Bimodular Antiparallel G-Quadruplex Nanoconstruct with Antiproliferative Activity
Abstract
:1. Introduction
2. Results
2.1. Oligonucleotide Design in Brief
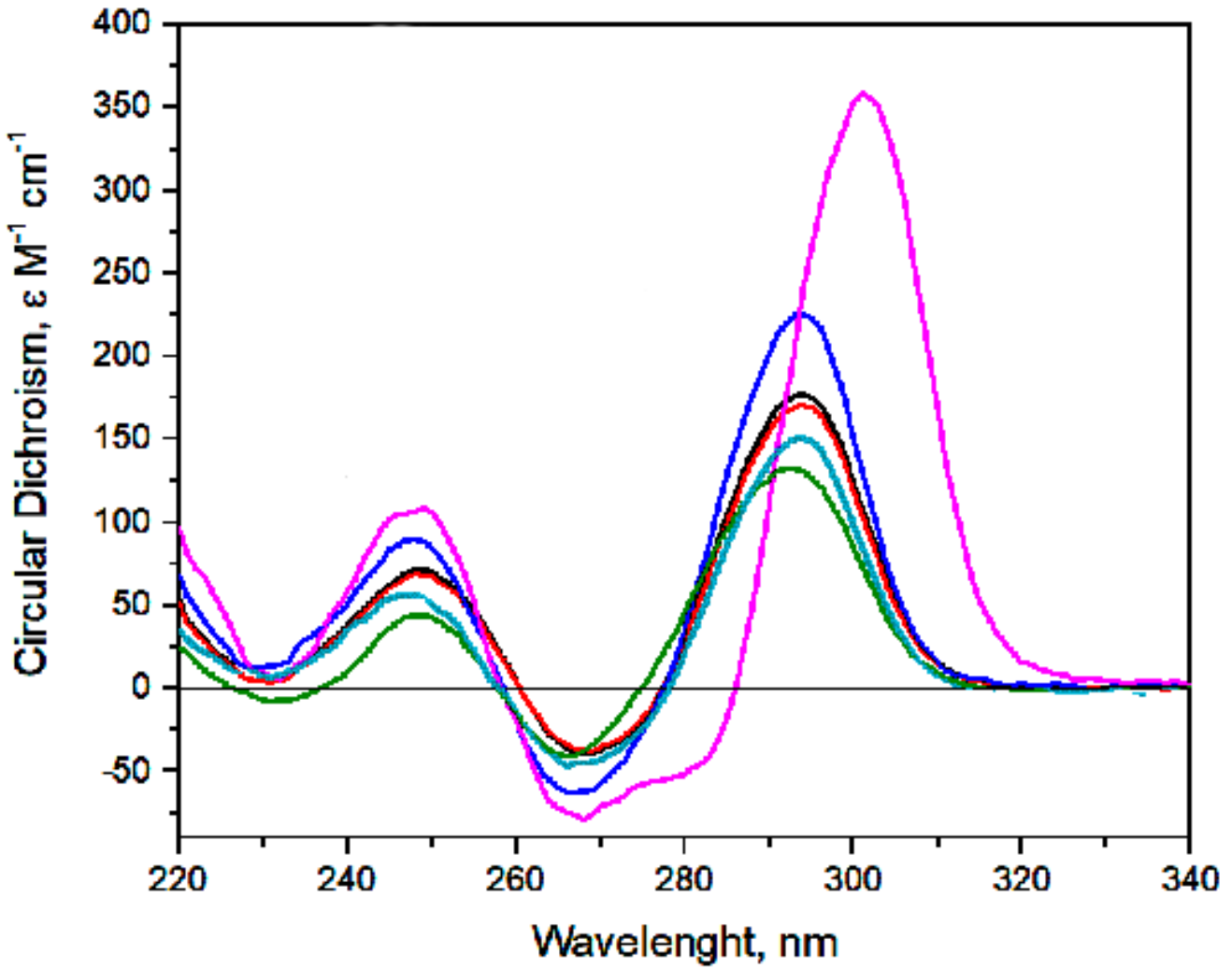
2.2. Monitoring of biHD1 and Derivatives Conformations
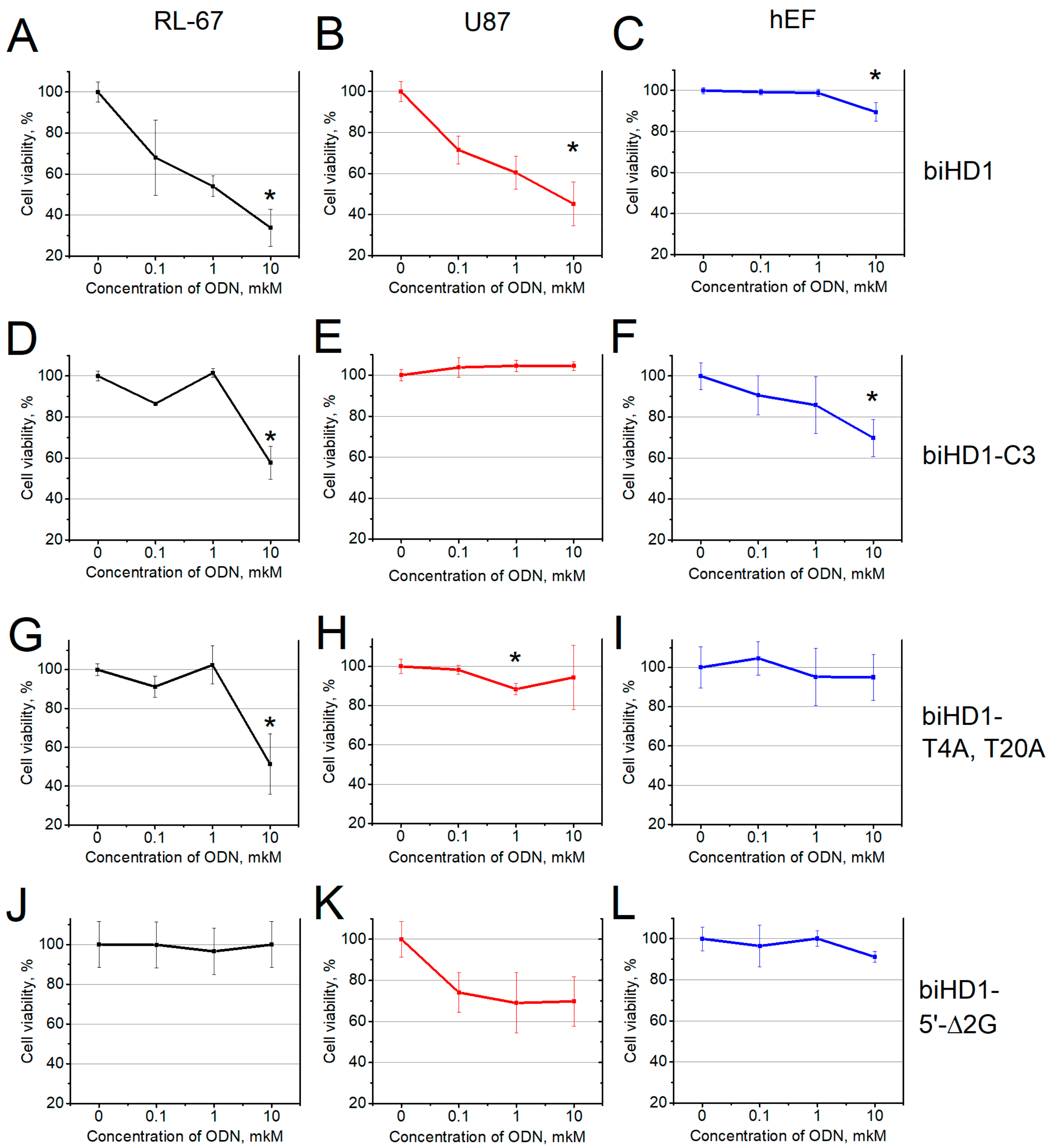
2.3. Antiproliferative Activity
2.3.1. Antiproliferative Activity of biHD1
2.3.2. Antiproliferative Activity of biHD1 Derivatives
3. Discussion
BiHD1
BiHD1-C3
HD1
BiHD1-T4A,T20A
BiHD1-5’-Δ2G
BiHD1+Ba
4. Materials and Methods
4.1. Oligodeoxyribonucleotides (ODN)
4.2. Circular Dichroism Spectroscopy
4.3. Cell Lines and MTT Assay
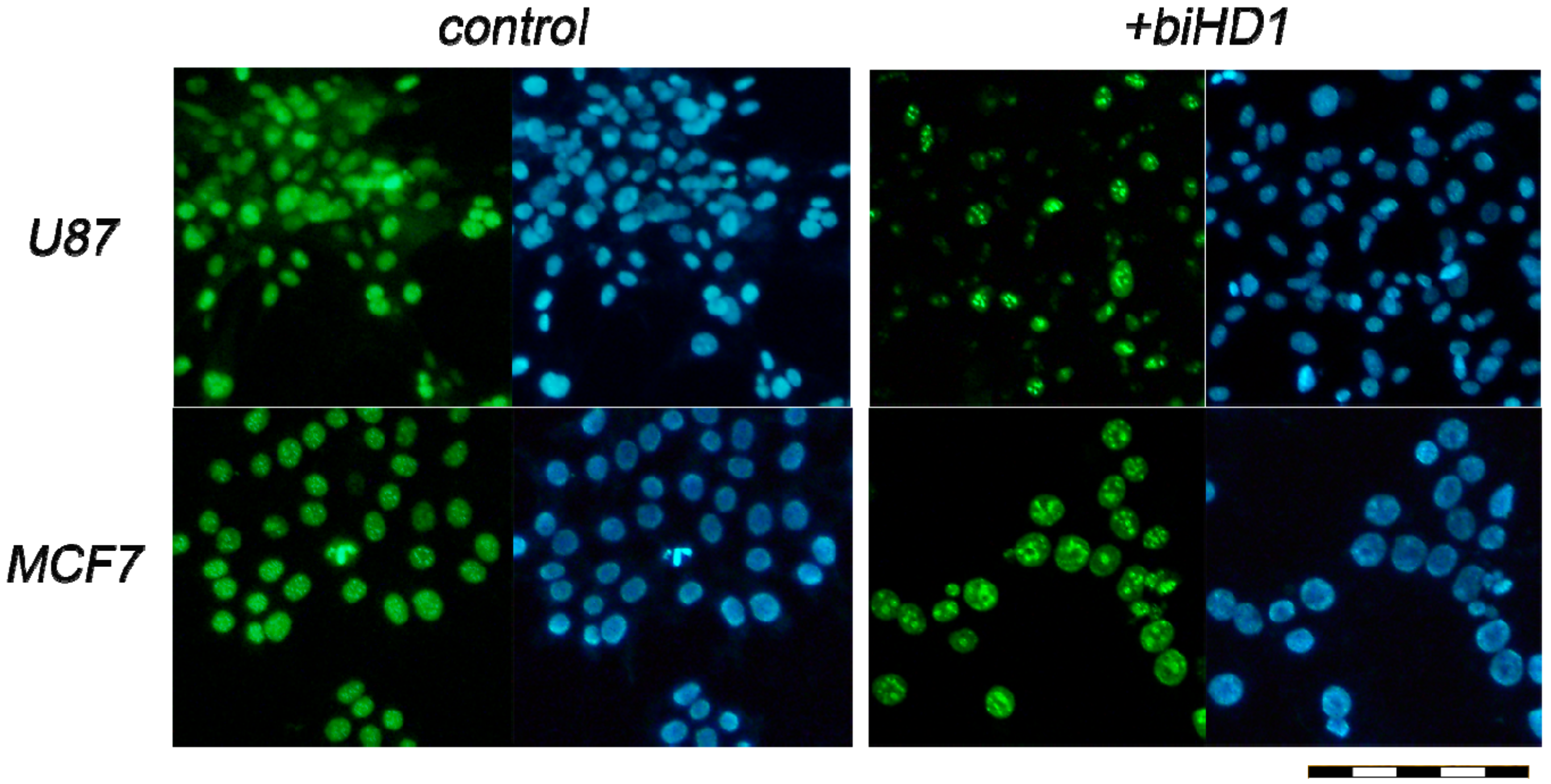
4.4. Immunohistochemistry
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bates, P.J.; Reyes-Reyes, E.M.; Malik, M.T.; Murphy, E.M.; O’Toole, M.G.; Trent, J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent, Uses and mechanisms. Biochim. Biophys. Acta Gen. Subj. 2017, 861, 1414–1428.2. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.; Qi, C.; Meng, J.; Zhang, N.; Bing, T.; Yang, X.; Cao, Z.; Shangguan, D. General cell-binding activity of intramolecular G-quadruplexes with parallel structure. PLoS ONE 2013, 8, e62348. [Google Scholar] [CrossRef] [PubMed]
- Ogloblina, A.M.; Khristich, A.N.; Karpechenko, N.Y.; Semina, S.E.; Belitsky, G.A.; Dolinnaya, N.G.; Yakubovskaya, M.G. Multi-targeted effects of G4-aptamers and their antiproliferative activity against cancer cells. Biochimie 2018, 145, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.J.; Laber, D.A.; Miller, D.M.; Thomas, S.D.; Trent, J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009, 86, 151–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N., Jr.; Harzstark, A.L.