Abstract
The reaction between [Cu(μ-OAc)(μ-Pip)(MeOH)]2 (1) (OAc = acetate; Pip = 1,3-benzodioxole-5-carboxylate) and isonicotinamide (Isn) in MeOH as solvent yielded two mixture pairs of three compounds: {(HPip)2(Isn) (2), [Cu(Pip)2(Isn)2] (3)} and {(3), {[Cu3(Pip)2(OAc)2(μ-Isn)2(Isn)2(μ-OCH3)2(MeOH)2]·2MeOH}n (4)}. Modifying the reaction conditions (t, T, molar ratio), 2 and 3 have been successfully isolated, whereas 3 and 4 had to be mechanically separated. The recrystallization of 3 in pentanol yielded single crystals of compound [Cu(Pip)2(Isn)2]·C5H11OH (3a). The X-ray crystal structure of 2, 3a, and 4 has been elucidated showing a cocrystal, a monomer, and an unusual coordination polymer, respectively. The Pip ligand exhibited a chelate (3a) or a monodentate (4) coordination mode, but the Isonicotinamide (Isn) ligand is the one that promoted the arrangement of different structures and also mainly directs the formation of the 2D and 3D supramolecular assemblies. All the structures have been analyzed by Hirshfeld surface. In addition, the energy frameworks and lattice energy values of 2 and 3a have been calculated.
1. Introduction
The design of new organic and inorganic materials linked by intermolecular forces is a topic of current interest as models for biological systems and in materials science [1,2]. In this sense, hydrogen bonds are the main directing motif and are raised as one of the strongest and most used tools for crystal engineering [3,4]. Their large electrostatic components facilitate its ordering as first suggested by Etter’s rules—the best hydrogen bond donor will associate with the best hydrogen bond acceptor [5] and subsequently superseded by supramolecular suitable directing motifs (synthons), which imply functional groups [6,7]. Synthons are the backbone to design novel structures based on hierarchical, and therefore predictable, associations of the parts involved in them.
Above all, carboxylic acids can form strong hydrogen bonds through their acidic moiety, which in combination with N-containing groups (amines, amides, or pyridines) are replaced by the corresponding N-H···O interactions. The strength of this association is governed by the acidic and basic character of both parties involved in it [8]. This has led to the formation of single and multi-component crystals made of organic molecules named cocrystals [9]. The importance of cocrystals lies in the possibility to modify the physical properties of their components (melting point, solubility, or conductivity, among others) while keeping their chemical properties unaltered. This goal is achieved by the rearrangement of the intermolecular interactions promoted by the components and, thus, the formation of a new crystal structure [10,11]. It should be noted that during the formation of these assemblies, and in presence of a metal ion, the Isonicotinamide (Isn) units can interact either with the acidic moiety or with the metal center, wherefore a competition with the final complex formation occurs [12,13].
Among all the possible coformers, Isn stands out from the rest for its potential acceptor ability and therefore, it is considered a valuable component in the formation of cocrystals [14]. The Isn synthon itself promotes the amide···amide hydrogen bond interaction, ordering in a linear fashion but pointing in opposite direction. The pKa rule allows to ascertain if the combination of the selected acid and base will form a salt or a cocrystal. If ΔpKa < 0, the carboxylic acid···pyridine (O–H···N) interaction will be neutral [15,16]. For this reason, 1,3-benzodioxole-5-carboxylic acid (HPip, pKa = 4.35) has raised as a prospective candidate, owing to the fact that it obeys this rule and, thus, should form cocrystals with Isn (pKb = 10.87).
Besides, Isn formed numerous coordination complexes promoting their assembly in supramolecular structures mainly directed by the amide···amide pattern, even in competitive interactions with additional directing motifs [17,18,19,20]. In turn, carboxylates have a large versatility of coordination modes and can be used as a building block including Isn [21,22]. Among them, Cu(II) nodes possess high affinity toward oxygen donor atoms, especially belonging to carboxylate motifs and formed thousands of complexes. For this reason, metal carboxylates have been raised as one of the major interesting research items concerning their structural versatility and labile nature [23,24].
Previously, our group evinced the solvent-dependent formation of five Cu(II) compounds—one monomer and four dimers—resulting from the reaction between [Cu(μ-OAc)(μ-Pip)(MeOH)]2 (1) and Isn in different solvents. It should be noted that, in all of them, the Pip ligands were displaced, and only acetate ligands remained coordinated [25]. As a continuation, in this contribution we have assayed the reactions between 1 and Isn in order to synthesize compounds containing both Pip and Isn ligands in their structures. With this aim, we have modified the reaction conditions (reaction time, temperature, concentration, etc.). This made it possible to isolate three compounds: one cocrystal ((HPip)2(Isn), 2), one Cu(II) monomer ([Cu(Pip)2(Isn)2], 3), and a Cu(II) coordination polymer ({[Cu3(Pip)2(OAc)2(μ-Isn)2(Isn)2(μ-OCH3)2(MeOH)2]·2MeOH}n, 4) (Scheme 1). Compound 3 was recrystallized in pentanol yielding [Cu(Pip)2(Isn)2]·C5H11OH (3a). Hirshfeld surfaces analysis as well as Energy frameworks and Lattice energy calculations facilitated the better understanding of the Isn role in these systems.
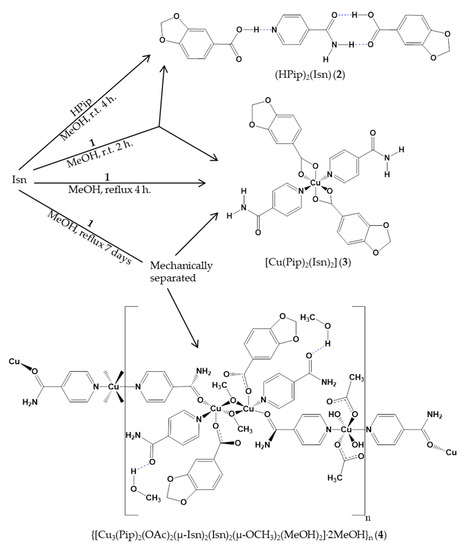
Scheme 1.
Outline of the synthesis of compounds 2–4.
2. Results and Discussion
2.1. Synthesis and Characterization of Cocrystal 2
The reaction of 1 [26,27] with Isn in MeOH as solvent at room temperature (r.t.) for 2h yielded a mixture of compounds (HPip)2(Isn) (2) and [Cu(Pip)2(Isn)2] (3). Otherwise, when the reaction was performed under reflux, only compound 3 was obtained. Following this, compound 2 was isolated by crystallizing a mixture of HPip:Isn (2:1 molar ratio) in MeOH at r.t. In this work, the reactions were performed without adding acetic acid, and the experimental conditions (t, T or concentration) have been modified in order to incorporate the Pip units in the structure of the complexes [28].
Compound 2 was characterized by EA, FTIR-ATR, 1H- and 13C{1H}-NMR spectroscopies, and single crystal X-ray diffraction method. The FTIR-ATR and 1H- and 13C{1H}-NMR spectra in dmso-d6 solution displays the characteristic bands of the two molecules (Figures S1–S3). In the FTIR-ATR spectrum (Figure S1a), the ν(N-H) bands appear at 3321 and 3140 cm−1 and are shifted with respect to the free Isn (3359 and 3179 cm−1, respectively) (Figure S1b). There is a broad signal at 1680 cm−1, assigned to the ν(C=O)Isn and ν(COOH)HPip, which is also shifted compared to the HPip- (Figure S1c) and Isn-free molecules (1667 and 1655 cm−1, respectively).
Additional peaks can be identified as δ(O=CN), δip(C-H) and δoop(C-H) [29]. The 1H- and 13C{1H}-NMR spectra show no displacement of the bands respect to the free molecules. The 1H-NMR spectrum confirms the 2:1 proportion of the coformers (Figures S2 and S3).
