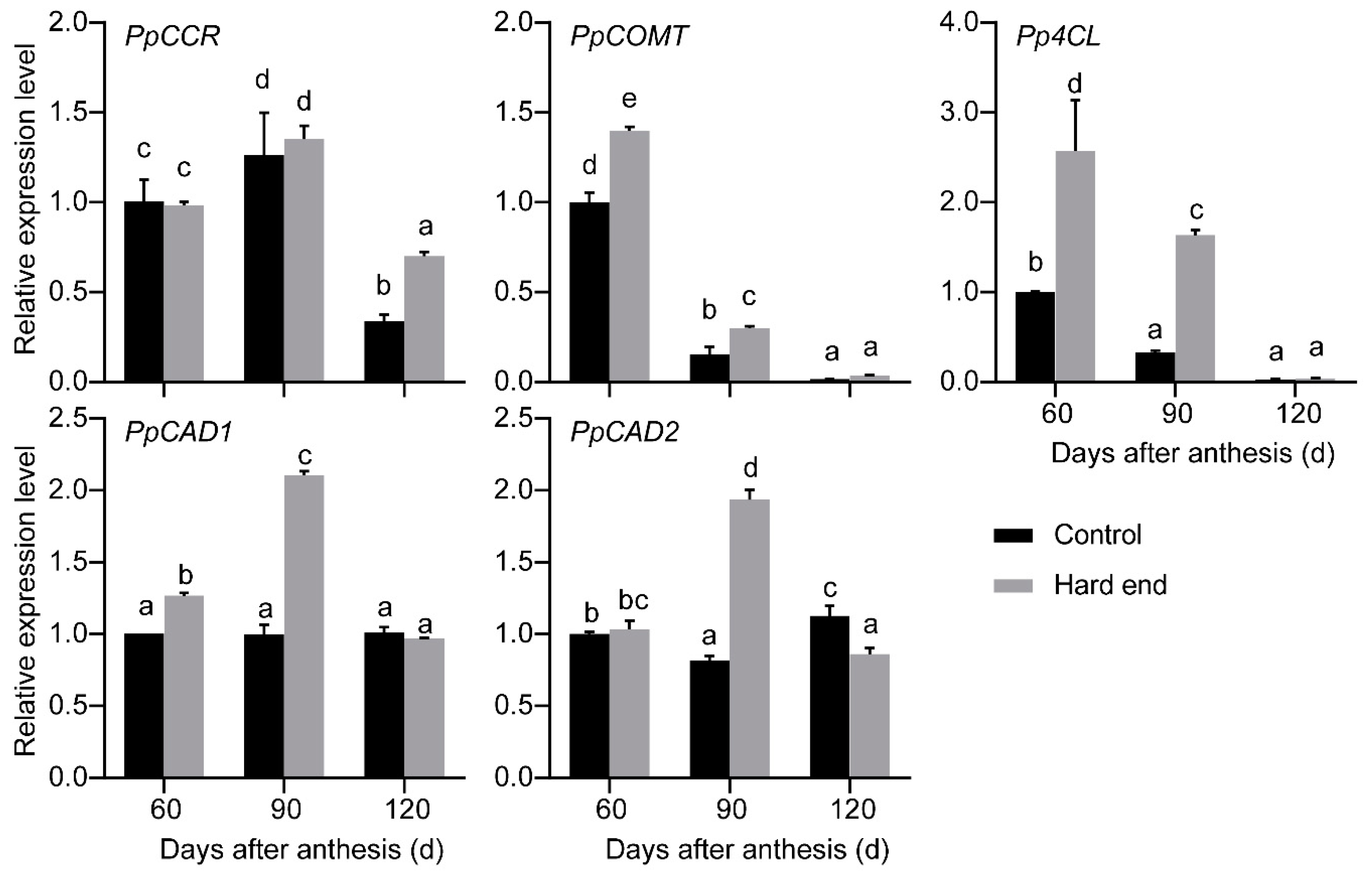
2.1. RNA-Seq Analysis
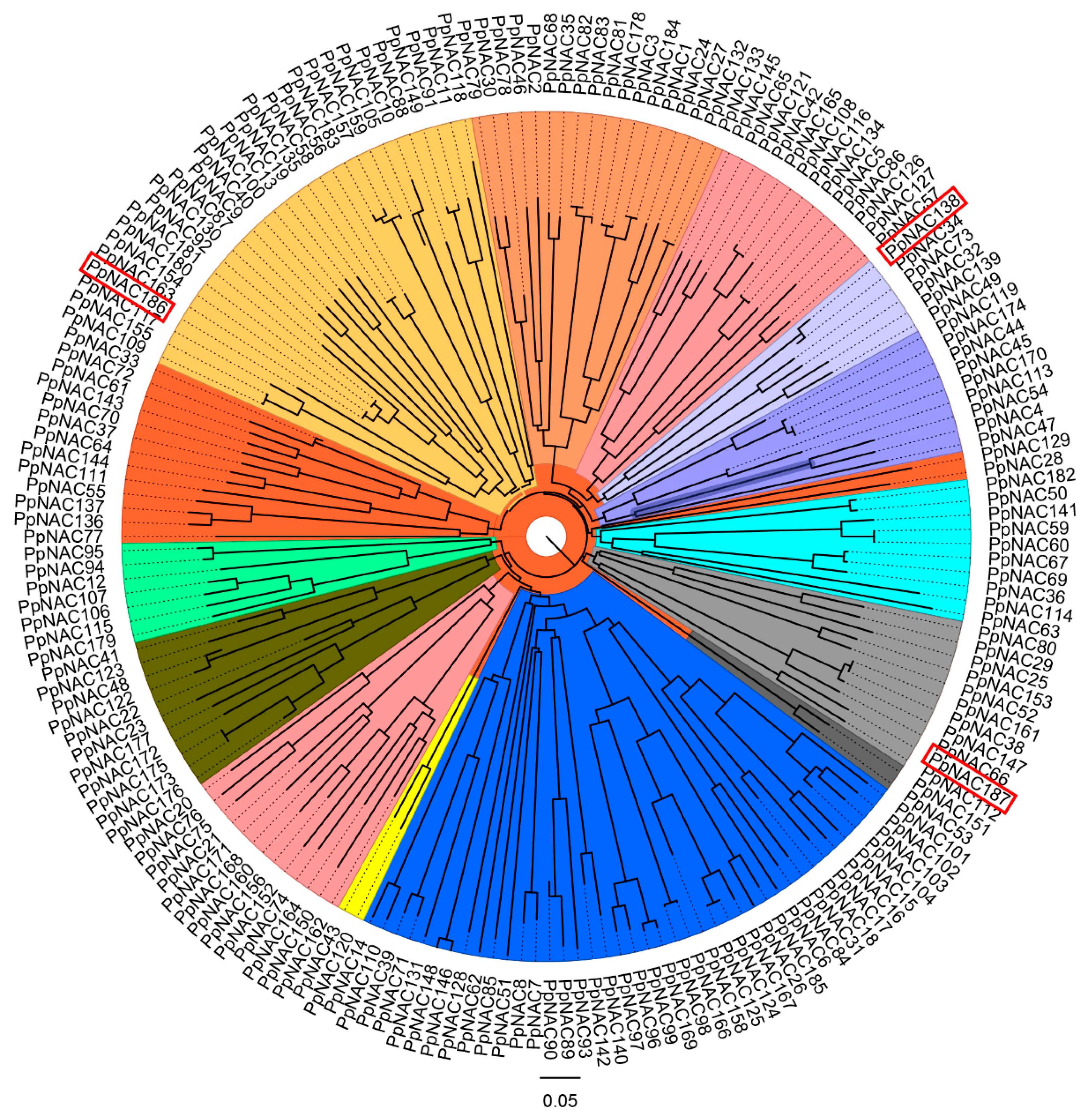
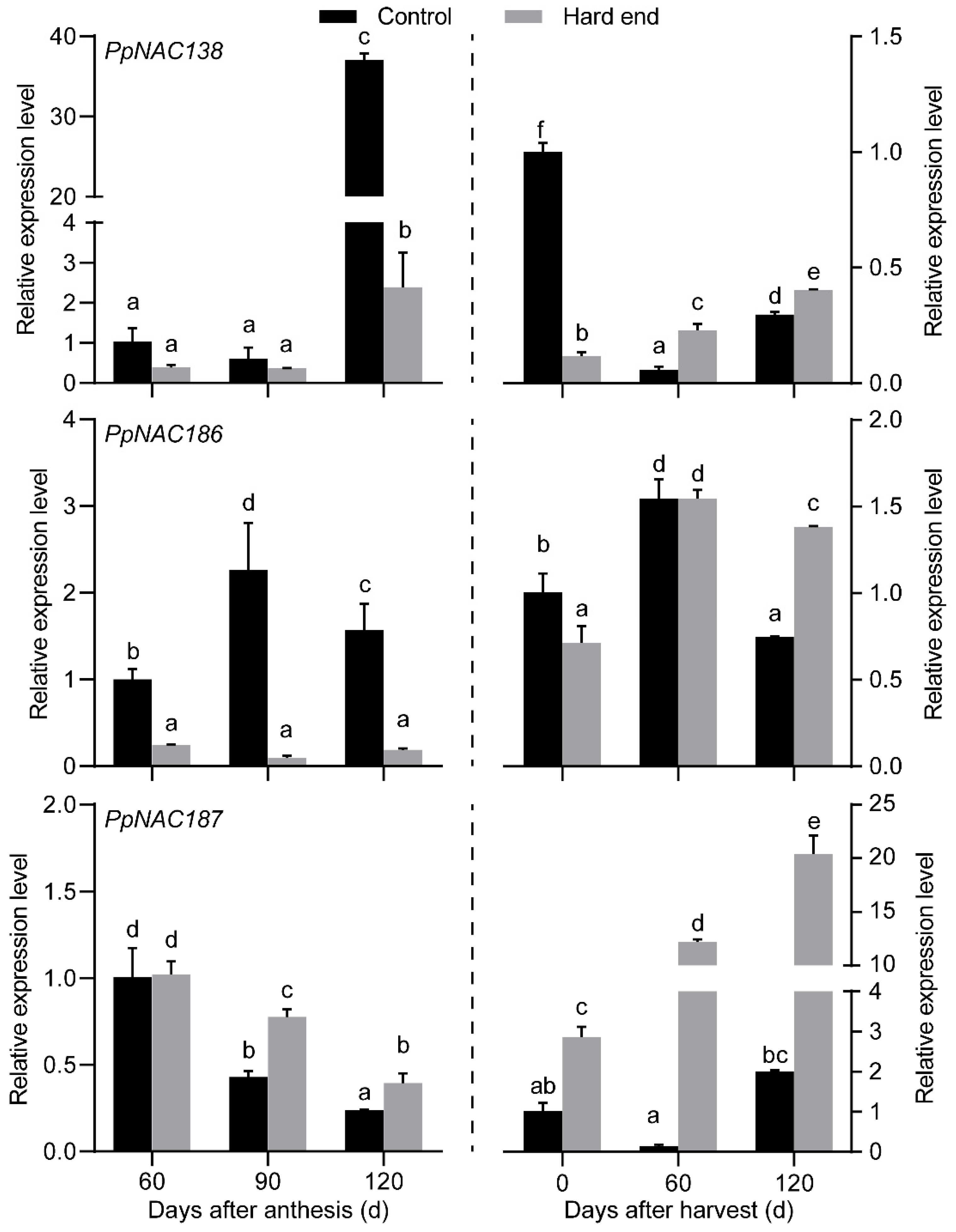
2.2. The Phylogenetic Analysis of PpNACs and Their Expression Pattern