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; Choueiri, T.; et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest. New Drugs 2014, 32, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Niu, N.; Wei, T.; Tozawa, H.; Chen, X.; Zhang, C.; Zhang, J.; Wada, Y.; Kapron, C.M.; Liu, J. The roles of signal transducer and activator of transcription factor 3 in tumor angiogenesis. Oncotarget 2017, 8, 69139–69161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, N.; Li, Y.; Xu, X.; Sha, W.; Li, P.; Feng, L.; Tweardy, D.J. Targeting Stat3 with G-Quartet Oligodeoxynucleotides in Human Cancer Cells. DNA Cell Biol. 2003, 22, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Li, D.; Sung, C.K.; Yim, H.; Troke, P.; Benjamin, T. DNA-binding and regulatory properties of the transcription factor and putative tumor suppressor p150Sal2. Biochim. Biophys. Acta Gene Regul. Mech. 2015, 1809, 276–283. [Google Scholar] [CrossRef]
- Hu, J.; Zhao, Z.; Liu, Q.; Ye, M.; Hu, B.; Wang, J.; Tan, W. Study of the Function of G-Rich Aptamers Selected for Lung Adenocarcinoma. Chem. Asian J. 2015, 10, 1519–1525. [Google Scholar] [CrossRef]
- Nonaka, Y.; Sode, K.; Ikebukuro, K. Screening and Improvement of an Anti-VEGF DNA Aptamer. Molecules 2010, 15, 215. [Google Scholar] [CrossRef]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Dapić, V.; Abdomerović, V.; Marrington, R.; Peberdy, J.; Rodger, A.; Trent, J.O.; Bates, P.J. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003, 31, 2097–2107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trajkovski, M.; Sket, P.; Plavec, J. Cation localization and movement within DNA thrombin binding aptamer in solution. Org. Biomol. Chem. 2009, 7, 4677–4684. [Google Scholar] [CrossRef]
- Russo Krauss, I.; Merlino, A.; Randazzo, A.; Novellino, E.; Mazzarella, L.; Sica, F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012, 40, 8119–8128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotkowiak, W.; Lisowiec-Wachnicka, J.; Grynda, J.; Kierzek, R.; Wengel., J.; Pasternak, A. Thermodynamic, Anticoagulant, and Antiproliferative Properties of Thrombin Binding Aptamer Containing Novel UNA Derivative. Mol. Ther. Nucleic Acids 2018. [Google Scholar] [CrossRef] [PubMed]
