1. Introduction
Fluorescent
Pseudomonas species possess a robust metabolic machinery with the inherent ability to produce multiple and diverse secondary metabolites, including antibiotics, rhamnolipids and cyclic lipopeptides (CLPs) [
1,
2]. CLPs are bioactive molecules which possess multiple functions in the producing bacteria, including swarming motility, biofilm formation, virulence, and can further mediate biological control against plant pathogens via direct antagonism and elicitation of induced systemic resistance (ISR) [
3,
4,
5,
6,
7]. The mode of action of CLPs is attributed to their capacity to penetrate the plasma membrane, and modify the membrane integrity of target microbes, cell and/or tissues [
2,
4], leading to hyphal leakage and extensive branching, among others [
6,
8,
9,
10].
Pseudomonas CLPs are categorized into fourteen different groups [
5], based on the oligopeptide length and macrocycle size and fatty acid length. CLP members belonging to these families have been described from
Pseudomonas strains isolated from diverse ecologies [
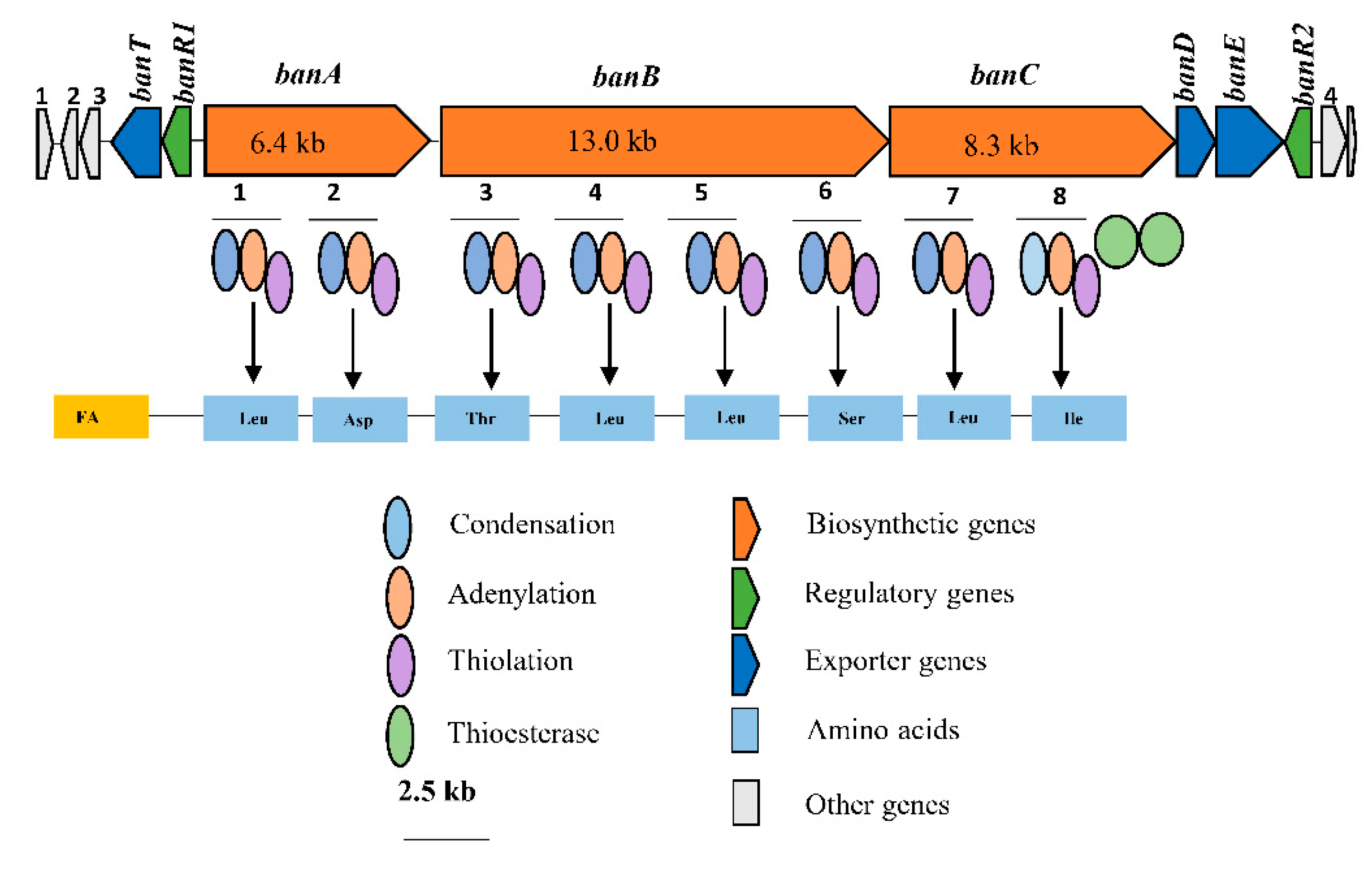
11]. These molecules are encoded by non-ribosomal peptide synthetases (NRPSs) via distinct modules which comprise adenylation (A), condensation (C) and thiolation (T) domains [
12,
13,
14,
15].
Genome mining has led to the discovery of NRPS clusters that encode diverse CLPs [
1,
16]. The phylogeny of NRPS domains are quite complex and demonstrate different evolutionary patterns. The A and C domains are the most conserved and have been shown to evolve independently in the same pathway [
17]. The A domain selects the cognate amino acid and generates the corresponding amino acyl adenylate. The specificity-conferring code used by the A domains has been deciphered [
18], and enables structural predictions of unknown CLPs from the primary sequence. Some positions within the signature sequence are variable, allowing some flexibility in amino acid selection. On the other hand, C domain phylogeny facilitates the stereochemistry determination of the amino acids which are added to the growing peptide chain [
17]. Within this phylogeny, six domain clades have been identified namely,
LC
L,
DC
L, Starter C, cyclization, epimerization, and dual E/C domains [
19].
Chemical structures of CLPs have been largely deciphered using ultraviolet-visible (UV), infrared (IR) spectrophotometry, mass spectrometry (MS) and nuclear magnetic resonance (NMR) [
20]. Novel reports of CLPs by specific strains have been accompanied by both nuclear magnetic resonance (NMR) characterization and/or genetic analyses of the NRPS gene clusters. Examples of CLPs that have been characterized using NMR, genetic analysis, or both methods include xantholysin [
21], WLIP [
7,
22,
23], viscosin [
24], poaeamide [
25], massetolide [
26,
27], lokisin [
7,
23], anikasin [
28], orfamide [
6,
29], arthrofactin [
30,
31], putisolvin [
32,
33], bananamides A-C [
34], cocoyamide [
9], and gacamide [
35], among others.
Concomitantly, chemical structure determination of certain CLPs, such as entolysin produced by
P. entomophilia L48T, was conducted via MS, using matrix-assisted laser desorption ionization (MALDI) mass spectrometry (MS) and tandem MS (MS/MS) analysis [
36]. This led to complementary results regarding CLP structure as those obtained via in silico analysis of the whole genome of L48T. Nevertheless, the use of high resolution mass spectrometry (HRMS) such as very high field Fourier transformation ion cyclotron resonance (FT-ICR) technologies or the Orbitrap and hybrid Q-TOF technologies are now commonplace.
