Design, Synthesis and Biological Evaluation of Pentacyclic Triterpene Derivatives: Optimization of Anti-ABL Kinase Activity
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
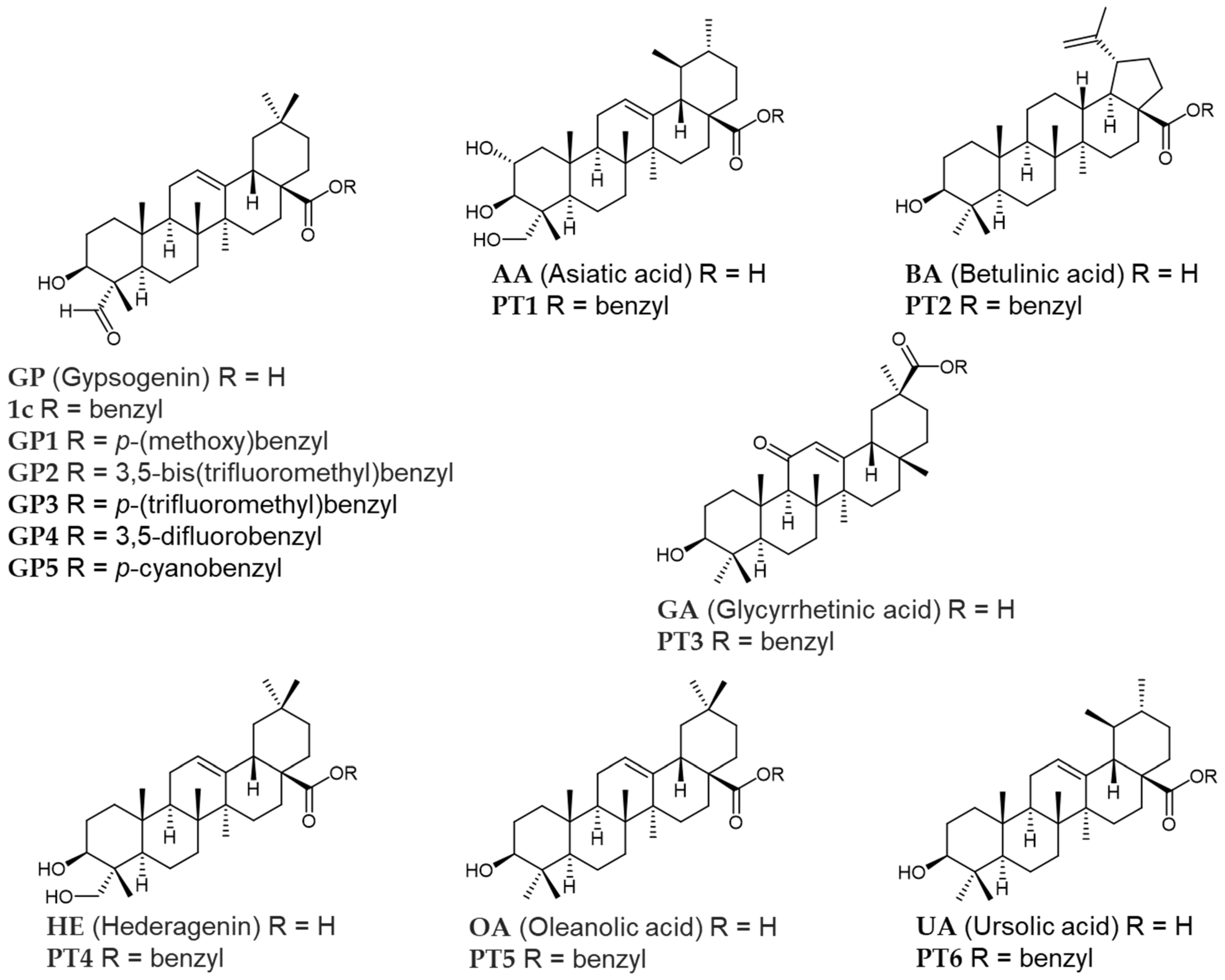
3.1. Chemistry
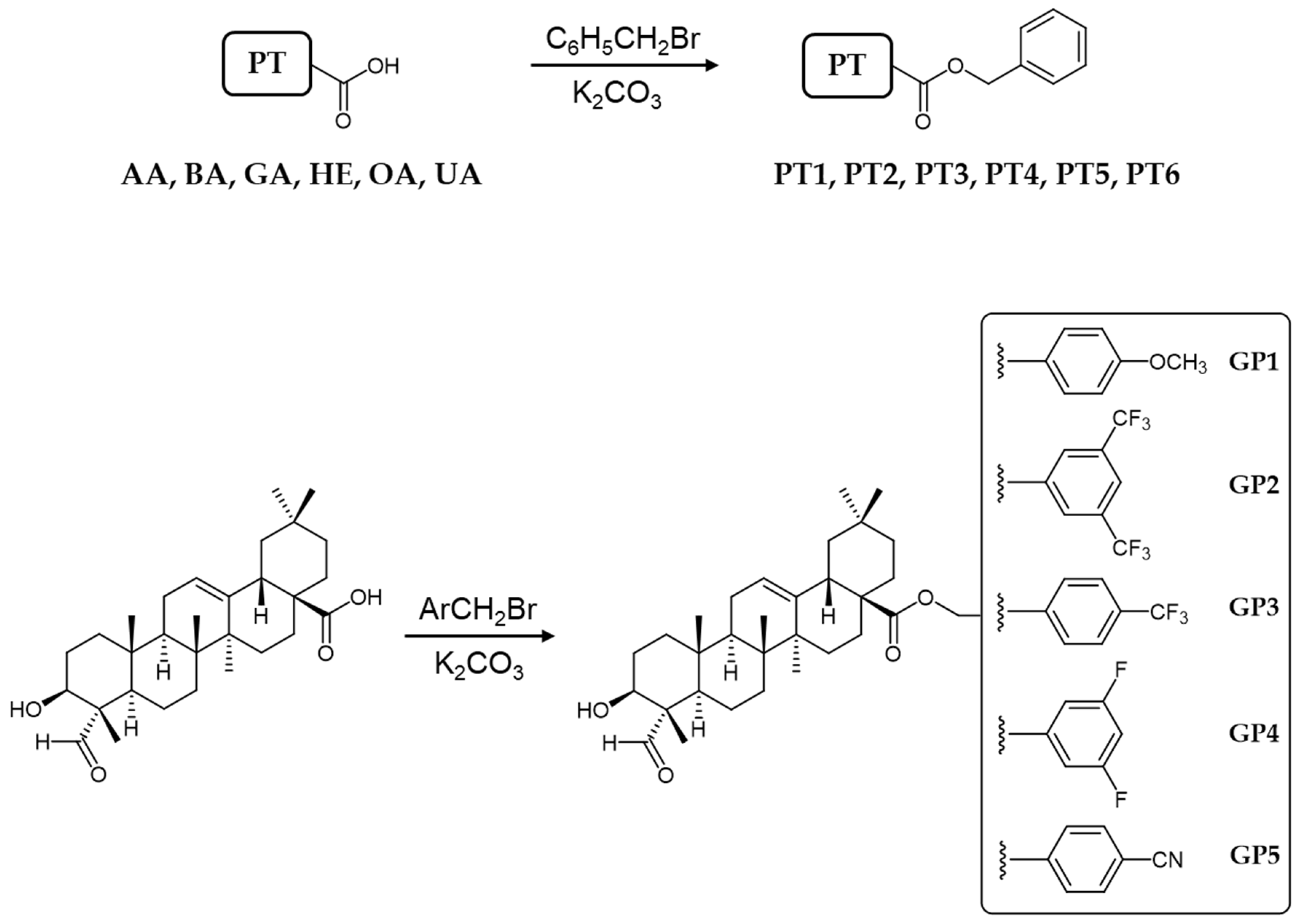
3.2. General Method of Preparation of PT1–PT5 and GP1–GP5 Compounds
3.3. Biochemistry