- Scuotto, M.; Rivieccio, E.; Varone, A.; Corda, D.; Bucci, M.; Vellecco, V.; Cirino, G.; Virgilio, A.; Esposito, V.; Galeone, A.; et al. Site specific replacements of a single loop nucleoside with a dibenzyl linker may switch the activity of TBA from anticoagulant to antiproliferative. Nucleic Acids Res. 2015, 43, 7702–7716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esposito, V.; Russo, A.; Amato, T.; Varra, M.; Vellecco, V.; Bucci, M.; Russo, G.; Virgilio, A.; Galeone, A. Backbone modified TBA analogues endowed with antiproliferative activity. Biochim. Biophys. Acta Gen. Subj. 2018, 1861, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Esposito, V.; Russo, A.; Amato, T.; Vellecco, V.; Bucci, M.; Mayol, L.; Russo, G.; Virgilio, A.; Galeone, A. The “Janus face” of the thrombin binding aptamer: Investigating the anticoagulant and antiproliferative properties through straightforward chemical modifications. Bioorg. Chem. 2018, 76, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Zavyalova, E.; Kopylov, A. DNA aptamer-based molecular nanoconstructions and nanodevices for diagnostics and therapy. In Nanostructures for the Engineering of Cells, Tissues and Organs; Grumezescu, A.M., Ed.; William Andrew Publishing: New York, NY, USA, 2018; pp. 249–290. [Google Scholar]
- Zavyalova, E.G.; Legatova, V.A.; Alieva, R.S.; Zalevsky, A.O.; Tashlitsky, V.N.; Arutyunyan, A.M.; Kopylov, A.M. Putative mechanisms underlying high inhibitory activities of bimodular DNA aptamers to Thrombin. Biomolecules 2019, 9, E41. [Google Scholar] [CrossRef] [PubMed]
- Kortylewski, M.; Nechaev, S. How to Train Your Dragon, Targeted Delivery of MicroRNA to Cancer Cells In Vivo. Mol. Ther. 2014, 22, 1070–1071. [Google Scholar] [CrossRef] [PubMed]