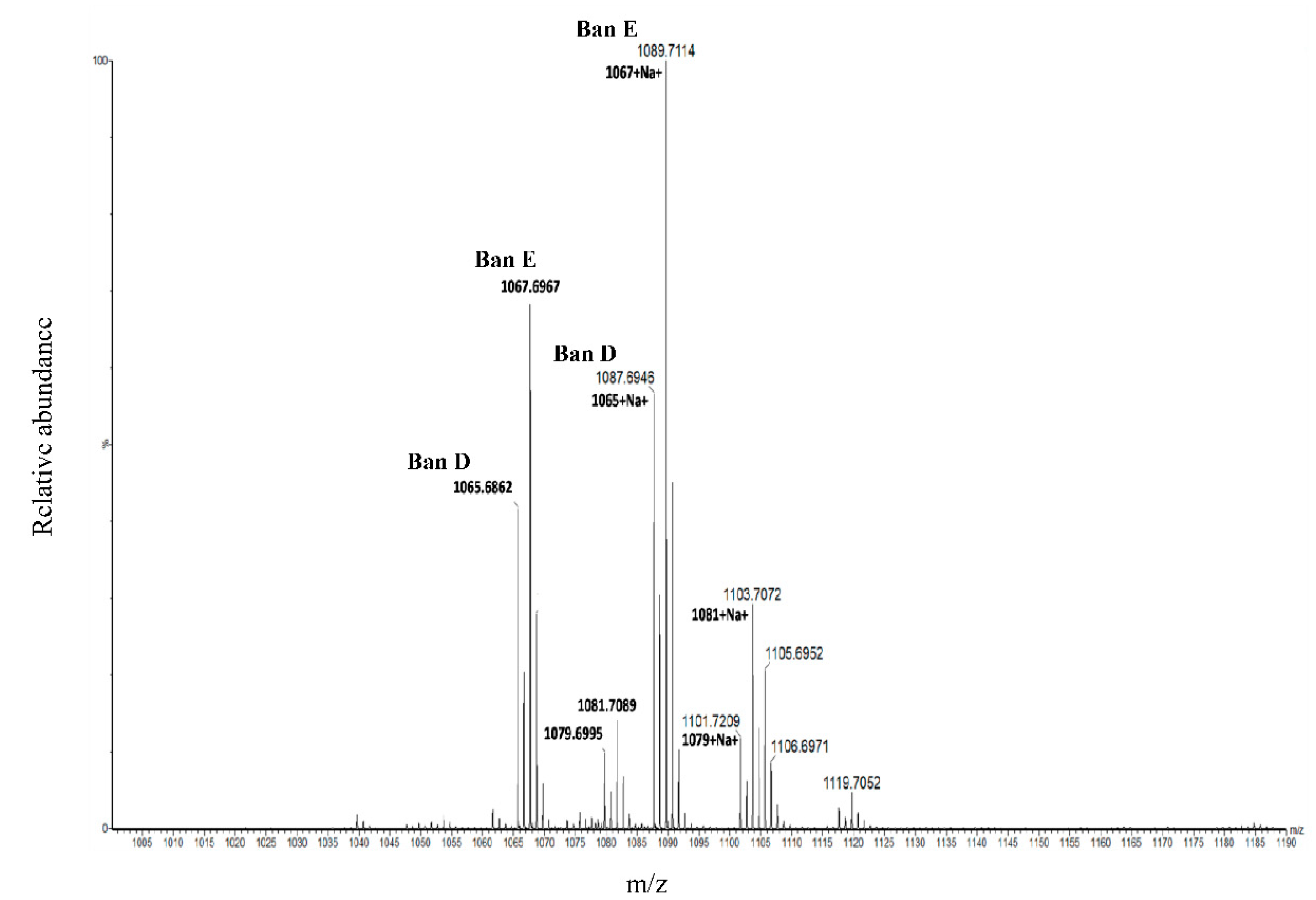
In this context of HRMS, the Kendrick mass defect (KMD) analysis has been employed as a proof of concept for the detection, identification and discrimination of chemically related compounds on the basis of their exact masses obtained from MS [
37]. The use of this method for molecular formula assignment from MS-derived exact masses is being explored for novel CLPs. The proof of concept of the KMD approach has been recently published and appears as a new tool for the elucidation of novel CLPs/CLP derivatives [
38].
The first bananamide-type CLP was elucidated from
P. granadensis F-278,770
T (=LMG27940
T), which produces a bananamide derivative named MDN-0066 with the following structure, 3-OH C10:0 –Leu1–Glu2–Thr3–Leu4–Leu5–Ser6–Leu7–Ile8 [
39]. In search of antitumor therapies, MDN-0066 was demonstrated to successfully induce apoptosis in renal (kidney) cancer cell lines [
39]. Furthermore, another bananamide-type CLP was isolated and characterized from a banana rhizosphere isolate from Sri Lanka,
Pseudomonas sp. BW11P2 [
34]. This CLP possesses an amino acid (AA) chain length of eight with six AA in the ring. Besides the production of bananamide 1, with the chemical structure 3-OH C12:0–Leu1–Asp2–Thr3–Leu4–Leu5–Gln6–Leu7–Ile8, BW11P2 produces two other derivatives designated bananamide 2, chemically elucidated as 3-OH C10:0–Leu1–Asp2–Thr3–Leu4–Leu5–Gln6– Leu7–Ile8, and 3, chemically determined as 3-OH C12:1–Leu1–Asp2–Thr3–Leu4–Leu5–Gln6–Leu7– Ile8. We previously isolated and characterized
Pseudomonas spp. COW3 and COW65 from the cocoyam rhizosphere in Cameroon [
23]. These strains produce a common CLP designated N3 and in in vivo experiments, COW3 effectively suppressed the cocoyam root rot disease caused by
Pythium myriotylum [
9]. Recent studies showed the capacity of COW3 to induce systemic resistance against
Pyricularia oryzae (syn.
Magnaporthe oryzae) VT5M1 on rice, although the CLP N3 appeared not to be involved in this process [
7]. However, crude N3 produced by COW3 inhibited appressoria formation and suppressed
P. oryzae VT5M1 on rice via direct antagonism [
7].
In this study, we elucidated the structure of N3 by performing chemical structural analyses, using the Kendrick mass defect-based molecular formula assignment and NMR. Also, we sequenced the genome of COW3 and conducted in silico analysis of the NRPS gene clusters. Next, we investigated the biological activity of purified novel bananamide derivatives produced by COW3 against P. myriotylum CMR1, a soil-borne pathogen of cocoyam (host plant of COW3 and COW65), and a foliar rice pathogen, P. oryzae VT5M1.
3. Discussion
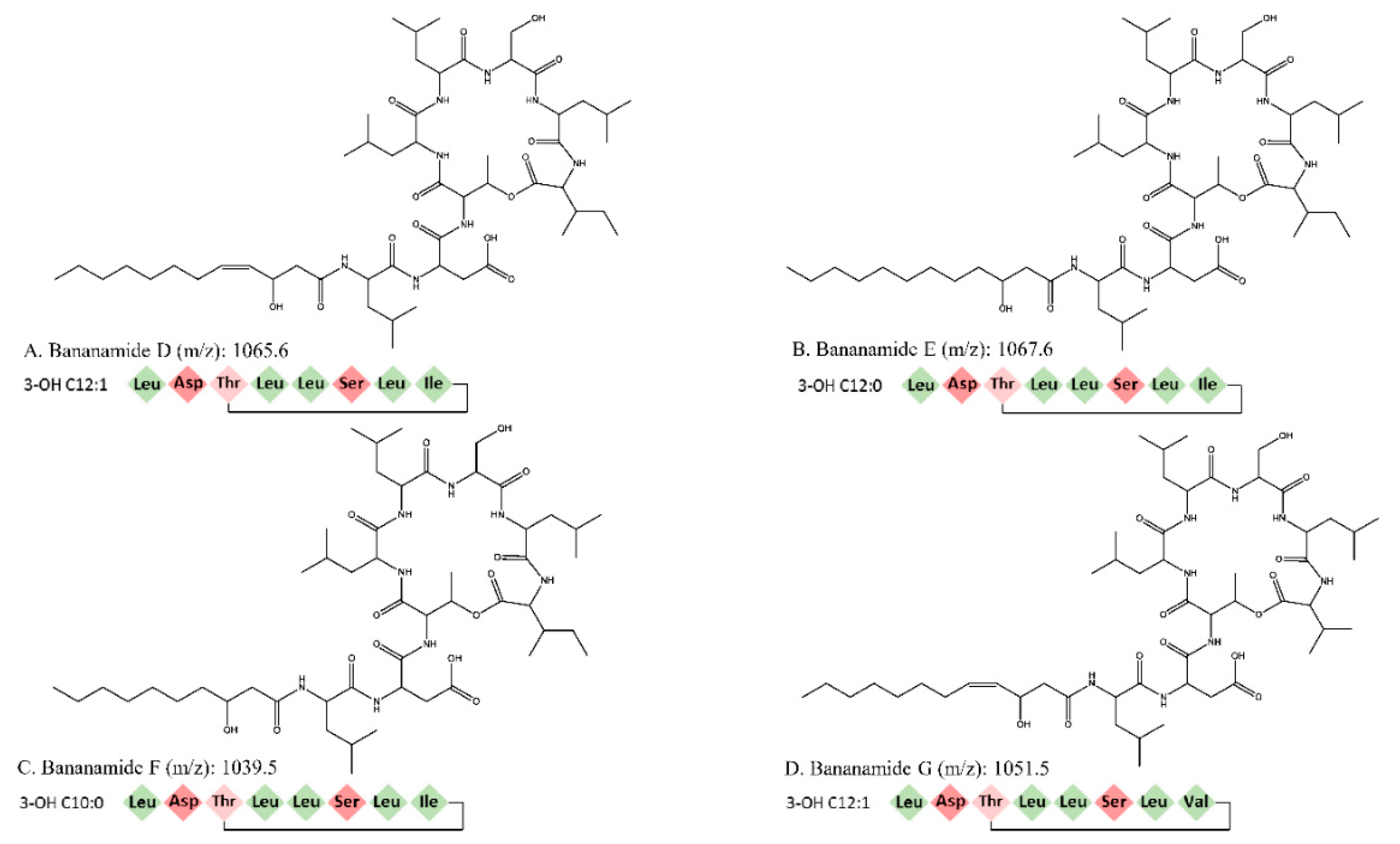
In this study we conducted the chemical characterization, genetic analysis and biological activity of novel bananamide derivatives, bananamides D-G, produced by Pseudomonas sp. COW3. A combination of genome mining and subsequent chemical analysis enabled the identification of the major CLPs produced by COW3.