Therefore, the EA, FTIR-ATR, and 1H- and 13C{1H}-NMR data are in agreement with the structures determined by single crystal X-ray diffraction method. The ΔpKa (pKa conjugate acid of pyridine—pKa carboxylic acid)) between the parties which form the association is a determining factor for the formation of a salt or a cocrystal. If ΔpKa < 0 or ΔpKa > 3.75, a cocrystal or a salt will be formed, respectively. If 0 < ΔpKa < 3.75, there is an intermediate interaction, and either cocrystal or salt can be found [16].
2.2. Crystal Structure of Compound 2
Compound 2 crystallizes in the triclinic P space group. Its crystal structure can be described as the sum of the interactions between its functional groups. The position and nature of each functionality generates the particular interactions, which dictate the crystal arrangement [4,29]. The association of HPip and Isn leads to the formation of a cocrystal [30], in which there is not any homomeric interaction and therefore, the assembly has grown mixing molecules. In this kind of multi-component solids, salts or cocrystals can be formed depending on where the proton is located and hence the drift in the bond distances of the parties involved. In this case, the C=O, C-N and C-O distances of Isn and HPip molecules have been analyzed and compared with the values from the anionic Pip ligand present in the complexes, which suggested for both of them a neutral charge (Table 1). In addition, the ΔpKa between both, the conjugated acid of Isn (pKa = 3.61), and HPip (pKa = 4.35) has a value of −0.74 and thus, they are prospective candidates for generate a cocrystal [16].

Table 1.
Comparative of bond lengths (C=O, C-N, C-N) between cocrystal and monomer and polymer compounds.
The basic structural motifs (BSMs) [31], also called long-range synthon Aufbau modules (LSAMs), are two crystallographically independent trimeric large synthons (A and B) constructed by three subunits—two HPip and one Isn. These BSMs are fastened through basic molecules (BSMs) [32]. Those are two small synthons: a type III dimeric square motif (N-HIsn···O=CHPip; O-HHPip···O=C-N-HIsn) and a type I dimeric chain motif (O-H···NIsn) with distances similar to other reported cocrystals (Table 2) (Figure 1a). In contrast, type II motifs (amide···amide) are not present, probably induced by the MeOH solvent, which is polar enough to break this interaction [33]. Moreover, Isn molecule holds together two adjacent HPip units, from contiguous BSMs, via the remaining amidic Hanti and the Hortho and Hmeta from opposite sides. Protons at the same flank merge with the same oxygen atom of the carbonyl, while the Hortho pyridil interacts with the protonated oxygen atom of other HPip molecule (Table 2) (Figure 1b).

Table 2.
Supramolecular interactions present in compound 2.
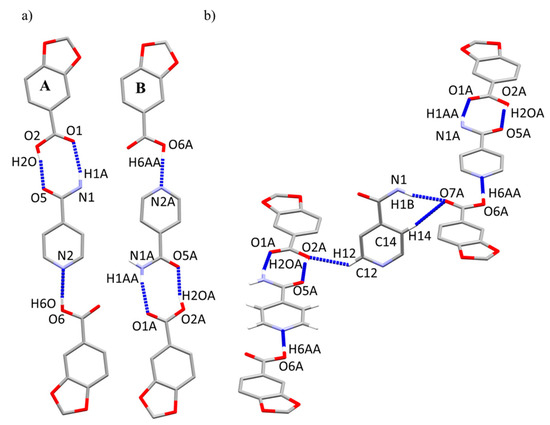
Figure 1.
Basic molecules (BSMs) composed by two HPip and one Isn subunits (left, A; right, B) present in compound 2. (a) interactions forming the trimeric BSM (A, B). (b) Intermolecular interactions between the BSMs.
BSMs are disposed along the (1 0 1/2) and (1/2 0 3/4) planes (A and B, respectively), and each one of them associates with a closely packed α-helix shape (Figure 2a). These chains are joined via the central Isn units, which forms 2D grid shape layers parallel to the ac plane (Figure 2b).
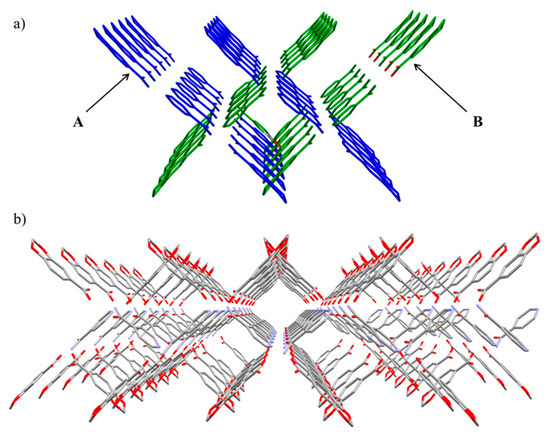
Figure 2.
(a) α-helix shape through the assembly of A and B chains. (b) b axis view of the 2D layers along the ac plane.
2.3. Synthesis and Characterization of the Coordination Complexes 3 and 4
The synthesis of [Cu(Pip)2(Isn)2] (3) and {[Cu3(Pip)2(OAc)2(μ-Isn)2(Isn)2(μ-OCH3)2(MeOH)2] 2MeOH}n (4) was performed mixing 1 and Isn in a 1:7 molar ratio with MeOH as solvent and under reflux conditions for a week. In these conditions, both compounds simultaneously crystallized in a 1.5:1 proportion (violet (3) and green (4) crystals) and they were mechanically separated. Compound 3 was successfully isolated under reflux conditions and modifying the compound 1:Isn (1:2) molar ratio. Compound 3 was recrystallized in a MeOH/pentanol (1:1) mixture yielding single crystals of compound [Cu(Pip)2(Isn)2]·C5H11OH (3a). Unfortunately, 4 could not be obtained as a pure product changing the reaction conditions. Compounds 3 and 4 were characterized by EA, FTIR-ATR, and UV-Vis spectroscopies. The violet crystals of 3 from the mixture were characterized by EA and FTIR-ATR spectroscopies, while single crystals of 3a were characterized by FTIR-ATR spectroscopy. Complex 4 unavoidably suffers a loss of the occluded MeOH molecules after manipulations required for the EA. In addition, the structure of 3a and 4 was elucidated by single crystal X-ray diffraction method.
The FTIR-ATR spectra of compounds 3–4 (Figures S4–S6) display two pairs of bands at 1557 cm−1 (3) and 1602 cm−1 (4) and at 1435 cm−1 (3) and 1387 cm−1 (4) assigned to [νas(COO)] and [νs(COO)], respectively, from the carboxylate groups. The difference between these bands (Δ = νas(COO) − νs(COO)) [34] is 121 (3) and 215 (4), attributable to a chelate and a monodentate coordination mode, respectively. The ν(C=O)Isn band is present at 1694 cm−1 in compound 3, while in 4, it is split in two at 1696 and 1631 cm−1, which suggests the presence of two different Isn ligands. Likewise, the [δ(O=CN)] vibrational mode is identified at 629 cm−1 (3) and at 627 and 582 cm−1(4). Additional bands as ν(N-H), δip(C-H) and δoop(C-H) are also assigned (see exp. section) [28]. If the FTIR-ATR of 3 is compared to 3a, the unique difference arises in the solvent region in which the alcohol group of the pentanol molecule promoted the formation of broader bands overlapped with those deriving from the amino group (Figure S5).
The electronic spectra of compounds 3 and 4 have been recorded in MeOH as solvent. All spectra show one band in the visible region, characteristic of d9 Cu(II) complexes which is assigned to the 2Eg → 2T2g electronic transition (Figure S7) [35]. Compound 3 has its λmax at 713 nm (ε = 77 M−1 × cm−1), while 4 displayed this d–d electronic transition single band at 706 nm (ε = 404 M−1 × cm−1). The high ε value of 4 respect to 3 could be explained by the larger number of Cu(II) centers in the polymeric array.