- Soldevilla, M.M.; Meraviglia-Crivelli de Caso, D.; Menon, A.P.; Pastor, F. Aptamer-iRNAs as Therapeutics for Cancer Treatment. Pharmaceuticals (Basel) 2018, 11, E108. [Google Scholar] [CrossRef] [PubMed]
- Golovin, A.V.; Kopylov, A.M.; Reshetnikov, R.V.; Zavyalova, E.G.; Pavlova, G.V.; Babiy, V.E. Anti-Thrombosis Aptamers and Method for Stabilizing the Structure Thereof. WO2011075004A1, 23 June 2011. [Google Scholar]
- Zavyalova, E.; Samoylenkova, N.; Revishchin, A.; Turashev, A.; Gordeychuk, I.; Golovin, A.; Kopylov, A.; Pavlova, G. The Evaluation of Pharmacodynamics and Pharmacokinetics of Anti-thrombin DNA Aptamer RA-36. Front. Pharmacol. 2017, 8, 922. [Google Scholar] [CrossRef] [Green Version]
- Zavyalova, E.; Tagiltsev, G.; Reshetnikov, R.; Arutyunyan, A.; Kopylov, A. Cation Coordination Alters the Conformation of a Thrombin-Binding G-Quadruplex DNA Aptamer That Affects Inhibition of Thrombin. Nucleic Acid Ther. 2016, 26, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Yuminova, A.V.; Smirnova, I.G.; Arutyunyan, A.M.; Kopylov, A.M.; Golovin, A.V. The structure of G-quadruplex thrombin binding DNA-aptamer RA-36. Moscow Univ. Chem. Bull. 2015, 56, 44–48. [Google Scholar]
- Amato, T.; Virgilio, A.; Pirone, L.; Vellecco, V.; Bucci, M.; Pedone, E.; Esposito, V.; Galeone, A. Investigating the properties of TBA variants with twin thrombin binding domains. Sci. Rep. 2019, 9, 9184. [Google Scholar] [CrossRef] [PubMed]
- Zavyalova, E.; Golovin, A.; Pavlova, G.; Kopylov, A. Module-activity relationship of G-quadruplex based DNA aptamers for human thrombin. Curr. Med. Chem. 2013, 20, 4836–4843. [Google Scholar] [CrossRef] [PubMed]
- Reshetnikov, R.V.; Sponer, J.; Rassokhina, O.I.; Kopylov, A.M.; Tsvetkov, P.O.; Makarov, A.A.; Golovin, A.V. Cation binding to 15-TBA quadruplex DNA is a multiple-pathway cation-dependent process. Nucleic Acids Res. 2011, 39, 9789–9802. [Google Scholar] [CrossRef] [Green Version]