Using NMR, we were able to identify two major CLPs, designated bananamide D and E, in combination with two minor derivatives called bananamides F and G. Chemical analysis revealed that these compounds are similar to the recently described bananamides [
34], MDN-0066 [
39], and the pseudofactins [
41]. However, bananamide D and variants produced by
Pseudomonas sp. COW3 differ from the previously described CLPs by amino acid substitution at certain positions, fatty acids tail and/or level of saturation. This is a common phenomenon with CLP-producing bacteria such as
Bacillus spp.,
Streptomyces spp. and
Pseudomonas spp. owing to their capacity to harness and incorporate amino acids in the environment to enable CLPs synthesis, thereby conferring a competitive advantage to them in relation to other microorganisms in the environment. For bananamide D and derivatives, the substitution of an amino acid at the eighth position, disparity in the length and presence of unsaturation at the fifth position in the fatty acid led to the confirmation of the novelty of the four structurally diverse bananamide CLPs from
Pseudomonas sp. COW3 strain. The Kendrick mass defect method for molecular formula assignment has been used for the detection and discrimination of chemically related compounds based on their exact masses [
37]. In this study, we present the first use of KMD in the discovery of novel bananamide variants. KMD identified the two major bananamides produced by
Pseudomonas sp. COW3 (D and E). KMD approach for molecular formula assignment to CLPs is therefore an effective and reliable method for the determination of the molecular formulae of CLPs using the experimentally determined high resolution mass (HRM). The HRM is often compared to masses of CLPs on NORINE database, thereby enabling the discovery and identification of novel CLPs.
Consequently, bioinformatics analyses revealed the eight amino acid composition of CLP produced by COW3 as follows: FA-D-Leu-D-Asp-D-Thr-D-Leu-D-Leu-D-Ser-L-Leu-L-Ile, which is consistent with the “colinearity rule” of nonribosomal peptides and chemical analysis. We discovered that bananamide D-G are novel bananamide derivatives, which differ from MDN-0066 (P. granadensis F-278,770T) and bananamides A-C (BW11P2) in the amino acids composition. Furthermore, our in silico analysis showed that similar to the NRPS backbone of the bananamide producer, BW11P2, our strain (COW3) possesses three NRPS genes
banA, banB and
banC. These results demonstrate that genome mining remains a powerful tool for the discovery of novel metabolites including CLPs [
16].
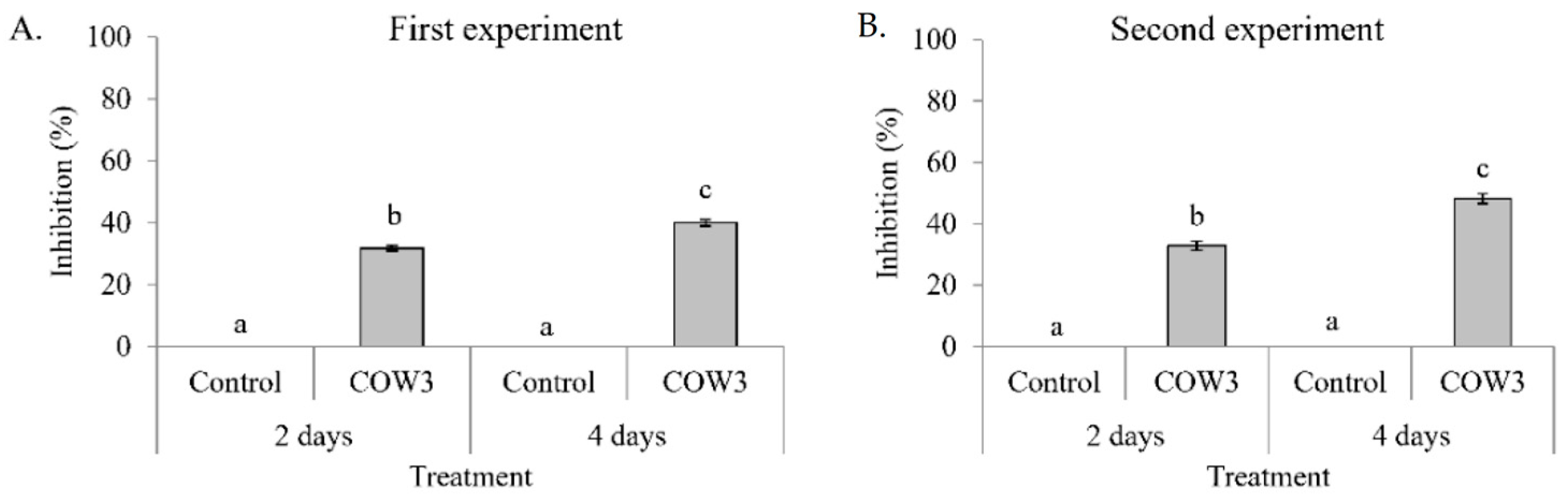
In order to obtain insights into the biological activities of COW3, we conducted whole cell co-cultivation assay against the soil-borne cocoyam pathogen,
P. myriotylum CMR1, and the rice blast pathogens,
P. oryzae VT5M1 and Guy11. In this study, whole cells of COW3 showed in vitro antagonistic activity against
P. myriotylum CMR1 with clear zones of inhibition after two days, coupled with mycophagy after four days of co-incubation with the pathogen. A recent study reports that
Trichoderma strains with in vitro antagonism against
P. myriotylum all produced cellulases, proteases, and xylanases [
42]. It remains to be investigated whether this is also the case for our bacterial strain
Pseudomonas sp. COW3. Our results are interesting considering the origin and context in which COW3 was isolated. COW3 is a cocoyam rhizosphere isolate obtained from the roots of cocoyam plants grown in a
Pythium root rot disease-suppressive Boteva soil [
9]. Our findings suggest that COW3 is a likely player, alongside other CLP producers, in carving a ‘safe haven’ for its cocoyam host plant.