2.4. Crystal and Extended Structure of Compound 3a
Compound 3a crystallizes in the triclinic P space group. It has a monomeric structure with a [CuO4N2] core that is composed by two pairs of Pip and Isn ligands (Figure 3). The average twist angle (ata) is a useful tool to differentiate between a trigonal prismatic and an octahedral geometry but it must be noted that ata value does not give information about how much the geometry is deviated. If this value is closer to 60° the geometry will be octahedral while for values proximate to 0° the geometry probe to be trigonal prismatic [36,37].
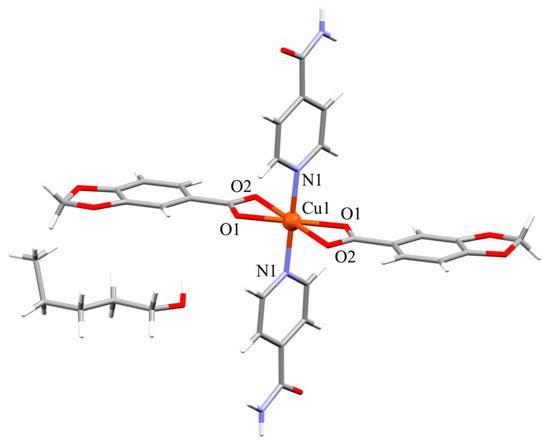
Figure 3.
Molecular structure of compound 3a.
The ata value for this complex is 60° being the three torsion angles: O2-Cg1-Cg2-O1, −79.26°; N1-Cg1-Cg2-O2, −46.55° and O1-Cg1-Cg2-N1, −54.19°. This six coordinated Cu(II) ion has a distorted octahedral geometry with two chelate Pip units and two monodentate Isn ligands. The axial positions of the octahedron are occupied by the oxygen chelate atoms with the longer Cu-O distance at 2.5069(16) Å, which is usually exhibited by d9 Cu(II) ions due to the Jahn-Teller effect [38,39], while the remaining oxygen atoms of the chelate Pip units and the nitrogen atoms of the Isn ligands sets the equatorial plane (Table 3). The chelate coordination mode of the Pip ligands promotes the distortion of the octahedral geometry resulting in angles ranging from 57.69(6)° to 122.31(6)°, and the metal node is out of the equatorial plane (0.522 Å) toward the apical site.

Table 3.
Selected bond lengths (Å) and bond angles (°) for compound 3a.
The monomeric units of 3a are held together via two double hydrogen bond interactions (Table 4). The main pair of interactions is the Isn···Isn pattern; the amide moieties engage in a head-to-head disposition, with carbonyl and amide groups pointing in opposite directions. This cooperation expands the structure through the [110] direction forming 1D chains. The other set of interactions are promoted by the furthest coordinated carboxylate oxygen atom of the Pip units that interacts with both hydrogen atoms from the same side of the Isn ligand throughout the [100] direction: one belongs to the amine moiety (N2-H2B···O2), and the other derives from the aromatic C-Hmeta (C12-H12···O2) (Figure 4a). These two groups of interactions form 2D layers along the ab plane (Figure 4b).

Table 4.
Selected intermolecular interactions for compound 3a.
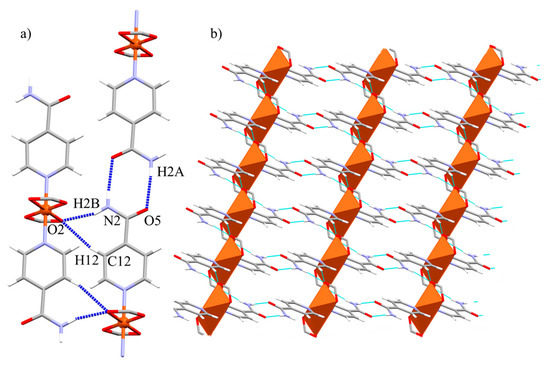
Figure 4.
(a) Representation of the intermolecular interactions promoted by 3a. Only carboxylate group of Pip units are shown for clarity. (b) View of the 2D nets generated by the Isn···Isn and Isn···Pip interactions along the ab plane.
2.5. Crystal and Extended Structure of Compound 4
Compound 4 crystallizes in the triclinic P space group. It presents a 1D polymeric structure composed of alternated monomeric and dimeric units, which expands through the [ 11] direction (Figure 5a). In this polymer, there are three Cu(II) centers with different arrays and coordination environments, displaying both [CuO4N2] and [CuO4N] cores. The first one pertains to monomeric units, in which the O atoms belong from two acetate and two MeOH molecules while the N atoms derive from two Isn ligands (Figure 5b). In the second core, the O atoms stem from one monodentate Pip unit, two methoxy and one carbonyl group from an Isn unit. The remaining N atoms pertain to two different Isn ligands. These methoxy groups frame the dimeric unit via a μ2-η1-O coordination mode (Figure 5c). The linkage of the monomeric and dimeric units for the polymeric growth, results from the bidentate bridging μ2-η1-η1-N, O coordination mode of the Isn ligand. It simultaneously coordinates to the monomeric Cu(II) ion by the pyridyl N atom and held together via coordination of the carbonyl OIsn atom to one Cu(II) center of the dimeric unit.
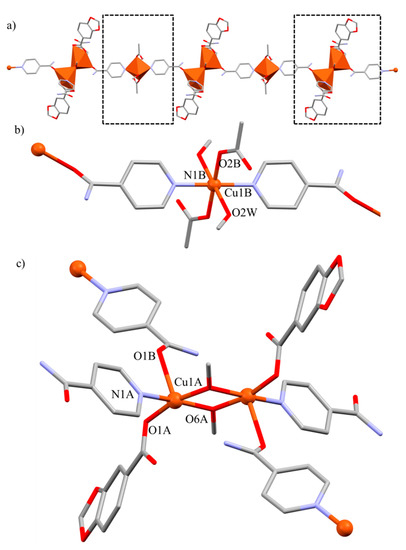
Figure 5.
Molecular structure of compound 4. (a) Polymeric 1D chain along the [ 11] direction. In detail, a view of (b) monomeric units and (c) dimeric units of the polymeric array. Hydrogen atoms are omitted for clarity.
All the lengths and angles (Table 5) are in range of other carboxylate and pyridil-containing Cu(II) compounds [20]. These dimeric units display a distorted square pyramidal geometry with a τ value of 0.03 [40]. The hindered disposition of the carbonyl OIsn atom in the dimeric arrays forces the geometry at the apical position. At the same time, the μ2-η1-O coordination modes of the methoxy groups drift the equatorial angles and promote its deviation from the perfect square pyramid (the apical-equatorial angles range from 84.80(10)° to 104.73(10)°). The Isn molecule has a more basic pKb value (10.87) respect to other common pyridine derivatives (2-benzylpyridine, 5.13; 3-phenylpyridine, 4.8; 4-acetylpyridine, 3.51), which can promote the formation of methoxide molecules that drive the arrangement of the polymeric array. The additional reflux period in combination with the basic pKb of the Isn ligand could drive the MeOH groups deprotonation, and their consequent coordination to the Cu(II) ions in 4.

Table 5.
Selected bond lengths (Å) and bond angles (°) for compound 4.
A further consideration is a large Cu···O bond length of 2.672 Å, which would provide a pseudo-octahedral geometry to the Cu(II) center of the dimeric unit. This distance between the metal ion and the uncoordinated carboxylate oxygen atom of the Pip unit seems to be too long to be considered as a Cu-O bond but not to the extent of being negligible. Other reported longer distances have been previously taken into consideration, as in the case of catena-(bis(μ-D-Hydrogen malato)-copper(II) dihydrate [41]. Different from most of complexes containing Cu(II) and Isn found in the literature, to the best of our knowledge, there is only one reference in which Isn synthon avoids the supramolecular amide···amide pattern at expense of the coordination of the amidic oxygen atom [42].