- Newsholme, P.; Adogu, A.A.; Soos, M.A.; Hales, C.N. Complement-induced Ca2+ influx in cultured fibroblasts is decreased by the calcium-channel antagonist nifedipine or by some bivalent inorganic cations. Biochem. J. 1993, 295, 773–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-Moreno, L.; Arenas, M.I.; Carmena, M.J.; Schally, A.V.; Sánchez-Chapado, M.; Prieto, J.C.; Bajo, A.M. Anti-proliferative and pro-apoptotic effects of GHRH antagonists in prostate cancer. Oncotarget 2016, 7, 52195–52206. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Kaufman, P.D. Ki-67: More than a proliferation marker. Chromosoma 2018, 127, 175–186. [Google Scholar] [CrossRef]
- Menon, S.S.; Guruvayoorappan, C.; Sakthivel, K.M.; Rasmi, R.R. Ki-67 protein as a tumour proliferation marker. Clin. Chim. Acta 2019, 491, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Bing, T.; Liu, X.; Qi, C.; Shen, L.; Wang, L.; Shangguan, D. Cytotoxicity of guanine-based degradation products contributes to the antiproliferative activity of guanine-rich oligonucleotides. Chem. Sci. 2015, 6, 3831–3838. [Google Scholar] [CrossRef] [PubMed]
- Cerofolini, L.; Amato, J.; Giachetti, A.; Limongelli, V.; Novellino, E.; Parrinello, M.; Fragai, M.; Randazzo, A.; Luchinat, C. G-triplex structure and formation propensity. Nucleic Acids Res. 2014, 42, 13393–13404. [Google Scholar] [CrossRef] [PubMed]
- Lopatina, K.A.; Razina, T.G.; Zueva, E.P.; Krylova, S.G.; Guryev, A.M.; Amosova, E.N.; Rybalkina, O.Y.; Safonova, E.A.; Efimova, L.A.; Belousov, M.V. Preclinical studies of α(1,2)-L-ramno-α(1,4)-D-galactopiranoziluronan from rhizomes Acorus calamus L. in cancer experiment. Sib. J. Oncol. 2015, 1, 59–63. [Google Scholar]