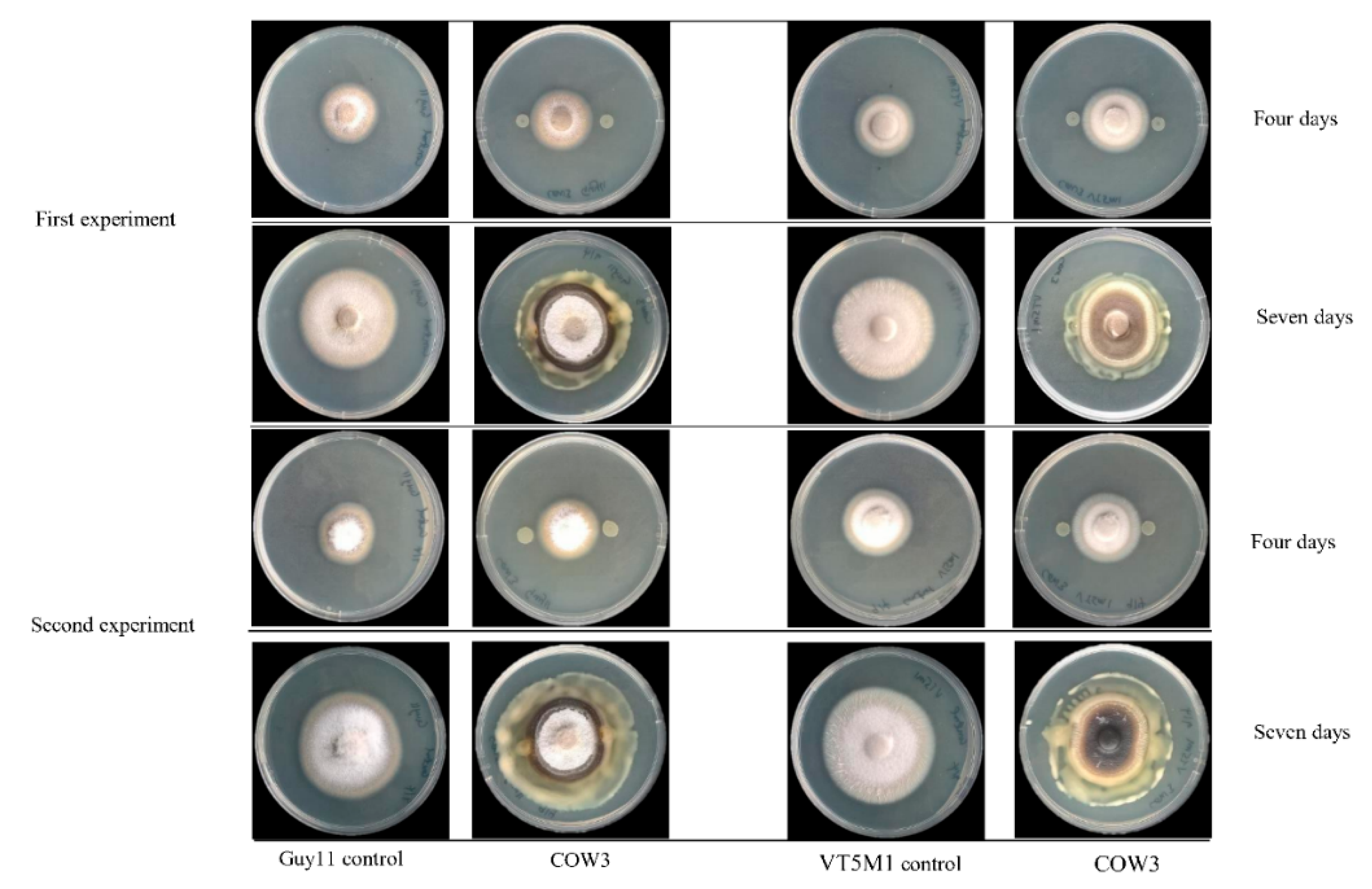
On the other hand, in co-cultivation experiments with
P. oryzae VT5M1 and Guy11, COW3 mainly mediates hyphal digestion and mycophagy after seven days. Strains with biocontrol capacities have been reported to demonstrate mycophagous behaviour against fungal plant pathogens via synthesis of proteases, chitinases, N-acetyl-beta-d-glucosaminidase and glucanases [
43]. The functional role of CLPs in direct antagonistic and mycophagous activities of COW3 needs to be studied further using site-directed mutagenesis.
In previous studies, crude bananamide D-G (previously named CLP N3) inhibited appressoria formation by
P. oryzae and reduced rice blast disease severity [
7]. Besides the sensitivity of renal carcinoma cell lines to MDN-0066 [
39], there has been no specific study aimed at investigating the biological activity of bananamides. In this study, we investigated the effect of structural diversity on the activity of purified bananamide D-G against our target pathogens,
P. myriotylum CMR1 and
P. oryzae. The goal was to decipher the effect of substitution of a valine for isoleucine at position 8 in bananamide G and to further investigate how differences in the fatty acids residue and level of saturation of bananamide D-G could affect their biological activity against these pathogens.
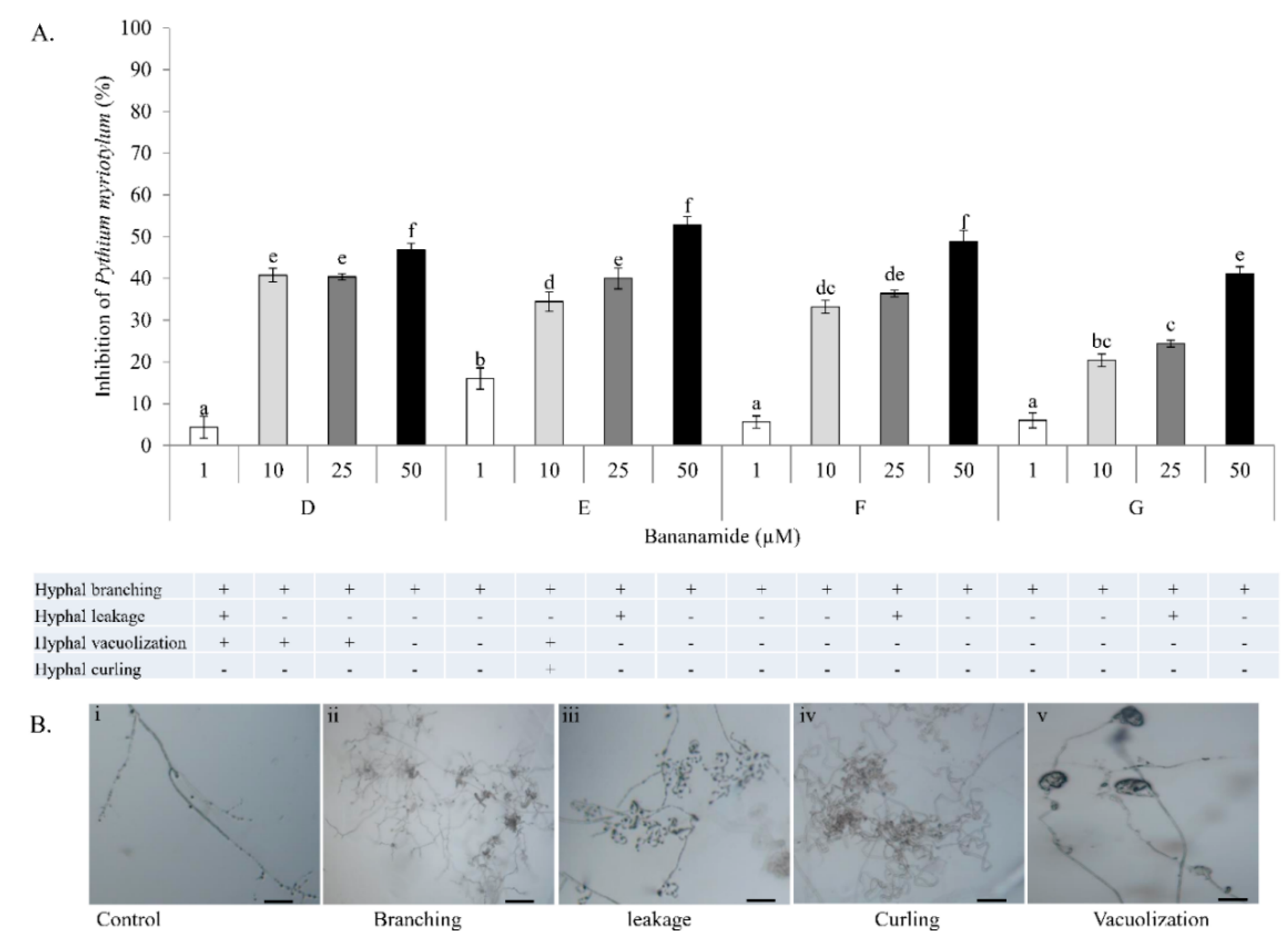
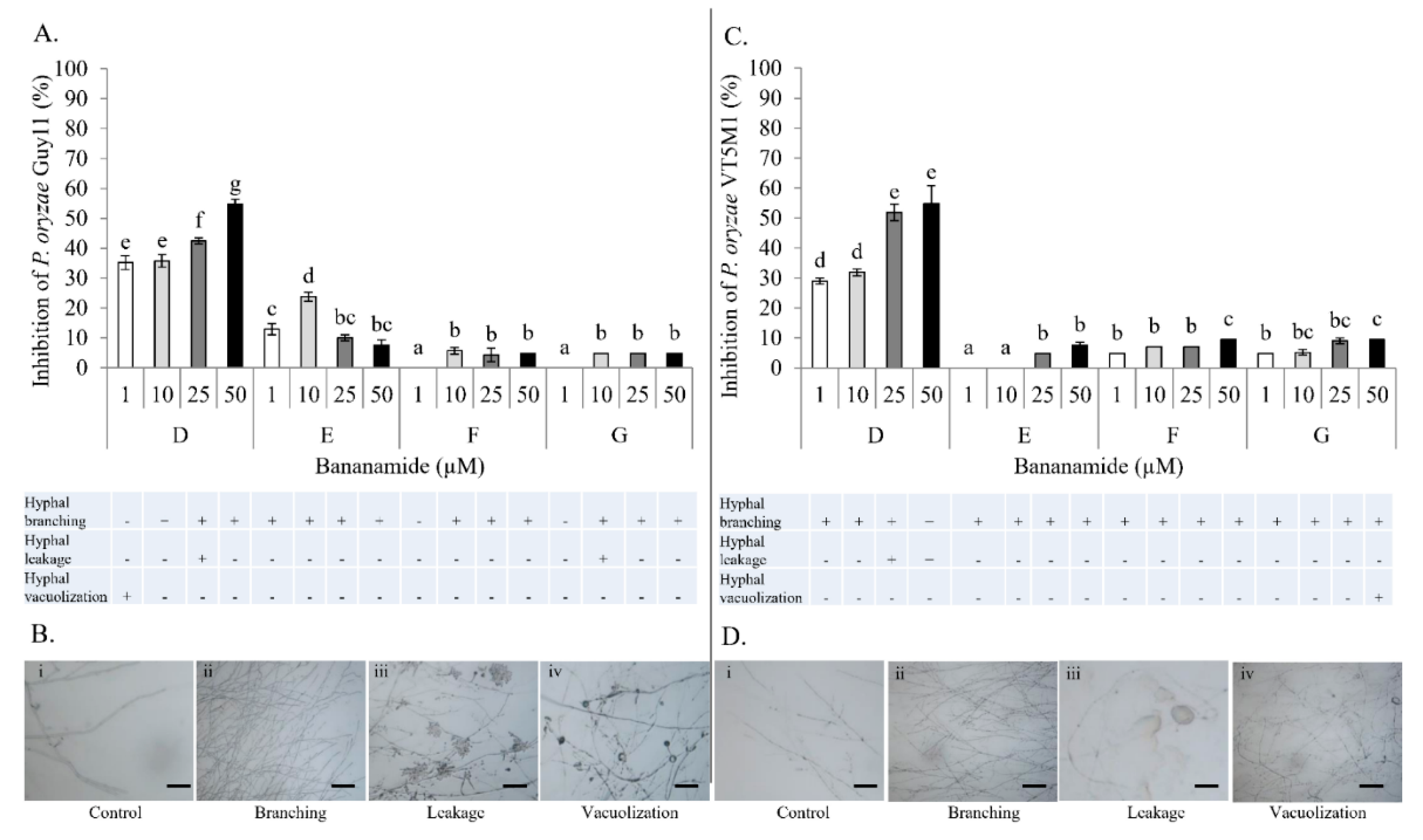
Our result showed that pure bananamide D, E, F and G produced by
Pseudomonas sp. COW3 displayed antagonistic activity against
P. myriotylum CMR1 which could be attributed to the detrimental effect of CLPs on the oomycete. Also, microscopic analysis revealed the capacity of bananamide D-G to cause hyphal branching in
P. myriotyum CMR1. It is known that CLPs act by compromising the membrane integrity of phytopathogens resulting in anti-oomycete activities, among others [
4,
19,
44]. This is in agreement with the observation of hyphal branching in
P. myriotylum NGR03 due to purified xantholysin, putisolvin, entolysin and white line inducing principle (WLIP) [
9]. Similar results of hyphal branching were observed with
R. solani AG 4-HGI and AG2-1, in interaction with orfamides [
3,
6], and viscosinamide-mediated branching in
R. solani [
4,
45]. Furthermore, structure-function activity was also displayed in the antagonism of purified bananamide D-G against
P. oryzae VT5M1 and GUY11. Treatments with bananamide D resulted in higher inhibition zones in both genetically different isolates, in a dose dependent manner. In comparison with other minor variants, bananamide E appeared to have a higher inhibitory efficacy on
P. oryzae at 10 µM. In addition to hyphal branching by the CLP variants, variable concentrations of bananamide variants produced by COW3 displayed hyphal vacuolization. It has been shown that increase in the number of carbon atoms in β-amino fatty acid chain of iturin-type lipopeptides produced by