The supramolecular structure of 4 is based on the Isn ligand, which acts as a pillar of the expansion joining 1D chains and forming the 3D net (Figure 6). These polymeric chains are non-aligned but consecutively disposed in an alternate monomeric-dimeric manner.
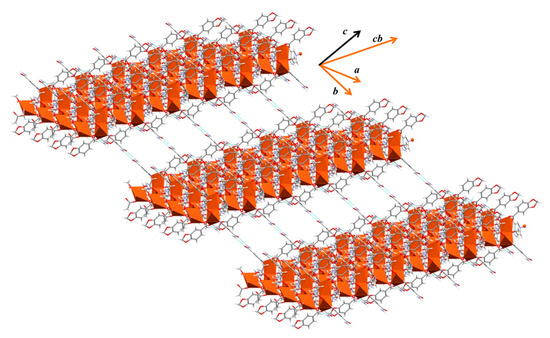
Figure 6.
Supramolecular 3D expansion of compound 4.
The monodentate Isn ligand of the dimeric arrays associate the chains by two hydrogen bond interactions. The mentioned amide···amide pattern holds together dimeric arrays through the b direction (Figure 7a), and this Isn units also participate in the expansion via a double N-Hanti/C-Hortho···O=C interaction with a monodentate Pip unit of other dimeric array in the bc direction. Besides, the carbonyl of this Isn ligand also forms an O···H-C interaction with the occluded MeOH molecule (Figure 7b). The final 3D net is supported by the bidentate bridged Isn linkers, which interact along the a axis by means of their N-Hanti with the non-coordinated carbonyl oxygen atom of the monodentate OAc units pertaining to the monomeric arrays (Table 6) (Figure 7c).
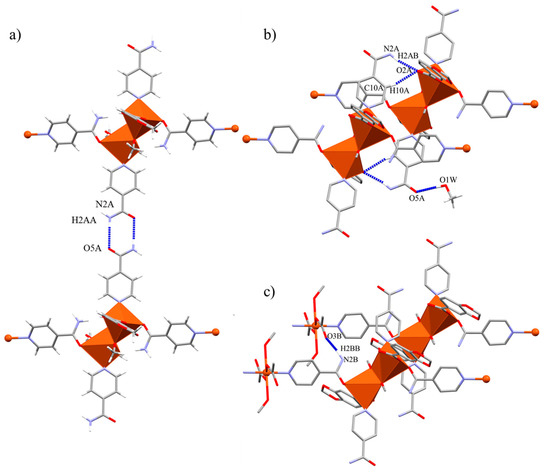
Figure 7.
Hydrogen bond interactions present in compound 4. (a) Amide-amide association along the b axis. (b) N-Hanti/C-Hortho···O=C(Pip) interaction between Isn and Pip ligands through the bc plane. (c) N-Hanti ···O=C(OAc) interaction promoted by bridging Isn ligand.

Table 6.
Selected intermolecular interactions for compound 4.
2.6. Hirshfeld Surfaces Analysis and Energy Frameworks Calculations
Hirshfeld surfaces analysis of all the structures, energy frameworks and lattice energy calculations of complexes 2 and 3a have been performed with CrystalExplorer 17.5 [43]. All the surfaces have been calculated at an isovalue of 0.5 e × au−3. The crystal structures of the (HPip)2(Isn) (2) adduct and 3a have been examined by energy framework based on TONTO, using CE-B3LYP(d,p) molecular wave-functions, calculated using .cif files obtained from the single crystal X-ray diffraction data. The disorder present in 3a has been identified, and the fractional occupancy has been corrected to 1 in the .cif file. It has not been possible to perform the same calculations with 4 owing to its polymeric structure.
Hirshfeld surface, in combination with 2D fingerprint plot, are powerful graphical tools to evaluate intra- and intermolecular interactions present in crystal structures. The surface mapping facilitates their identification while the fingerprint plot outlines the distances between the atoms involved in these interactions. All these calculations required the generation of the Hirshfeld surface with electron density mapping to construct the wave-function of each selected molecule. Owing to 2 being a multi-component crystal, this surface mapping has been performed for each unique molecule in the unit cell (four HPip and two Isn), verifying that there is one computed wave-function for each unique molecule. Hence, energies were calculated separately for each unique molecule in the unit cell (those from the asymmetric unit), and the energy framework was constructed based on crystal symmetry.
In the same way, Hirshfeld surface with electron density mapping and energy calculation of 3a have been performed for each unique molecule in the unit cell (one monomer and one pentanol molecule), and the energy frameworks were constructed based on crystal symmetry. The selection of the appropriate radius for the energy calculation depends on the polarity of the parts involved in the interactions. More polar groups will need a bigger radius, while less polar molecules require a smaller one. Pairwise interaction energies between molecules were computed considering radius of 20.0 Å for 2 and 3a from the centroid of the selected molecule to an atom belonging to its nearest neighbor. Energy framework is a unique tool to visualize the supramolecular architecture of crystal structures. The total energy (Etot) is divided into electrostatic (Eele), polarization (Epol) and dispersion (Edis) components in which cylinders represent the relative strength of the molecular packing based on a scale factor. The comparison between these components is only possible if the scale factor is the same. For 2 and 3a, the scale factor has been fixed at 240.
The main interactions present in compound 2 are identified and highlighted by Hirshfeld surface analysis (Figure S8a). The shape of the 2D fingerprint plot as well as the percentage of surface implied in the interactions suggests a difference between those which held the A and B BSMs (Figure S8b). This data was not sufficiently clear, so a further study including energy calculations was required (vide infra). By comparison, the main interactions and the percentage of surface implied present in compounds 3a and 4 are pinpointed in the Figure S9. In both, the majority involved belongs to the Isn ligand (amide···amide and N-Hanti/C-H···O interactions) and therefore have similar values of surface area implied in them.
Energy frameworks of 2 exhibited that the most important energy component is electrostatic (Figure 8a). These attractive forces derive from the amide···acid heterosynthon, while the dispersion energy is comparatively low and is mainly generated by the interaction between HPip units itself (Figure 8b). The total energy framework shows the association of BSMs in pairs that are stacked along the b axis (Figure 8c). The assembly of this cocrystal is dominated by O-H···N and N-H···O hydrogen bonds from the heterosynthons. The strongest associations in both A and B BSMs arise from the amide···acid dimeric square motifs (−68.3 kJ·mol−1 for B and −68.0 kJ·mol−1 for A) with similar but not identical energy values. Followed by the N···H-O chain motifs with energy values of −47.9 kJ·mol−1 (B) and −33.9 kJ·mol−1 (A) (Figure 8d).
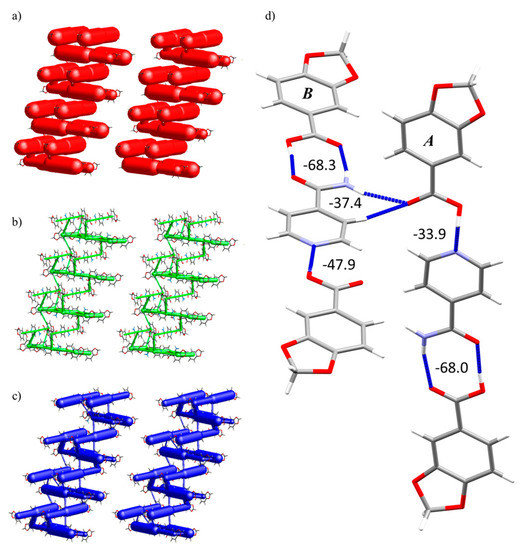
Figure 8.