- Allen, M.; Bjerke, M.; Edlund, H.; Nelander, S.; Westermark, B. Origin of the U87MG glioma cell line, Good news and bad news. Sci. Transl. Med. 2016, 8, 354re3. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.R. Re-evaluation of HeLa, HeLa S3, and HEp-2 karyotypes. Cytogenet. Genome Res. 1988, 4, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Brattain, M.G.; Fine, W.D.; Khaled, F.M.; Thompson, J.; Brattain, D.E. Heterogeneity of malignant cells from a human colonic carcinoma. Cancer Res. 1981, 41, 1751–1756. [Google Scholar]
- Soule, H.D.; Vazguez, J.; Long, A.; Albert, S.; Brennan, M. A human cell line from a pleural effusion derived from a breast carcinoma. J. Nat. Cancer Inst. 1973, 51, 1409–1416. [Google Scholar] [CrossRef]
- Stromskaya, T.P.; Filippova, N.A.; Rybalkina, E.Yu.; Egudina, S.V.; Shtil, A.A.; Eliseenkova, A.V.; Stavrovskaya, A.A. Alterations of melanin synthesis in human melanoma cells selected in vitro for multidrug resistance. Exp. Toxicol. Pathol. 1995, 47, 157–166. [Google Scholar] [CrossRef]
- Chen, T.R. Chromosome identity of human prostate cancer cell lines, PC-3 and PPC-1. Cytogenet. Cell Genet. 1993, 62, 183–184. [Google Scholar] [CrossRef]
- Kaighn, M.E.; Narayan, K.S.; Ohnuki, Y.; Lechner, J.F.; Jones, L.W. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest. Urol. 1979, 17, 16–23. [Google Scholar] [PubMed]
- Amit, M.; Margulets, V.; Segev, H.; Shariki, K.; Laevsky, I.; Coleman, R.; Itskovitz-Eldor, J. Human Feeder Layers for Human Embryonic Stem Cells. Biol. Reprod. 2003, 68, 2150–2156. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |
| Number | Name | ODN Nucleotide Sequence |
|---|---|---|
| 1 | HD1 | GGTTGGTGTGGTTGG |
| 2 | biHD1 | GGTTGGTGTGGTTGGTGGTTGGTGTGGTTGG |
| 3 | biHD1-C3 | GGTTGGTGTGGTTGGprGGTTGGTGTGGTTGG |
| 4 | biHD1-T4A,T20A | GGTAGGTGTGGTTGGTGGTAGGTGTGGTTGG |
| 5 | biHD1-5′-∆2G | TTGGTGTGGTTGGTGGTTGGTGTGGTTGG |
| 6 | biHD1+Ba | GGTTGGTGTGGTTGGTGGTTGGTGTGGTTGG |
| Cell Line | Cell Viability, % | ||||
|---|---|---|---|---|---|