Bacillus amyloliquefaciens SD-32, enhanced biological activity against fungal pathogens [
46]. More so, a decrease in defense-inducing activity was observed after the metabolic engineering of
Bacillus lipopeptides involving various substitutions of Val/Leu, Leu/Val, Leu/Ile and Val/Ile in the peptide moiety. Reduction in the length of fatty acid chains of these lipopeptides from 14 carbons, to 12 or 13 carbons led to loss of activity [
47]. However, the substitution of valine with isoleucine at position 4, and difference in the length of fatty acid chain had no effect on the activity of orfamides on
Rhizoctonia solani AG 4-HGI [
6]. Thus, our findings corroborate earlier reports by [
46,
47], that structural changes/differences in a lipopeptide could influence its biological activity. Furthermore, our result showed that bananamide E, which has a 3OH-C12:0 lipid tail, caused hyphal curling in
P. myriotylum CMR1 unlike bananamide D, F and G. Bananamide G with valine at position eight triggered vacuolization in
P. oryzae VT5M1 mycelia. Bananamide D with Ile8 displayed higher inhibition and mycelial distortion in comparison to bananamide G with the same fatty acid tail (3OH - C12:1), but Val8. Based on this, we therefore posit that specific amino acids at certain position(s) and length of fatty acids could influence the structural-function activity of
Pseudomonas-derived CLPs.
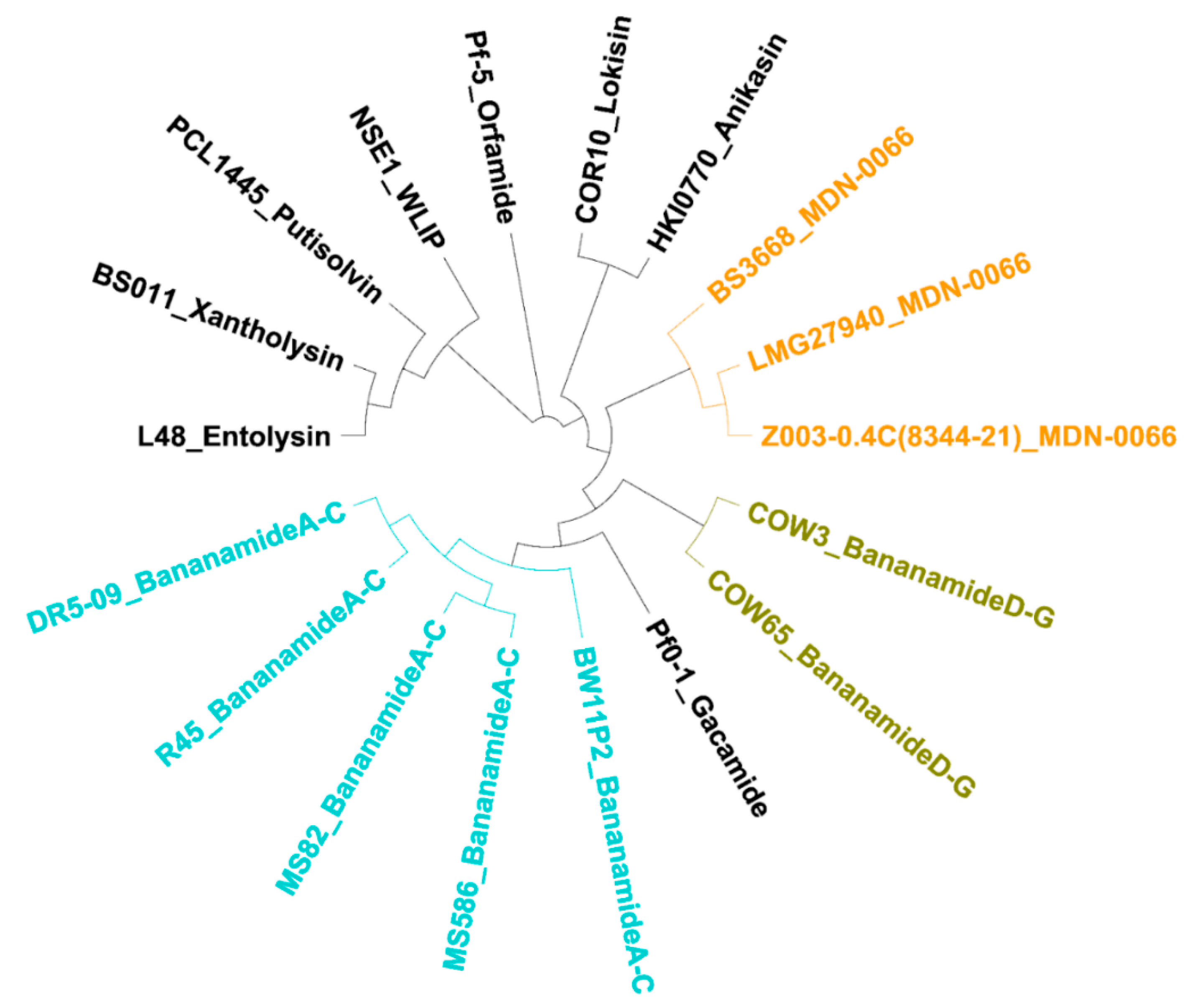
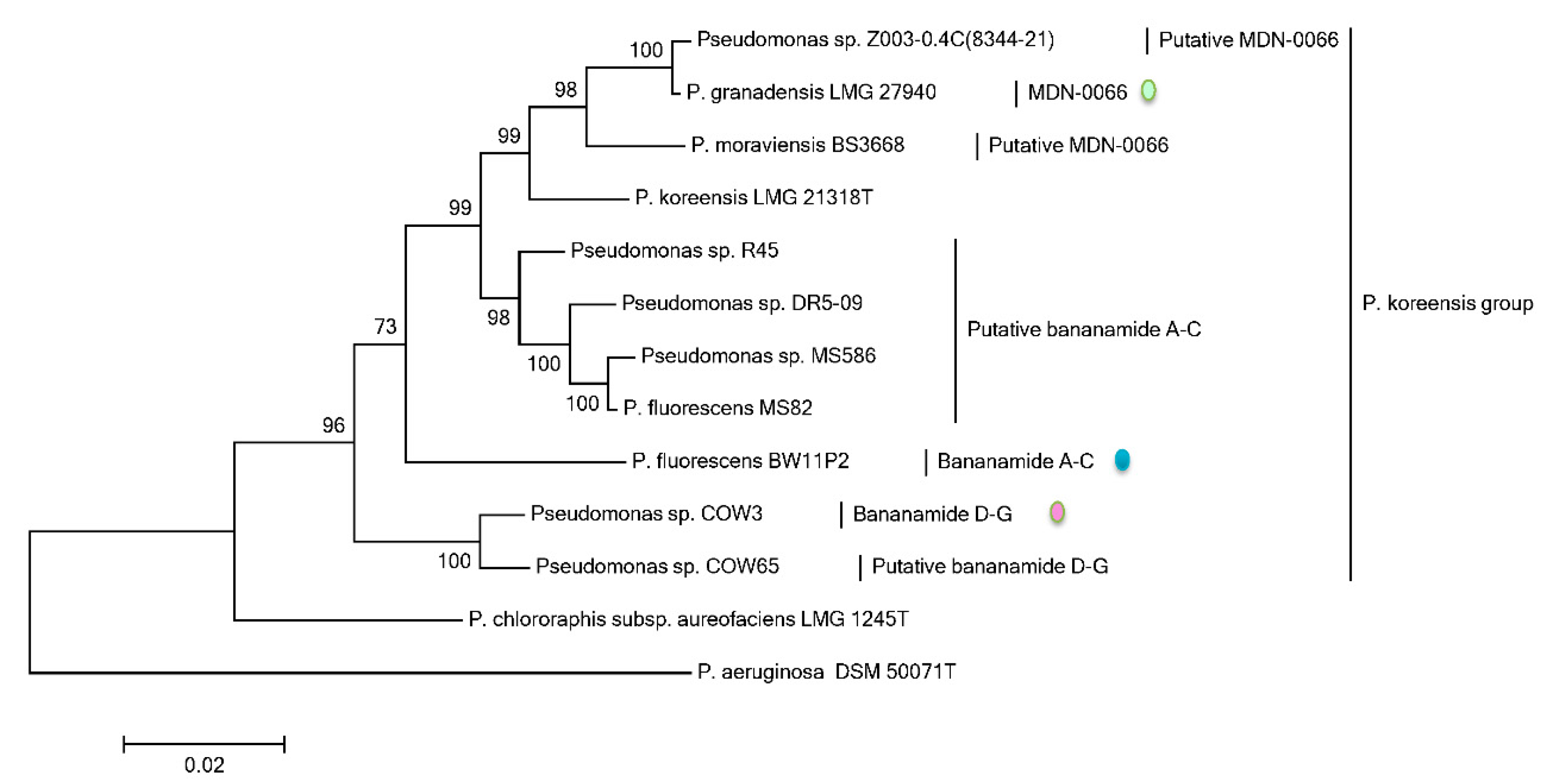
Phylogenetic MLSA analyses using
rpoD, gyrB, 16S rDNA and
rpoB partial sequences further positioned all bananamide-producers and (novel) derivatives in the
P. fluorescens complex - a conglomerate group comprising nine separate groups whose members are characterized by enormous secondary metabolite diversity including CLPs [
48,
49]. Specifically, the bananamide producers are situated in the
P. koreensis group - a group which also situates strains producing CLPs such as lokisin, amphisin, cocoyamide and other amphisin group members. Thus, the discovery of bananamide D-G and putative bananamide producers further expands the metabolic reputation of the
P. koreensis group and at large, that of the
P. fluorescens complex. The result of the phylogenetic analysis of the housekeeping genes is in perfect agreement with the phylogenetic analysis based on NRPS protein sequences and A domains and chemical analyses. Continuous genome mining of secondary metabolites will further unravel what novel chemistry nature has in store.
The diverse origins of bananamide-like producers are intriguing. Bananamide producers appear to be principally associated with the rhizosphere of plants in the tropical regions of the world which are characterized by high humidity (rainy season) and dry periods (dry season).