Energy framework diagram (Eele, Edis and Etot) for compound 2. All diagrams use the same energy cylinder scale of 240, and the energy threshold is 6.00 kJ·mol−1. (a) Electrostatic energy. (b) Dispersion energy. (c) Total energy. (d) BSMs A and B pinpointing the main interactions and their energy values in kJ·mol−1.
As suggested by the 2D fingerprint plot with percentage values, there was a difference in the interaction that associates the BSMs. The energy of the N···H-O interaction in A and B presented a difference of ΔEB-A(N···H-O) = 14.0 kJ·mol−1, which is not negligible and is probably driven by the neighboring interaction in which the carbonyl group participates. These two BSMs are fastened by the central Isn unit, which forms this mentioned C-H/N-H···O double interaction with an acid of the neighboring BSMs with an energy value of −37.4 kJ·mol−1.
The computed lattice energy (Elat) value of each unique molecule has been calculated according to Equation (1), in which Ni is the number of molecule pairs in the cluster with that particular interaction energy and Ei is the interaction energy value.
The average Elat value of each HPip and Isn coformers has been calculated, and the crystal Elat is computed considering the 2:1 (HPip:Isn) molar ratio. The crystal structure Elat value of 2 is −362.8 kJ·mol−1, which is comparable to other reported energies of multicomponent cocrystals [44].
The energy frameworks of 3a are also driven by the electrostatic component, in which weaker destabilizing energies (1.0 and 2.2 kJ·mol−1) belong to the occluded pentanol molecules (Figure S10a), while the most remarkable dispersion energy is promoted by the parallel Pip ligands along the c axis (Figure S10b). When compared to 2, the dispersion energy of 3a is more significant and plays a relevant role in this system. This is induced by the restriction on the orientation of the Pip ligands caused by the coordination to the metal center.
The total energy frameworks confirm that the main interactions are those from Isn as mentioned in the structural description (Figure S10c). The amide···amide pattern stacks the monomeric units along the [110] with an energy value of −63.5 kJ·mol−1 and these chains are assembled through the N-H/C-H···O interactions with two energy values of −23.3 and −21.6 kJ·mol−1. The crystal structure Elat value is −414.6 kJ·mol−1 and it has also been calculated according to Equation (1).
3. Conclusions
We successfully isolated three different isonicotinamide based compounds by modifying the reaction conditions. The crystal structure elucidation of 2, 3a, and 4 allowed ascertaining the formation of a cocrystal (2), a monomer (3a), and an unusual coordination polymer (4). Isn ligand can acts as either hydrogen bond donor or acceptor or both by strong N···H-O and N-H···O interactions. Likewise, it has a potential role as mono- or bidentate linker coordinating to the Cu(II) center via N- or N,O-atoms, and these tendencies could promote the formation of the mixtures.
Compound 2 is constructed by type I and type III synthons instead of type II, which is the most common association in Isn-containing systems. This could be driven by the strong hydrogen bond donor solvent (MeOH) used in the crystallization process which probably dissociated the weaker type II homomeric synthon present in Isn and facilitated the formation of the stronger type III interaction. The union of both, Isn and HPip, fulfill the pKa rule and supports the formation of a cocrystal instead of a salt.
In the two complexes 3a and 4, both Pip and Isn ligands participated in the growth of the final scaffolds, but Isn outstands as the keystone in the formation of the products. In the precursor structure (1), the Pip units act as bidentate bridged while in 3a and 4, they exhibited a chelate and a monodentate coordination mode, respectively. Likewise, the Isn unit also adopts two coordination modes (monodentate (3a) or bidentate bridged (4)). The additional reflux period, in combination with the basic pKb of the Isn ligand, could drive the MeOH groups deprotonation and their consequent coordination to the Cu(II) ions in 4. It is noteworthy that Isn ligand has been the driving unit of both monomeric and coordination polymer, designing different molecular and supramolecular networks and downgrading Pip to an auxiliary ligand.
Hirshfeld surface analyses pinpointed the role of Isn as synthon. Furthermore, energy framework calculations distinguished Isn as the driving cohesive force, while HPip itself mainly participates in the London dispersion forces. Lattice energy calculation showed a notable value as a consequence of the large number of interactions promoted by the Isn ligand.
The formation of the cocrystal has enabled us to easily recognize the role of Isn as synthon and facilitated the better understanding of the driving forces in those systems.
4. Experimental Section
Cu(II) acetate monohydrate (Cu(OAc)2·H2O), 1,3-benzodioxole-5-carboxylic acid (piperonylic acid, HPip), and isonicotinamide (Isn); methanol (MeOH) and pentanol (C5H11OH) for recrystallization were used as solvents and were purchased from Sigma-Aldrich (Darmstadt, Germany). Deuterated dimethylsulfoxide (dmso-d6) was used for the NMR experiments and was purchased from Eurisotop (Saint-Aubin, France). All of them were used without further purification. The [Cu(μ-OAc)(μ-Pip)(MeOH)]2 (1) was previously synthesized in our research group [26,27].
Melting point was measured on an MPM-H2 KLEINFELD (Gehrden, Germany) melting point meter apparatus. Elemental analysis (C, H, N) were carried out on a Thermo Scientific (Waltham, MA, USA) Flash 2000 CHNS Analyzer. The FTIR-ATR spectra were recorded on a Perkin Elmer (Waltham, MA, USA) spectrometer, equipped with a universal attenuated total reflectance (ATR) accessory with diamond window in the range 4000–550 cm−1. 1H-NMR and 13C{1H}-NMR spectra were recorded on an NMR-FT Bruker400 (Karlsruhe, Germany) MHz spectrometer in dmso-d6 solution at room temperature (r.t.). All chemical shifts (δ) are given in ppm relative to TMS as internal standard. The electronic spectra in solution of MeOH (≈1.0 × 10−3 M) were run on an Agilent HP 8453 UV-Vis spectrophotometer with a quartz cell having a path length of 1 cm in the range of 500–900 nm. Spectroscopic characterization details of all compounds are in the Supplementary Materials (Figures S1–S7).
Synthesis of (HPip)2(Isn) (2) and [Cu(Pip)2(Isn)2] (3): A methanolic solution (25 mL) of 1 (98.9 mg, 0.155 mmol) was added dropwise to a methanolic solution (15 mL) of Isn (38.1 mg, 0.312 mmol) and was stirred for 2 h at r.t. A mixture of colorless and violet crystals was obtained by slow evaporation of the solution on air for 10 days.
Synthesis of (HPip)2(Isn) (2): A methanolic solution (15 mL) of HPip (100 mg, 0.602 mmol) was added dropwise to a methanolic solution (10 mL) of Isn (36.7 mg, 0.300 mmol) and was stirred for 4h at r.t. After that, reaction was concentrated until half of the volume. Suitable colorless crystals were obtained by slow evaporation of the solution on air for three days. Single crystals were filtered and washed with cold diethyl ether.