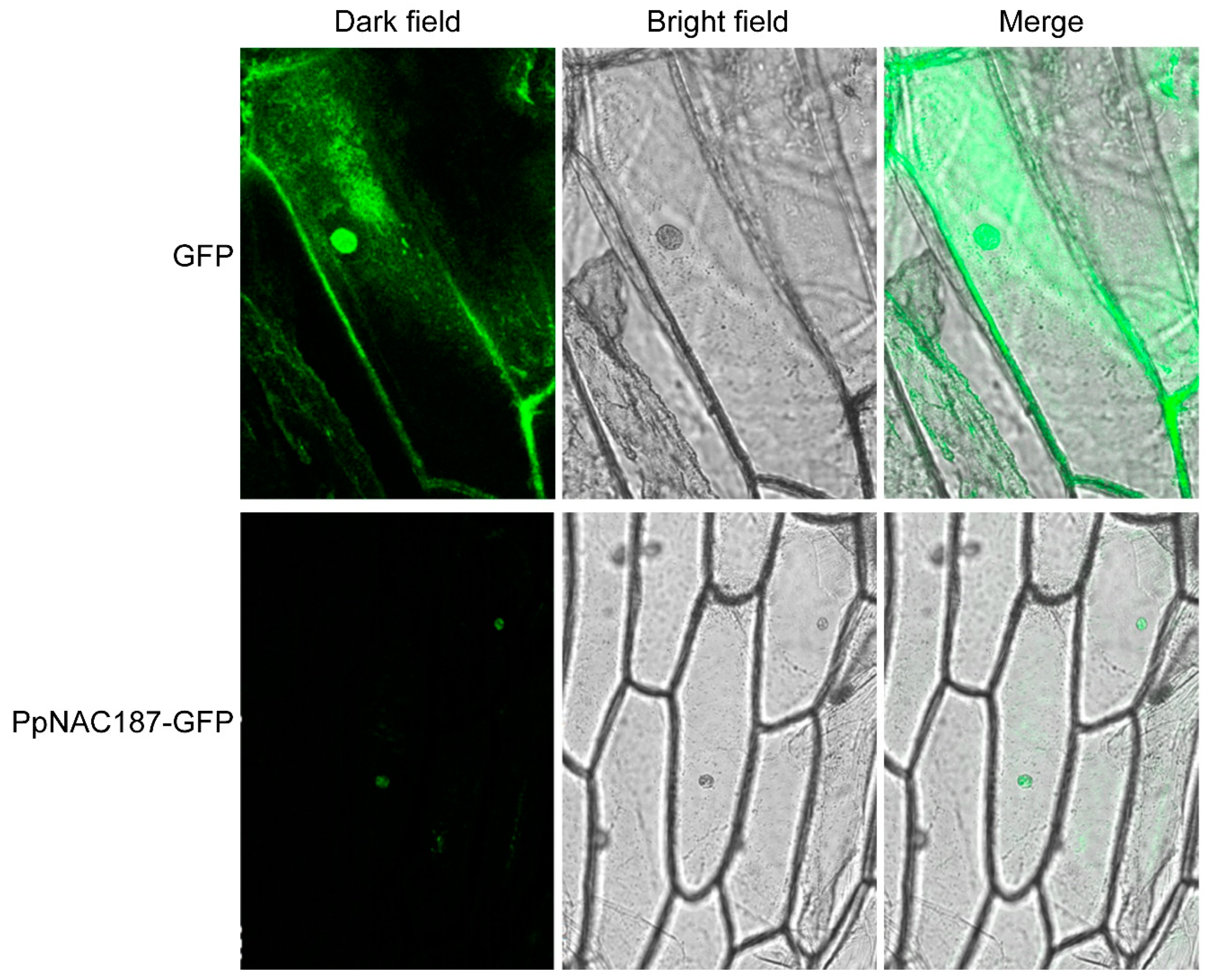
2.3. Subcellular Localization of PpNAC187
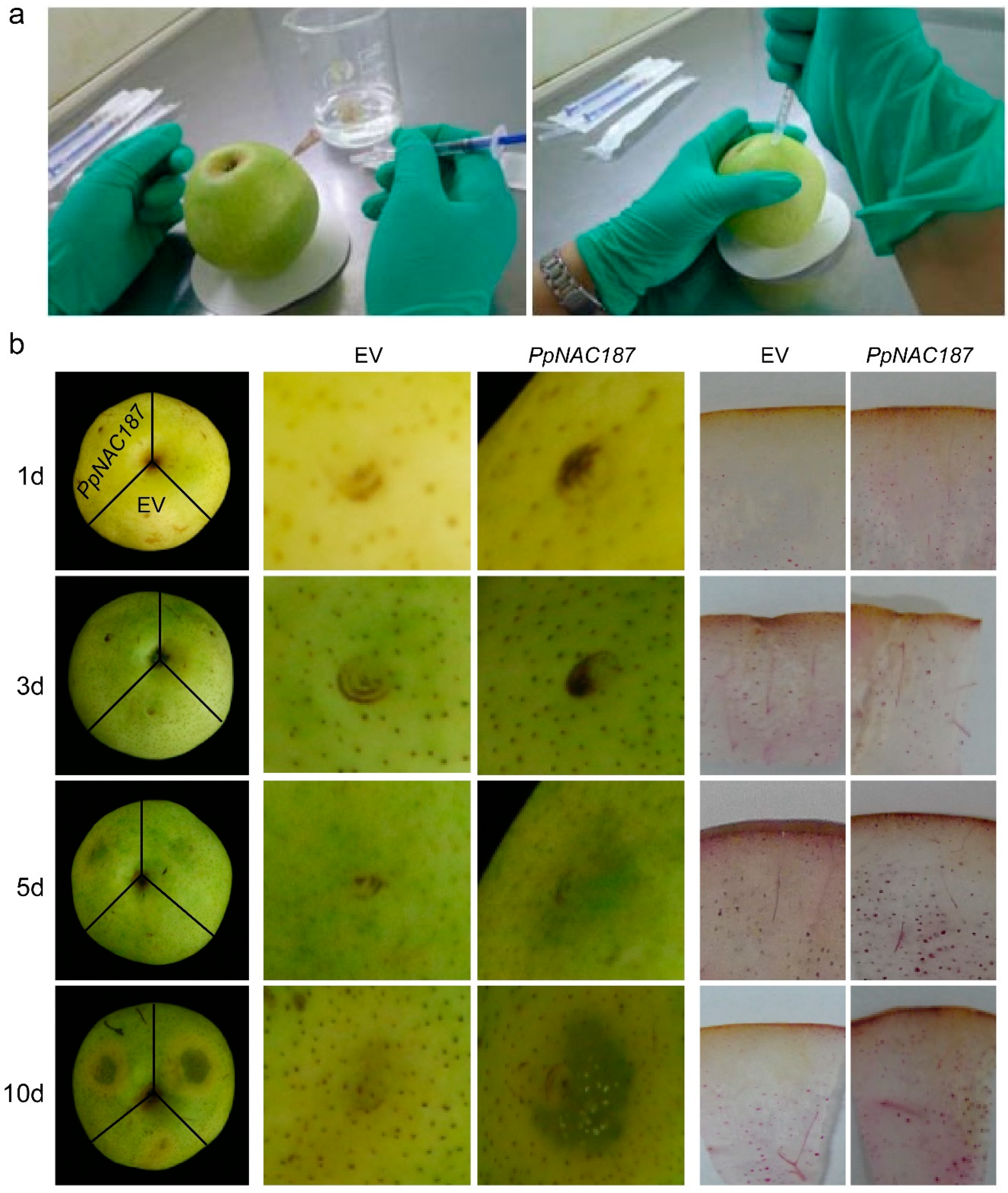
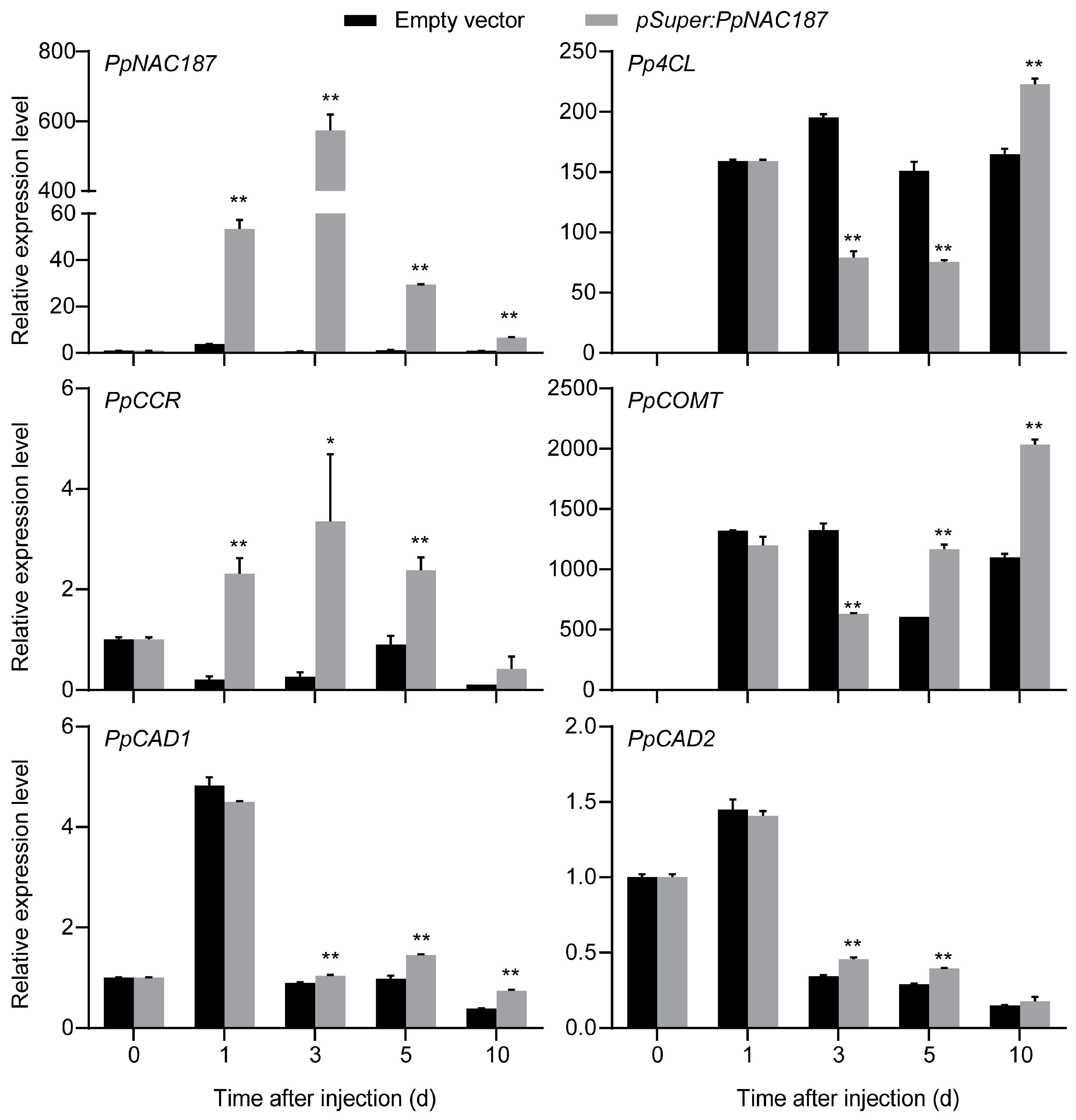
2.4. Transient Expression of PpNAC187 in ‘Whangkeumbae’ Pear Flesh
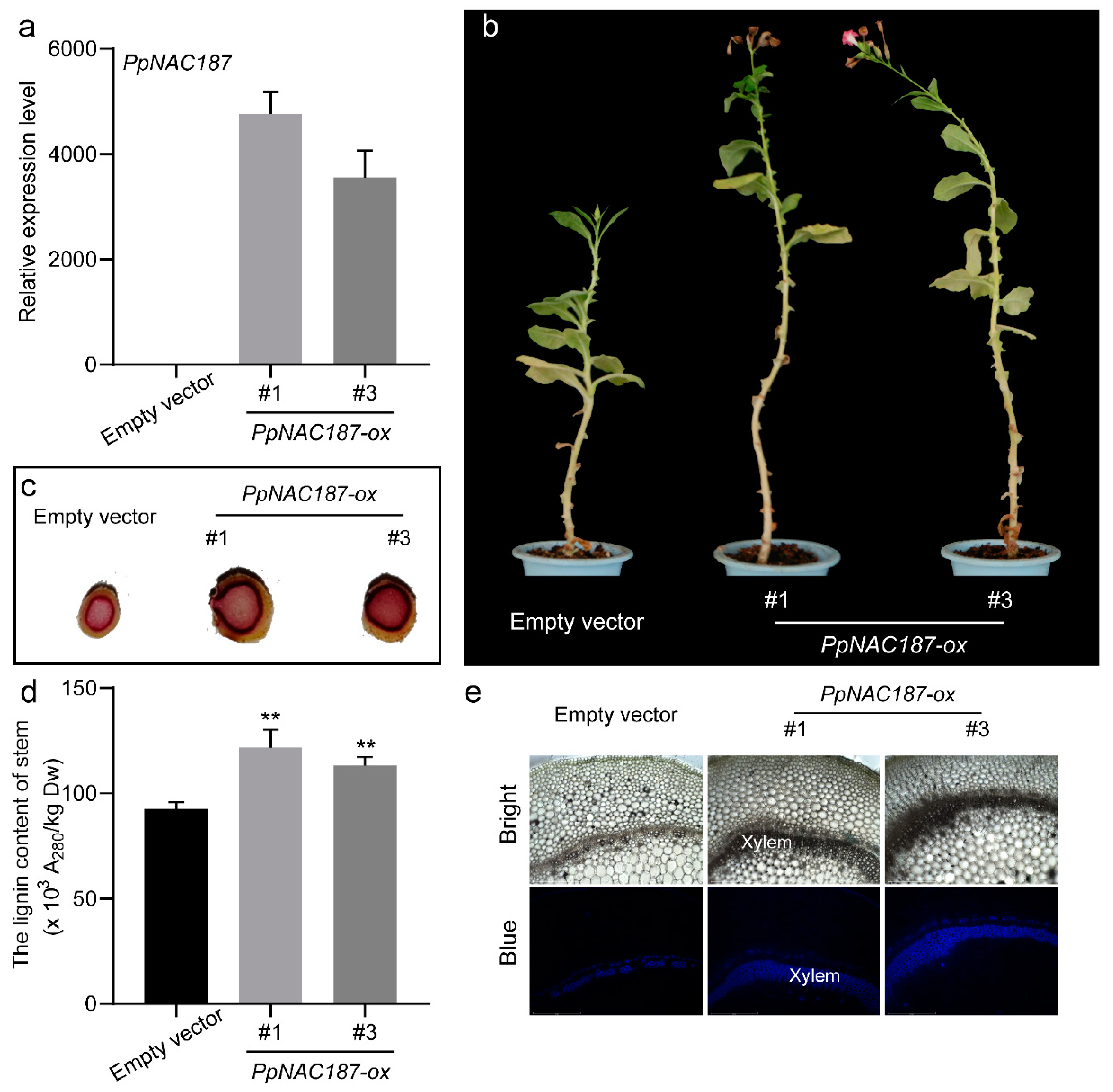