Pseudomonas spp. COW3 and COW65 were isolated from the cocoyam rhizosphere in tropical rain forests of Cameroon [
9]. Moreover, the bananamide A-C producer,
P. fluorescens BW11P2, was isolated from the banana rhizoplane in the tropical wetlands of Galgadera, Sri Lanka [
34,
50], while the putative bananamide-producing strain
Pseudomonas sp. R45 was isolated from the sugar cane rhizosphere soil sample in Piracicaba-SP, Brazil [
51,
52]. Other putative-bananamide producing strains
Pseudomonas spp. DR 5-09, MS586 and MS82 were isolated from plant roots at Daecheong, South Korea, a cotton field in the USA and from soybean roots in Mississippi, USA, respectively. The MDN-0066 producer
P. granadensis F-278,770T originates from a soil sample obtained from the Tejeda, Almijara and Alhama Natural Park, Granada, Spain [
39,
53]. In contrast, the two putative MDN-0066 producers, Z003-0.4C(8344-21) and BS3668 appear to be associated with entirely different ecologies. The isolation metadata on the Genomes Online Database reports that Z003-0.4C(8344-21) is a human-associated strain which was collected from the human airways (respiratory system); BS3668 was isolated from a waste treatment plant in Idaho, USA. The occurrence of these CLPs in different hosts and diverse ecologies suggest that these compounds can demonstrate various ecological roles.
4. Materials and Methods
4.1. Strains, Media and Growth Conditions
Pseudomonas and
P. oryzae strains used in this study are presented in
Table 3.
Pseudomonas strains were cultured on King’s B (KB) agar [
54] at 28 °C.
P. myriotylum CMR1 [
55] was grown on glucose casamino acid and yeast extract agar (GCY) [
56].
P. oryzae isolates VT5M1 [
57] and Guy11 [
58] were grown on complete medium (CM) [
59], at 28 °C for five days.
4.2. Extraction of Crude CLPs from Pseudomonas sp. COW3
Crude CLPs were extracted from
Pseudomonas sp. COW3 using an established protocol [
7,
9].
4.3. Crude CLP Analysis of COW3 via the Kendrick Mass Defect Method for Molecular Formula Assignment
Crude CLP extract from Pseudomonas sp. COW3 was re-solubilized in methanol and directly injected at 15 µL/min in a SYNAPT-G2-Si mass spectrometer (Waters, Manchester, UK), operating in positive and sensitivity modes. The sample was electrosprayed at an applied voltage of 3 kV, using a desolvation gas (N2) flow of 500 L/h, a nebuliser gas flow of 6.5 bar and desolvation temperatures of 150 and 300 °C, respectively. The full MS survey scans were acquired at a 20,000 (FWHM) resolving power, over the mass range of 200–2000 m/z. Peaks were analyzed using Mass Lynx software (ver.4.1; Waters). Mass spectra were externally calibrated in positive mode using a solution of 5 mM sodium formate (Sigma-Aldrich, Saint-Quentin-Fallavier, France) in 50/50 (v/v) acetonitrile (ACN)/H2O.