2. Yield: 49.5 mg (57%). Mp 193–194 °C. Elem. Anal. Calc. for C22H18N2O9 (454.39 g/mol): C 58.15; H 3.99; N 6.17. Found: C 58.02; H 3.84; N 6.14. FTIR-ATR (cm−1): 3321(m) [ν(N-H)], 3140(m) [ν(O-H)], 3075(w), 3072(w) [νar(C-H)], 2907(w) [νal(C-H)], 2791(w), 2650-2283(w), 1680(s) [ν(C=O)], 1620(m), 1551(m) [ν(C=C/C=N)], 1500(m), 1482(w), 1431(w), 1409(s), 1361(m) [δ(C=C/C=N)], 1280(w), 1254(w), 1233(m), 1164(m) [ν(C-O-C)], 1108(m), 1032(m), 1023(m) [δip(C-H)], 906(m), 763(m) [δoop(C-H)], 707(w), 674(w), 643(s) [δ(O=CN)], 583(w). 1H-NMR (360 MHz, DMSO-d6, Me4Si, 298 K): δ = 8.71 [2H, d, 3J = 5.8 Hz, CH-CH-N]Isn, 8.24 [1H, br, OCNHH-C]Isn, 7.76 [2H, dd, 3J = 4.5 Hz; 4J = 1.5 Hz, CH-CH-N]Isn, 7.73 [1H, br, OCNHH-C]Isn, 7.54 [2H, dd, 3J = 8.1 Hz; 4J = 1.5 Hz, CH-CH-C-O]HPip, 7.35 [2H, d, 4J = 1.5 Hz, CH-C-O]HPip, 6.99 [2H, d, 3J = 7.9 Hz, CH-CH-C-O]HPip, 6.11 [4H, s, O-CH2-O]HPip. 13C{1H}-NMR (360 MHz, DMSO-d6, Me4Si, 298 K): δ = 166.89 [O2C-C]HPip, 166.62 [OCNH2-C]Isn, 151.29 [CH-CH-C-O]HPip, 150.34 [CH-CH-N]Isn, 147.63 [CH-C-O]HPip, 141.46 [OCNH2-C]Isn, 125.15 [CH-CH-C-O]HPip, 124.82 [O2C-C]HPip, 121.65 [CH-CH-N]Isn, 108.99 [CH-C-O]HPip, 108.19 [CH-CH-C-O]HPip, 102.10 [O-CH2-O]HPip.
Synthesis of [Cu(Pip)2(Isn)2] (3): A methanolic solution (25 mL) of 1 (100 mg, 0.156 mmol) was added dropwise to a methanolic solution (15 mL) of Isn (39.0 mg, 0.319 mmol) and was stirred for 4h under reflux conditions. Solution turned dark blue, and a blue powder precipitated. The solid was filtered, washed with 5 mL of cold MeOH, and dried under vacuum. The powder was recrystallized in pentanol, and the resulting solution remained exposed to air until violet crystals of [Cu(Pip)2(Isn)2]·C5H11OH (3a) were grown by slow evaporation after four days.
3. Yield: 87.3 mg (88%). Elem. Anal. Calc. for C28H22CuN4O10 (638.05 g/mol): C 52.71; H 3.48; N 8.78. Found: C 52.82; H 3.64; N 8.64. FTIR-ATR (cm−1): 3333(br) [ν(N-H)], 3165(br) [ν(N-H)], 3072(w) [νar(C-H)], 2964-2860(br) [νal(C-H)], 1694(m) [ν(C=O)], 1630(m), 1617(m), 1557(m) [νas(COO)], 1543(m) [ν(C=C/C=N)], 1498(m), 1435(m) [νs(COO)], 1384(s) [δ(C=C/C=N)], 1343(m), 1247(s), 1165(m) [ν(C-O-C)], 1108(m), 1067(m), 1035(s) [δip(C-H)], 938(m), 924(m), 805(m) [δoop(C-H)], 772(s) [δoop(C-H)], 722(m), 682(m), 651(m), 629(m) [δ(O=CN)], 590(m). UV-Vis (MeOH, 1.10·10−3 M) λmax (ε) = 713 (77) nm.
Compound 3a presented the same FTIR-ATR spectrum as 3 with small differences in the 3400–2900 cm−1 region due to the presence of ν(O-H) signals attributable to pentanol molecules.
Synthesis of [Cu(Pip)2(Isn)2] (3) and {[Cu3(Pip)2(OAc)2(μ-Isn)2(Isn)2(μ-OCH3)2(MeOH)2] ·2MeOH}n (4): A methanolic solution (25 mL) of 1 (100 mg, 0.156 mmol) was added dropwise to a methanolic solution (15 mL) of Isn (127 mg, 1.04 mmol) and was stirred vigorously under reflux conditions for a week. Solution was evaporated until half of the volume and kept at 4 °C for a month until simultaneous violet (3, Yield: 51.4 mg (26%)) and green (4, Yield: 34.1 mg (52%)) crystals were formed. These crystals were mechanically separated.
4. Elem. Anal. Calc. for C48H54Cu3N8O20 (1253.62 g/mol): C 45.99; H 4.34; N 8.94. Found: C 45.67; H 4.29; N 9.18. FTIR-ATR (cm−1): 3435(w), 3333(br) [ν(N-H)], 3276(w), 3211 (br) [ν(N-H)], 3071(w) [νar(C-H)], 2992-2830(w) [νal(C-H)], 1696(m) [ν(C=O)], 1631(m), 1602(s) [ν(C=O)], 1554(m) [νas(COO)], 1504(m) [ν(C=C/C=N)], 1490(m), 1436(s), 1418(s) [δ(C=C/C=N)], 1387(s) [νs(COO)], 1258(s), 1243(m), 1221(m), 1170(m) [ν(C-O-C)], 1113(m), 1079(m), 1067(m), 1033(s) [δip(C-H)], 1015(m) [δip(C-H)], 935(m), 920(m), 889(m), 856(m), 805(m), 774(s) [δoop(C-H)], 765(m) [δoop(C-H)], 732(m), 683(s), 627(s) [δ(O=CN)], 582(s) [δ(O=CN)], 565(m). UV-Vis (MeOH, 9.90·10−4 M) λmax (ε) = 706 (404) nm.
5. Crystallographic Data
A colorless (2), a violet (3a), and a green (4) prism-like specimens were used for the X-ray crystallographic analysis. The X-ray intensity data were measured on a D8 Venture system equipped with a multilayer monochromator and a Mo microfocus (λ = 0.71073 Å). For 2–4, the frames were integrated with the Bruker SAINT software package using a narrow-frame algorithm. All hydrogen atoms were refined using a riding model (AFIX) with an isotropic temperature factor equal to 1.2, the equivalent temperature factor of the atom to which are linked and thus, the bond lengths of X–H were fixed. The structures were solved and refined using the Bruker SHELXTL Software package and refined using SHELX (version 2018/3) [45]. For 2–4, the final cell constants and volumes are based upon the refinement of the XYZ-centroids of reflections above 20 σ(I). Data were corrected for absorption effects using the Multi-Scan method (SADABS). Crystal data and relevant details of structure refinement for compounds 2–4 are reported in Table 7. CCDC 1944986 (2), 1944988 (3a), and 1944987 (4) contain the Supplementary Data of this paper. Molecular graphics were generated using Mercury 4.1.2 software [46,47] with POV-Ray package [48]. Color codes for all molecular graphics: orange (Cu), red (O), blue (N), dark grey (C), and white (H).

Table 7.
Crystal and structure refinement data for compounds 2–4.
Supplementary Materials
The following are available online at www.mdpi.com/xxx/s1, Figure S1. FTIR-ATR spectra of a. compound (HPip)2(Isn) (2); b. Isn and c. HPip.; Figure S2. 1H-NMR spectra of a. compound (HPip)2(Isn) (2); b. HPip and c. Isn.; Figure S3. 13C{1H}-NMR spectrum of a. compound (HPip)2(Isn) (2); b. HPip and c. Isn., Figure S4. FTIR-ATR spectrum of compound [Cu(Pip)2(Isn)2] (3).; Figure S5. FTIR-ATR spectrum of compound [Cu(Pip)2(Isn)2]·(C5H11OH) (3a).; Figure S6. FTIR-ATR spectrum of compound {[Cu3(Pip)2(OAc)2(μ-Isn)2(Isn)2(μ-OCH3)2(MeOH)2]·2MeOH}n (4).; Figure S7. Electronic spectra of compounds 3 and 4 between 500 nm and 900 nm in MeOH solution.; Figure S8. Hirshfeld surface representation and fingerprint plot of 2. In detail view of a. each molecule which forms the BSMs and b. each intra-BSM interaction with the surface area implied in it.; Figure S9. Hirshfeld surface representation and fingerprint plot of a. 3a and b. 4.; Figure S10. Energy frameworks diagram of a. Eele, b. Edis, c. Etot for compound 3a. All diagrams use the same energy cylinder scale of 240 and the energy threshold was fixed at 3.0 kJ·mol−1 for Eele and 30.0 kJ·mol−1 for Edis and Etot. Color codes of energy frameworks are red (Eele); green (Edis); blue (Etot) and yellow (destabilizing energies). Complete information about the crystal structure and molecular geometry is available in .cif format as Supporting Information.