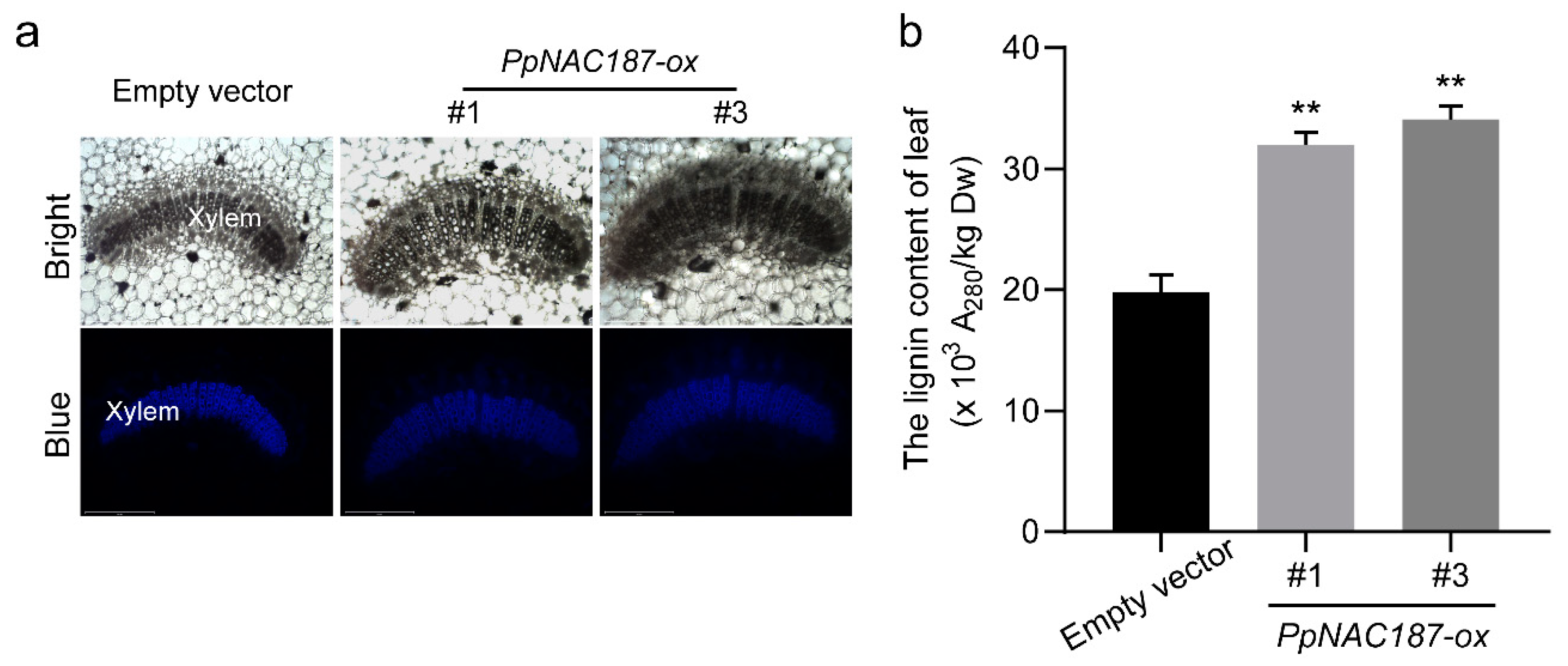
2.5. Functional Verification of PpNAC187 in Transgenic Tobacco
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. RNA-Seq Analysis
4.3. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
4.4. Cloning of PpNAC187
4.5. Sequence Alignment and Phylogenetic Analysis
4.6. Construction of the Expression Vector
4.7. Subcellular Localization of the PpNAC187 Transcription Factor
4.8. Transient Expression of PpNAC187 in ‘Whangkeumbae’ Pear
4.9. Agrobacterium-Mediated Transformation of Tobacco with PpNAC187
4.10. PCR Verification of Transformed PpNAC187 Tobacco
4.11. Wiesner Staining and Microscopy
4.12. Lignin Assay
4.13. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Charles, F.P.; Michael, J.C.; Lacy, P.M.; Dean, H.R. Market Diseases of Apples, Pears, and Quince; US Agricultural Research Service, United States Department of Agriculture: Washington, DC, USA, 1971.