4.4. Molecular Formula Assignment with Kendrick Mass Defect
The Kendrick mass (KM) related to the CH
2 pattern is calculated from the molecular formula of NORINE-referenced compounds and from the experimentally measured masses. A dealiasing, which corresponds to a mathematical shift that leads to the mass to fit the spectral width without aliasing, was performed by shifting the KM value by −0.28, equation according to [
38]:
Subsequently, the dealiased KM value is rounded to the nearest whole number and defines the nominal Kendrick mass (NKM), equation:
The NKM is then subtracted from Kendrick mass to obtain the regular Kendrick mass defect (RKMD), equation:
RKMD and the NKM values were calculated for each known and curated molecular formulae extracted from the NORINE database. Subsequently, a RKMD/NKM 2D-plot (without aliasing) was generated using these values. A vectorial mesh enables the assignment of the molecular formulae and a web application (named Kendrick Formula Predictor) implementing the method can be run on-line with any list of mass-to-charge ratios at the following URL:
http://bioinfo.cristal.univ-lille.fr/kendrick-webapp/.
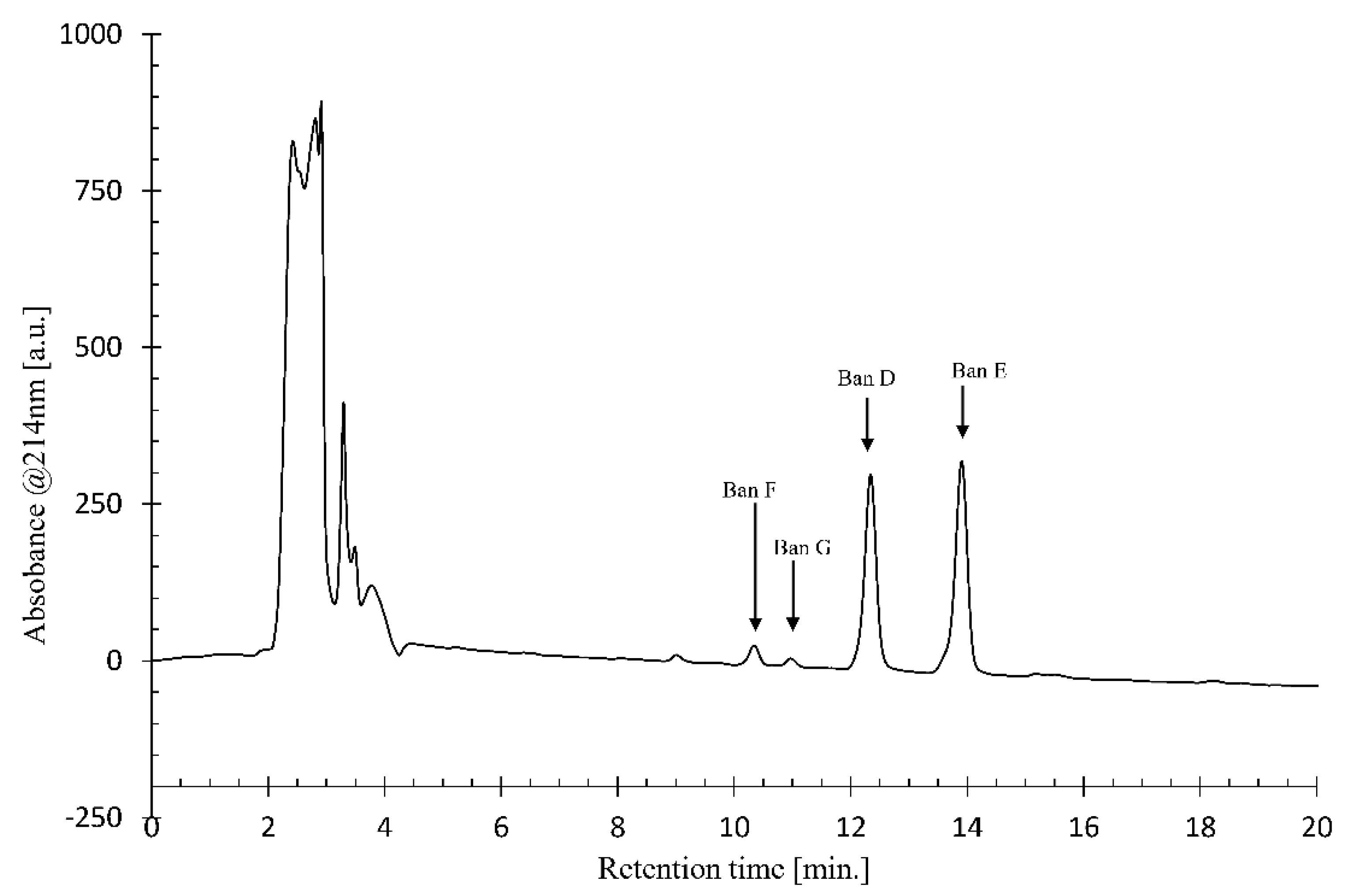
4.5. Analytical and Preparative Liquid Chromatography (LC) from Crude Extract
Using crude CLP extract from Pseudomonas sp. COW3, analytical LC-MS data were collected on a 1100 Series HPLC (Agilent Technologies, Santa Clara, CA, United States) equipped with an ESI ionisation source, coupled to a type SL MS detector. The HPLC was equipped with an analytical Kinetex C18 reversed-phase column (150 × 4.6 mm, 5 μm particle size; Phenomenex, Torrance, CA, USA). An elution gradient of H2O/CH3CN (25:75 to 0:100, v/v) over 20 min was applied at a flow rate of 1 mL.min-1. The crude CLP extract was dissolved in methanol and the purification conditions of each CLP were optimized by using an Agilent Technologies 1100 Series HPLC device equipped with a Luna C-18 analytical reversed phase HPLC column (250 × 4.6 mm, 5 μm). The signal was detected using a diode array detector at a wavelength of 214 nm. Different gradient elutions of H2O/CH3CN were tested to find the optimal separation conditions while the column temperature was kept at 35°C. Large scale purification of CLPs was subsequently performed by injection of the methanol solution into a Prostar HPLC device (Agilent Technologies) equipped with a Luna C-18(2) preparative RP-HPLC column (250 × 21.2 mm, 5 μm particle size) for separation of the individual CLP analogues. The optimal elution gradient (25:75 to 0:100, v/v) of H2O/CH3CN was applied over 20 min at a flow rate of 17.5 mL.min-1, while the column temperature was kept at 35 °C.
4.6. Chemical Characterization of Novel Bananamide Derivatives via Nuclear Magnetic Resonance (NMR)
All NMR measurements were performed on an Avance III spectrometer (Bruker, (Billerica, MA, USA) operating at a respective 1H and 13C frequency of 500.13 MHz and 125.76 MHz equipped with a BBI-Z probe. The sample temperature was set to 298.0 K. Standard pulse sequences as present in the Bruker library were consistently used unless otherwise stated. Spectra that were used for NMR assignment included 1H-1H COSY (cosygpqf), 1H-1H TOCSY (mlevph), 1H-1H NOESY (noesygpph),1H-1H ROESY (offroesygpprphpp), 1H-13C HSQC (hsqcedetgpsisp2.4) and 1H-13C HMBC (hmbcgplpndqf). High precision 5 mm NMR tubes (Norell, Landisville, NJ, USA) were used. Dimethylformamide-d7 (DMF) (99.50%) was purchased from Eurisotop (Saint-Aubin, France). 1H and 13C chemical shift scales were calibrated by using the residual solvent signal employing Tetramethylsilane (TMS) as secondary reference.
2D spectra measured for structure elucidation include a 2D 1H-1H DQF-COSY, 2D 1H - 1H TOCSY with a 90 ms MLEV-17 spinlock, 2D 1H-1H off-resonance ROESYs with 200 ms mixing time and gradient-selected 1H-13C gHSQC and gHMBC. Typically, 2048 data points were sampled in the direct dimension for 512 data points in the indirect dimension, with the spectral width set to 11 ppm and 110 ppm along the 1H and 13C dimension, respectively. The 1H-13C HMBC was measured with a 210 ppm 13C spectral width. For 2D processing, the spectra were zero filled to a 2048 × 2048 real data matrix. Prior to Fourier transformation, all spectra were multiplied with a squared cosine bell function in both dimensions or sine bell in the direct dimension for the gHMBC.