Author Contributions
For research articles with several authors, a short paragraph specifying their individual contributions must be provided. The following statements should be used “conceptualization, J.P.; methodology, F.S.-F and D.E..; software, F.S.-F; validation, J.P. and T.C.; formal analysis, F.S.-F and M.F.-B; investigation, F.S.-F and D.E.; resources, J.P. and T.C.; data curation, F.S.-F., M.F.-B. and T.C.; writing—original draft preparation, F.S.-F. and D.E.; writing—review and editing, J.P. and T.C.; visualization, F.S.-F; supervision, J.P.; project administration, J.P.; funding acquisition, J.P. and T.C.”, please turn to the CRediT taxonomy for the term explanation. Authorship must be limited to those who have contributed substantially to the work reported.
Funding
This work was financed by CB615921 project, the Spanish National Plan of Research MAT2015-65756R, and 2017SGR1687 project from the Generalitat de Catalunya. The authors also acknowledge “Fundació La Caixa” for CB616406 endowment. F.S.-F. acknowledges the PIF pre-doctoral fellowship from the Universitat Autònoma de Barcelona.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Swiegers, G.F.; Malefetse, T.J. New Self-Assembled Structural Motifs in Coordination Chemistry. Chem. Rev. 2000, 100, 3483–3538. [Google Scholar] [CrossRef]
- Desiraju, G.R. Hydrogen bonds and other intermolecular interactions in organometallic crystals. J. Chem. Soc. Dalton Trans. 2000, 21, 3745–3751. [Google Scholar] [CrossRef]
- Cambridge Structural Database (CSD) (Version 5.37). 2016. Available online: https://libguides.caltech.edu/crystallography/CSD#Applications (accessed on 14 November 2019).
- Ye, B.-H.; Tong, M.-L.; Chen, X.-M. Metal-organic molecular architectures with 2,2′-bipyridyl-like and carboxylate ligands. Coord. Chem. Rev. 2005, 249, 545–565. [Google Scholar] [CrossRef]
- Etter, M.C. Encoding and decoding hydrogen-bond patterns of organic compounds. Acc. Chem. Res. 1990, 23, 120–126. [Google Scholar] [CrossRef]
- Desiraju, G.R. Supramolecular Synthons in Crystal Engineering—A New Organic Synthesis. Angew. Chem., Int. Ed. 1995, 34, 2311–2327. [Google Scholar] [CrossRef]
- Nangia, A.; Desiraju, G.R. Supramolecular Synthons and Pattern Recognition. In Design of Organic Solids; Topics in Current Chemistry, 198; Weber, E., Aoyama, Y., Caira, M.R., Desiraju, G.R., Glusker, J.P., Hamilton, A.D., Meléndez, R.E., Nangia, A., Eds.; Springer: Berlin/Heidelberg, Germany, 1998; pp. 57–95. [Google Scholar]
- Wang, Y.-Y.; Shi, Q.; Shi, Q.-Z.; Gao, Y.-C.; Zhou, Z.-Y. Syntheses, characterization and crystal structure of copper(II) α,β-unsaturated carboxylate complexes with imidazole. Polyhedron 1999, 18, 2009–2015. [Google Scholar] [CrossRef]
- Choudhury, A.R.; Desiraju, G.R.; Dikundwar, A.G.; Dubey, R.; Duggirala, N.; Ghogale, P.P.; Ghosh, S.; Goswami, P.K.; Goud, N.R.; Jetti, R.R.K.R.; et al. Polymorphs, Salts, and Cocrystals: What’s in a Name? Cryst. Growth Des. 2012, 12, 2147–2152. [Google Scholar]
- Germán-Acacio, J.M.; Hernández-Ortega, S.; Aakeröy, C.B.; Valdés-Martínez, J. Using Lewis acidity differences in chelating ligands to control molecular structure and supramolecular assembly of Cu(II) complexes. Inorg. Chim. Acta 2009, 362, 4087–4090. [Google Scholar] [CrossRef]
- Ɖakovic, M.; Jagličić, Z.; Kozlevčar, B.; Popović, Z. Association of copper(II) isonicotinamide moieties via different anionic bridging ligands: Two paths of ferromagnetic interaction in the azide coordination compound. Polyhedron 2010, 29, 1910–1917. [Google Scholar] [CrossRef]
- Ataç, A.; Yurdakul, Ş.; Ide, S. Synthesis and vibrational spectroscopic studies of isonicotinamide metal(II) halide complexes. J. Mol. Struct. 2006, 783, 79–87. [Google Scholar] [CrossRef]
- Mukherjee, A. Building upon Supramolecular Synthons: Some Aspects of Crystal Engineering. Cryst. Growth Des. 2015, 15, 3076–3085. [Google Scholar] [CrossRef]
- Yenikaya, C.; Poyraz, M.; Sari, M.; Demirci, F.; İlkimen, H.; Büyükgüngör, O. Synthesis, characterization and biological evaluation of a novel Cu(II) complex with the mixed ligands 2,6-pyridinedicarboxylic acid and 2-aminopyridine. Polyhedron 2009, 28, 3526–3532. [Google Scholar] [CrossRef]
- Cruz-Cabeza, A.J. Acid–base crystalline complexes and the pKa rule. CrystEngComm. 2012, 14, 6362–6365. [Google Scholar] [CrossRef]
- Bhogala, B.R.; Basavoju, S.; Nangia, A. Tape and layer structures in cocrystals of some di- and tricarboxylic acids with 4,4′-bipyridines and isonicotinamide. From binary to ternary cocrystals. CrystEngComm. 2005, 7, 551–562. [Google Scholar] [CrossRef]
- Liu, C.-S.; Wang, J.-J.; Yan, L.-F.; Chang, Z.; Bu, X.-H.; Señudo, E.C.; Ribas, J. Copper(II), Cobalt(II), and Nickel(II) Complexes with a Bulky Anthracene-Based Carboxylic Ligand: Syntheses, Crystal Structures, and Magnetic Properties. Inorg. Chem. 2007, 46, 6299–6310. [Google Scholar] [CrossRef]
- Moulton, B.; Zaworotko, M.J. From Molecules to Crystal Engineering: Supramolecular Isomerism and Polymorphism in Network Solids. Chem. Rev. 2001, 101, 1629–1658. [Google Scholar] [CrossRef]
- Arici, M.; Yeşilel, O.Z.; Acar, E.; Dege, N. Synthesis, characterization and properties of nicotinamide and isonicotinamide complexes with diverse dicarboxylic acids. Polyhedron 2017, 127, 293–301. [Google Scholar] [CrossRef]
- Moncol, J.; Mudra, M.; Lönnecke, P.; Hewitt, M.; Valko, M.; Morris, H.; Svorec, J.; Melnik, M.; Mazur, M.; Koman, M. Crystal structures and spectroscopic behavior of monomeric, dimeric and polymeric copper(II) chloroacetate adducts with isonicotinamide, N-methylnicotinamide and N,N-diethylnicotinamide. Inorg. Chim. Acta 2007, 360, 3213–3225. [Google Scholar] [CrossRef]
- Bhogala, B.R.; Thallapally, P.K.; Nangia, A. 1:2 and 1:1 Ag(I)-Isonicotinamide coordination compounds: Five-fold interpenetrated CdSO4 network and the first example of (pyridine)N-Ag-O(amide) bonds. Crystal Growth & Design, 4, 215-218. Cryst. Growth Des. 2004, 4, 215–218. [Google Scholar]