- Yamaguchi, M.; Kubo, M.; Fukuda, H.; Demura, T. Vascular-related NACDOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008, 55, 652–664. [Google Scholar] [CrossRef] [PubMed]
- Rose, D.H.; Mccolloch, L.P.; Fisher, D.F. Market Diseases of Fruits and Vegetables: Apples, Pears, Quinces; US Department of Agriculture: Washington, DC, USA, 1951.
- Raese, J.; Drake, S. Calcium foliar sprays for control of alfalfa greening, cork spot, and hard end in ‘anjou’ pears. J. Plant Nutr. 2006, 29, 543–552. [Google Scholar] [CrossRef]
- Lu, G.L.; Li, Z.J.; Zhang, X.F.; Wang, R.; Yang, S.L. Expression analysis of lignin associated genes in hard end pear (Pyrus pyrifolia Whangkeumbae) and its response to calcium chloride treatment conditions. J. Plant Growth Regul. 2015, 34, 251–262. [Google Scholar] [CrossRef]
- Wang, Y.L.; Zhang, X.F.; Wang, Y.Z.; Yang, S.L.; Qu, H.Y. The changes of intracellular calcium concentration and distribution in the hard end pear (Pyrus pyrifolia cv. ‘Whangkeumbae’) fruit. Cell Calcium 2018, 71, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Watanabe, S. Initial time of development of hard end disorder in ‘Bartlett’ pear. J. Japan Soc. Hort. Sci. 1982, 51, 42–151. [Google Scholar] [CrossRef][Green Version]
- Fumio, T. Recent advances in research on Japanese pear rootstocks. J. Japan. Soc. Hort. Sci. 2012, 81, 1–10. [Google Scholar]
- Zhao, Q.; Dixon, R.A. Transcriptional networks for lignin biosynthesis: More complex than we thought. Trends Plant Sci. 2011, 16, 227–233. [Google Scholar] [CrossRef]
- Cheng, X.; Li, M.L.; Li, D.H.; Zhang, J.Y.; Jin, Q.; Sheng, L.L.; Cai, Y.P.; Lin, Y. Characterization and analysis of CCR and CAD gene families at the whole-genome level for lignin synthesis of stone cells in pear (Pyrus bretschneideri) fruit. Biol. Open. 2017, 6, 1602–1613. [Google Scholar] [CrossRef]
- Vermerris, W.; Abril, A. Enhancing cellulose utilization for fuels and chemicals by genetic modification of plant cell wall architecture. Curr. Opin. Biotech. 2015, 32, 104–112. [Google Scholar] [CrossRef]
- Wang, W.Q.; Zhang, J.; Ge, H.; Li, S.J.; Li, X.; Yin, X.R.; Grierson, D.; Chen, K.S. EjMYB8 transcriptionally regulates flesh lignification in loquat fruit. PLoS ONE 2016, 11, e0154399. [Google Scholar] [CrossRef]
- Akiyoshi, K.; Pulla, K.; Kazuya, Y.; Saori, E.; Keiko, Y.; Hiroyasu, E. Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J. 2000, 22, 289–301. [Google Scholar]
- Cassan-Wang, H.; Goué, N.; Saidi, M.N.; Legay, S.; Sivadon, P.; Goffner, D.; Grima-Pettenati, J. Identification of novel transcription factors regulating secondary cell wall formation in Arabidopsis. Front. Plant Sci. 2013, 4, 189. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wang, W.Q.; Zeng, J.K.; Zhang, J.; Donald, G.; Li, X.; Yin, X.R.; Chen, K.S. A NAC transcription factor, EjNAC1, affects lignification of loquat fruit by regulating lignin. Postharvest Biol. Tec. 2015, 102, 25–31. [Google Scholar] [CrossRef]
- Vroemen, C.W.; Mordhorst, A.P.; Albrecht, C.; Kwaaitaal, M.A.; deVries, S.C. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoots meristem formation in Arabidopsis. Plant Cell 2003, 15, 1563–1577. [Google Scholar] [CrossRef]
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 2007, 19, 270–280. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Ye, Z.H. Regulation of cell wall biosynthesis. Curr. Opin. Plant Biol. 2007, 10, 564–572. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Ye, Z.H. Transcriptional regulation of lignin biosynthesis. Plant Signal. Behav. 2009, 4, 1028–1034. [Google Scholar] [CrossRef]
- Ge, H.; Zhang, J.; Zhang, Y.J.; Li, X.; Yin, X.R.; Grierson, D.; Chen, K.S. EjNAC3 transcriptionally regulates chilling-induced lignification of loquat fruit via physical interaction with an atypical CAD-like gene. J. Exp. Bot. 2017, 68, 5129–5136. [Google Scholar] [CrossRef]
- Ahmad, M.; Yan, X.H.; Li, J.Z.; Yang, Q.S.; Jamil, W.; Teng, Y.W.; Bai, S.L. Genome wide identification and predicted functional analyses of NAC transcription factors in Asian pears. BMC Plant Biol. 2018, 18, 214. [Google Scholar] [CrossRef]