| biHD1 | biHD1-C3 | biHD1-T4A, T20 | biHD1-5′-∆2G | biHD1+Ba | |
| RL-67 | 34 | 58 | 51 | 100 | 103 |
| U87 | 45 | 104/103 * | 94 | 66 | 96 |
| HeLa | 57 | 70 | 91 | 74 | 86 |
| HCT116 | 58 | 96 | 105 | 74 | 100 |
| MCF7 | 64 | 90 | 102 | 85 | 87 |
| mS | 77 | 96 | 93 | 93 | 98 |
| PC3 | 78 | 131 | 91 | 85 | 76 |
| hEF | 90 | 70/87 * | 95 | 91 | 101 |
| Number | Name | Cell Line | Reference |
|---|---|---|---|
| 1 | RL-67 | Lung cancer | [36] |
| 2 | U87 | Central nervous system cancer | [37] |
| 3 | HeLa | Epithelioid cervix carcinoma | [38] |
| 4 | HCT116 | Colon cancer | [39] |
| 5 | MCF7 | Breast adenocarcinoma | [40] |
| 6 | mS | Melanoma | [41] |
| 7 | PC3 | Prostate cancer | [42,43] |
| 8 | hEF | Human embryonic fibroblasts | [44] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antipova, O.; Samoylenkova, N.; Savchenko, E.; Zavyalova, E.; Revishchin, A.; Pavlova, G.; Kopylov, A. Bimodular Antiparallel G-Quadruplex Nanoconstruct with Antiproliferative Activity. Molecules 2019, 24, 3625. https://doi.org/10.3390/molecules24193625
Antipova O, Samoylenkova N, Savchenko E, Zavyalova E, Revishchin A, Pavlova G, Kopylov A. Bimodular Antiparallel G-Quadruplex Nanoconstruct with Antiproliferative Activity. Molecules. 2019; 24(19):3625. https://doi.org/10.3390/molecules24193625
Chicago/Turabian StyleAntipova, Olga, Nadezhda Samoylenkova, Ekaterina Savchenko, Elena Zavyalova, Alexander Revishchin, Galina Pavlova, and Alexey Kopylov. 2019. "Bimodular Antiparallel G-Quadruplex Nanoconstruct with Antiproliferative Activity" Molecules 24, no. 19: 3625. https://doi.org/10.3390/molecules24193625
APA StyleAntipova, O., Samoylenkova, N., Savchenko, E., Zavyalova, E., Revishchin, A., Pavlova, G., & Kopylov, A. (2019). Bimodular Antiparallel G-Quadruplex Nanoconstruct with Antiproliferative Activity. Molecules, 24(19), 3625. https://doi.org/10.3390/molecules24193625