4.7. Genome Sequencing and Assembly
For genome sequencing,
Pseudomonas sp. COW3 (N3 producer) and COW65 (a putative N3 producer, [
9]), were grown in Luria Bertani (LB) broth for 24 h at 28 °C with continuous shaking at 150 rpm. Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA) according to manufacturer’s instructions.
Single-end or paired-end sequence reads were generated using the Illumina HiSeq2500 or MiSeq system at the BASECLEAR B. V. (Leiden, The Netherlands). FASTQ read sequence files were generated using bcl2fastq2 version 2.18. Initial quality assessment was based on data passing the Illumina Chastity filtering. Subsequently, reads containing PhiX control signal were removed using an in-house filtering protocol. In addition, reads containing (partial) adapters were clipped (up to a minimum read length of 50 bp). The second quality assessment was based on the remaining reads using the FASTQC quality control tool version 0.11.5 (Babraham Institute, Cambridge, UK)
The quality of Illumina reads was improved using the error correction tool BayesHammer [
60]. Error-corrected reads were assembled into contigs using SPAdes version 3.10 [
61]. The order of contigs, and the distances between them, was estimated using the insert size information derived from an alignment of the paired-end reads to the draft assembly. Consequently, contigs were linked together and placed into scaffolds using SSPACE version 2.3 [
62]. Using Illumina reads, gapped regions within scaffolds were (partially) closed using GapFiller version 1.10 [
63]. Finally, assembly errors and the nucleotide disagreements between the Illumina reads and scaffold sequences were corrected using Pilon version 1.21 [
64].
4.8. Genome Annotation, Genome Mining and Bioinformatics Analyses
Genome sequences were automatically annotated using the RAST annotation pipeline [
65,
66]. Furthermore, genomes of previously sequenced strains (
Supplementary Table S1) were re-annotated using the RAST annotation pipeline and also submitted to antiSMASH v5.0 [
67]. Genome mining was conducted on the annotated genomes, and comparison of NRPS proteins with other protein sequences in GenBank database was done by BLAST search (
https://blast.ncbi.nlm.nih.gov/Blast.cgi). The adenylation (A) and condensation (C) domains sequences of the non-ribosomal peptide synthase (NRPS) genes were extracted. Sequence alignment was carried out using MUSCLE [
68] in the software package Geneious Prime, 2019, and the cladograms were inferred by Neighbour Joining. The thioesterase (TE) domain sequences of the non-ribosomal peptide synthase (NRPS) gene were extracted. Sequence alignment was carried out using MUSCLE [
68], and the phylogenetic tree was constructed by maximum likelihood with 1000 bootstrap replication, in the software package MEGA6 [
69]. The antiSMASH v5.0 enabled the prediction of the amino acid composition of the peptide moiety. Moreover, proteins were translated from the
nrps and flanking genes of bananamide D-G-and other CLP-producing strains (
Supplementary Table S1). The protein sequences were concatenated, aligned and used to construct a phylogenetic tree in Geneious Prime, 2019 (Biomatters, Auckland, New Zealand).
4.9. Gap Filling of NRPS Scaffold Sequences in Pseudomonas sp. COW3
Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit from Promega. The region of interest was amplified by PCR using primers: COW3G_F (GTTCGGTTTCGATGCGATGG) and COW3G_R (GATTTCCATTTCGCCGACCG) designed with Geneious 11.1.5 software (
https://www.geneious.com). PCR reaction was carried in a 50 µL mixture containing 10 µL 5× Go Taq Reaction buffer, 0.2 µL Go Taq DNA Polymerase (Promega), 1 µL of 10 mM dNTP mixture, 2 µL of 10 µM COW3G_F primer, 2 µL of 10 µM COW3G_R primer, 2 µL genomic DNA, and 32.8 µL double-distilled water. Amplification was performed using a FlexCycler Block Analytikjena 2010 with the following program: 94 °C for 2 min, 30 cycles of 94 °C for 30 s, 65 °C for 1 min, 72 °C for 1 min, and extension at 72 °C for 10 min, and finally kept at 4 °C. Amplicons were visualized by 1.5% gel electrophoresis after staining with ethidium bromide. Purified PCR product was sequenced at LGC Genomics GmbH (Berlin, Germany).
4.10. Phylogenetic Analyses of Pseudomonas Strains
For multi-locus sequence analyses (MLSA),
rpoD, rpoB, 16S rDNA and
gyrB sequences of our bananamide D-G-producing
Pseudomonas strains (COW3 and COW65) were extracted from their corresponding draft genome sequences. In addition, draft genomes of putative bananamide-producing strains, and sequences of selected
Pseudomonas type strains were retrieved from GenBank (
Table A5). The sequences were aligned using MUSCLE [
68] in MEGA6 [
69]. A phylogeny tree was constructed by maximum likelihood with 1000 bootstrap replication, and
P. aeruginosa was used as outgroup.
rpoD, rpoB, 16S rDNA and
gyrB trees were generated separately after which a concatenated tree was obtained by combining the aligned sequences of the four genes.
4.11. In Vitro Direct Antagonism of Pseudomonas sp. COW3 against P. myriotylum CMR1
In vitro antagonism of the bananamide-producing strain COW3, against the soil-borne oomycete
P. myriotylum CMR1, which causes the cocoyam root rot disease, was investigated by Petri dish assay, as described by [
70]. Three (3) µL of overnight King B [
54] broth culture of the test bacteria were spotted on either side of GCY agar plates spanning 2 cm from the center. The plates were incubated for 24 h at 25 °C, after which a mycelium plug of CMR1 (5 mm in diameter) was placed at the center of the plate. The plates were incubated at 28 °C, and pictures were taken after two and four days.
4.12. In Vitro Antagonism of Pseudomonas sp. COW3 against P. oryzae
The Petri dish assay was used to investigate the in vitro antagonism of COW3, against two genetically different isolates of the rice blast pathogen,
P. oryzae VT5M1 and
P. oryzae Guy11, as described by [
70]. Three (3) µL of overnight LB broth culture of the test bacteria were spotted on either side of complete medium (CM) plates spanning 2 cm from the center. The plates were incubated for 24 h at 25 °C, after which a mycelium plug of VT5M1 or Guy11 (5 mm in diameter) was placed at the center of the plate. Plates were re-incubated at 25 °C for 4 to 10 days.
4.13. In Vitro Microscopic Inhibition of P. myriotylum, and P. oryzae using Purified Bananamides D-G
The antagonistic activity of purified bananamides D-G was studied, in interaction with
P. myriotylum CMR1, and
P. oryzae VT5M1 and Guy11. The experiment was conducted in in vitro microscopic conditions as described by [
9], with minor modifications. Sterile microscopic glass slides were covered with a thin film of water agar (Bacto agar; Difco) and placed in a plastic Petri dish containing sterile filter paper moistened with 2 mL of sterile distilled water. A single agar plug (diameter = 5 mm) of
P. myriotylum CMR1 and
P. oryzae isolates (VT5M1 and Guy11) was obtained from a five day old culture grown on GCY agar and CM agar, respectively. Plugs were inoculated at the center of each glass slide. Stock solutions (5 mM) of bananamides D, E, F and G were initially made by dilution with DMSO while subsequent dilutions were made using sterile MilliQ water. Fifteen (15) µL of each bananamide CLP solution at concentration 1 µM, 10 µM, 25 µM and 50 µM, was added separately at either side of the glass slide (2 cm from the fungal plug).
P. myriotylum CMR1 assay was incubated for 3 days, while
P. oryzae plates were incubated for six days at 28 °C. Microscopic slides were assessed for hyphal distortion phenotypes under an Olympus BX51 microscope. The efficacy of treatments of each bananamide variant was evaluated by measurement of mycelial growth diameter compared with those obtained with the
P. myriotylum and
P. oryzae control. Percentage inhibition of the fungal pathogens by bananamide variants and for each concentration was calculated relative to the mycelial growth in the control. Percentage inhibition was calculated according to the following formula:
Data obtained were analyzed using SPSS 25 statistical software. To compare across treatments, univariate ANOVA followed by Tukey’s posthoc tests, were used. Results were considered to be statistically different when p < 0.05. Figures were generated to represent the percentage inhibition of the fungi. Representative pictures of mycelial damage due to CLP-pathogen interaction are shown.