- Wang, Y.L.; Zhang, X.F.; Yang, S.L.; Wang, C.H.; Lu, G.L.; Wang, R.; Yang, Y.J.; Li, D.L. Heterogenous expression of Pyrus pyrifolia PpCAD2 and PpEXP2 in tobacco impacts lignin accumulation in transgenic plants. Gene 2017, 181–189. [Google Scholar] [CrossRef]
- Olsen, K.M.; Lea, U.S.; Slimestad, R.; Verheul, M.; Lillo, C. Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 2008, 165, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Meyer, K.; Chapple, C.; Douglas, C.J. Antisense suppression of 4-coumarate: Coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell 1997, 9, 1985–1998. [Google Scholar]
- Jean, C.L.; Rebecca, D.; Kris, M.; Vronique, S.; Catherine, L.; Brigitte, P.; Annette, N. Downregulation of cinnamoyl-coenzyme A reductase in poplar: Multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 2007, 19, 3669–3691. [Google Scholar]
- Guo, D.; Chen, F.; Inoue, K.; Blount, J.W.; Dixon, R.A. Downregulation of Caffeic Acid 3-O-Methyltransferase and Caffeoyl CoA 3-O-Methyltransferase in transgenic alfalfa: Impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 2001, 13, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.S.; Stewart, C.N. Plant synthetic promoters and transcription factors. Curr. Opin. Biotech. 2016, 37, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Demura, T.; Ye, Z.H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef] [PubMed]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription switches for protoxylem and metaxylem vessel formation. Gene Dev. 2005, 19, 1855–1860. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, R.; Gu, C.; Wu, J.; Wan, H.; Zhang, S.; Zhang, S. Evaluation of candidate reference genes for real time quantitative PCR normalization in pear fruit. Afr. J. Agric. Res. 2012, 7, 3701–3704. [Google Scholar]
- Niu, S.; Li, Z.; Yuan, H.; Fang, P.; Chen, X.; Li, W. Proper gibberellin localization in vascular tissue is required to regulate adventitious root development in tobacco. J. Exp. Bot. 2013, 64, 3411–3424. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Method 2011, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Glazebrook, J. Transformation of Agrobacterium using the freeze-thaw method. CSH Protoc. 2006, 7, 1031–1036. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Cao, Z.; Li, Y.; Zhao, Y.X.; Zhang, H. A simple and effective method for protein subcellular localization using Agrobacterium-mediated transformation of onion epidermal cells. Biologia 2007, 62, 529–532. [Google Scholar] [CrossRef]
- Spolaore, S.; Casadoro, G.; Trainotti, L. A simple protocol for transient gene expression in ripe fleshy fruit mediated by Agrobacterium. J. Exp. Bot. 2001, 52, 845–850. [Google Scholar] [CrossRef]
- Zheng, L.; Liu, G.F.; Meng, X.N.; Li, Y.B.; Wang, Y.C. A versatile Agrobacterium-mediated transient gene expression system for herbaceous plants and trees. Biochem. Genet. 2012, 50, 761–769. [Google Scholar] [CrossRef]
- Bruce, R.J.; West, C.A. Elicitation of lignin biosynthesis and isoperoxidase activity by pectic fragments in suspension cultures of castor bean. Plant Physiol. 1989, 91, 889–897. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Cheng, C.; Zhang, X.; Zhou, S.; Wang, C.; Ma, C.; Yang, S. PpNAC187 Enhances Lignin Synthesis in ‘Whangkeumbae’ Pear (Pyrus pyrifolia) ‘Hard-End’ Fruit. Molecules 2019, 24, 4338. https://doi.org/10.3390/molecules24234338
Li M, Cheng C, Zhang X, Zhou S, Wang C, Ma C, Yang S. PpNAC187 Enhances Lignin Synthesis in ‘Whangkeumbae’ Pear (Pyrus pyrifolia) ‘Hard-End’ Fruit. Molecules. 2019; 24(23):4338. https://doi.org/10.3390/molecules24234338
Chicago/Turabian StyleLi, Mingtong, Chenxia Cheng, Xinfu Zhang, Suping Zhou, Caihong Wang, Chunhui Ma, and Shaolan Yang. 2019. "PpNAC187 Enhances Lignin Synthesis in ‘Whangkeumbae’ Pear (Pyrus pyrifolia) ‘Hard-End’ Fruit" Molecules 24, no. 23: 4338. https://doi.org/10.3390/molecules24234338
APA StyleLi, M., Cheng, C., Zhang, X., Zhou, S., Wang, C., Ma, C., & Yang, S. (2019). PpNAC187 Enhances Lignin Synthesis in ‘Whangkeumbae’ Pear (Pyrus pyrifolia) ‘Hard-End’ Fruit. Molecules, 24(23), 4338. https://doi.org/10.3390/molecules24234338