- Lynch, M.B.; Lawrence, S.E.; Nolan, M. Predicting Nucleation of Isonicotinamide from the Solvent–Solute Interactions of Isonicotinamide in Common Organic Solvents. J. Phys. Chem. A 2018, 122, 3301–3312. [Google Scholar] [CrossRef]
- Vishweshwar, P.; Nangia, A.; Lynch, V.M. Molecular complexes of homologous alkanedicarboxylic acids with isonicotinamide: X-ray crystal structures, hydrogen bond synthons, and melting point alternation. Cryst. Growth Des. 2003, 3, 783–790. [Google Scholar] [CrossRef]
- Mohamed, S.; Tocher, D.A.; Vickers, M.; Karamertzanis, P.G.; Price, S.L. Salt or Cocrystal? A New Series of Crystal Structures Formed from Simple Pyridines and Carboxylic Acids. Cryst. Growth Des. 2009, 9, 2881–2889. [Google Scholar] [CrossRef]
- Sánchez-Férez, F.; Bayés, L.; Font-Bardia, M.; Pons, J. Solvent dependent formation of Cu(II) complexes based on isonicotinamide ligand. Inorg. Chim. Acta 2019, 494, 112–122. [Google Scholar] [CrossRef]
- Soldevila-Sanmartín, J.; Ayllón, J.A.; Calvet, T.; Font-Bardía, M.; Domingo, C.; Pons, J. Synthesis, crystal structure and magnetic properties of a Cu(II) paddle-wheel complex with mixed bridges. Inorg. Chem. Commun. 2016, 71, 90–93. [Google Scholar] [CrossRef]
- Sánchez-Férez, F.; Guerrero, M.; Ayllón, J.A.; Calvet, T.; Font-Bardía, M.; Giner Planas, J.; Pons, J. Reactivity of homoleptic and heteroleptic core paddle wheel Cu(II) compounds. Inorg. Chim. Acta 2019, 487, 295–306. [Google Scholar]
- Williams, D.H.; Fleming, I. Spectroscopic Methods in Organic Chemistry; McGrawHill: London, UK, 1995. [Google Scholar]
- Dey, A.; Kirchner, M.T.; Vangala, V.R.; Desiraju, G.R.; Mondal, R.; Howard, J.A.K. Crystal Structure Prediction of Aminols: Advantages of a Supramolecular Synthon Approach with Experimental Structures. J. Am. Chem. Soc. 2005, 127, 10545–10559. [Google Scholar] [CrossRef]
- Bavishi, D.D.; Borkhataria, C.H. Spring and parachute: How cocrystals enhance solubility. Prog. Cryst. Growth Charact. Mater. 2016, 62, 1–8. [Google Scholar] [CrossRef]
- Shishkin, O.V.; Zubatyuk, R.I.; Shishkina, S.V.; Dyakonenko, V.V.; Medviediev, V.V. Role of supramolecular synthons in the formation of the supramolecular architecture of molecular crystals revisited from an energetic viewpoint. Phys. Chem. Chem. Phys. 2014, 16, 6773–6786. [Google Scholar] [CrossRef]
- Seo, M.; Park, J.; Kim, S.Y. Self-assembly driven by an aromatic primary amide motif. Org. Biomol. Chem. 2012, 10, 5332–5342. [Google Scholar] [CrossRef]
- Tothadi, S.; Desiraju, G.R. Unusual co-crystal of isonicotinamide: The structural landscape in crystal engineering. Philos. Trans. R. Soc. A 2012, 370, 2900–2915. [Google Scholar] [CrossRef]
- Nakamoto, K. Infrared and Raman Spectra of Inorganic and Coordination Compounds. Applications in Coordination, Organometallic, and Bioinorganic Chemistry, 6th ed.; Wiley Interscience: New York, NY, USA, 2009. [Google Scholar]
- Sutton, D. Electronic Spectra of Transition Metal Complexes, 1st ed.; McGraw-Hill: New York, NY, USA, 1968. [Google Scholar]
- Morse, P.M.; Girolami, G.S. Are d0 ML6 Complexes Always Octahedral? The X-ray Structure of Trigonal-Prismatic [Li(tmed)]2[ZrMe6]. J. Am. Chem. Soc. 1989, 111, 4114–4116. [Google Scholar] [CrossRef]
- Friese, J.C.; Krol, A.; Puke, C.; Kirschbaum, K.; Giolando, D.M. Trigonal Prismatic vs Octahedral Coordination Geometry: Syntheses and Structural Characterization of Hexakis(arylthiolato) Zirconate Complexes. Inorg. Chem. 2000, 39, 1496–1500. [Google Scholar] [CrossRef] [PubMed]
- Falvello, L.R. Jahn–Teller effects in solid-state co-ordination chemistry. J. Chem. Soc. Dalton Trans. 1997, 1, 4463–4475. [Google Scholar] [CrossRef]
- Nimmermark, A.; Öhrström, L.; Reedijk, J. Metal-ligand bond lengths and strengths: Are they correlated? A detailed CSD analysis. Z. Kristallogr. 2013, 228, 311–317. [Google Scholar] [CrossRef]
- Adison, A.W.; Rao, T.N. Synthesis, structure, and spectroscopic properties of copper(II) compounds containing nitrogen–sulphur donor ligands; the crystal and molecular structure of aqua[1,7-bis(N-methylbenzimidazol-2′-yl)-2,6-dithiaheptane]copper(II) perchlorate. J. Chem. Soc. Dalton. Trans. 1984, 1, 1349–1356. [Google Scholar] [CrossRef]
- Gör, K.; Kürkçüoğlu, G.S.; Yeşilel, O.Z.; Büyükgüngör, O. One-dimensional bimetallic cyano complexes with nicotinamide and isonicotinamide ligands. J. Mol. Struct. 2014, 1060, 166–175. [Google Scholar] [CrossRef]
- Gudavarthy, R.V.; Burla, N.; Kulp, E.A.; Limmer, S.J.; Sinn, E.; Switzer, J.A. Epitaxial electrodeposition of chiral CuO films from copper(II) complexes of malic acid on Cu(111) and Cu(110) single crystals. J. Mater. Chem. 2011, 21, 6209–6216. [Google Scholar] [CrossRef]
- Wolff, S.K.; Grimwood, D.J.; McKinnon, J.J.; Jayatilaka, D. and Spackman, M.A. CrystalExplorer17.5; University of Western, Australia: Crawley, Australia, 2017. [Google Scholar]
- Évora, A.O.L.; Bernades, C.E.S.; Piedade, M.F.M.; Conceição, A.C.L.; Minas da Piedade, M.E. Energetics of Glycine Cocrystal or Salt Formation with Two Regioisomers: Fumaric Acid and Maleic Acid. Cryst. Growth Des. 2019, 19, 5054–5064. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.; Taylor, R.; Towler, M.; Van de Streek, J.J. Mercury: Visualization and analysis of crystal structures. Appl. Crystallogr. 2006, 39, 453–457. [Google Scholar] [CrossRef]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, I.; Taylor, R.; Towler, M.; van de Streek, J.; et al. Mercury CSD 2.0—New features for the visualization and investigation of crystal structures. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Persistence of Vision Pty. Ltd. Persistence of Vision (TM) Raytracer; Persistence of Vision Pty. Ltd.: Williamstown, Australia, 2004; Available online: http://www.povray.org/ (accessed on 14 November 2019).
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).