3.3.1. Cell Cultures
3.3.2. Cell Viability Assay
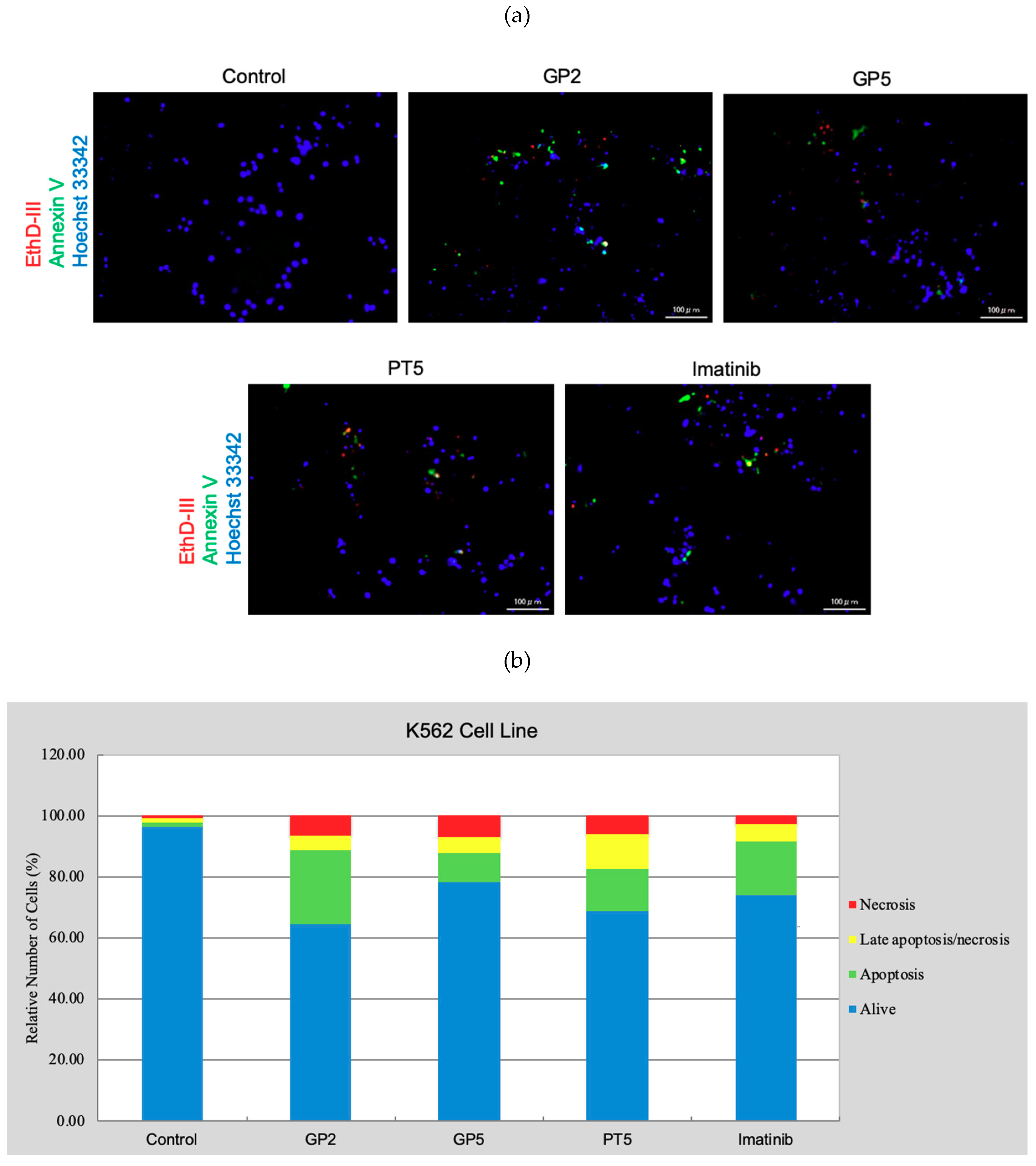
3.3.3. Cell Death Detection
3.3.4. Determination of ABL1 Kinase Inhibition
3.3.5. Immunoblot Analysis
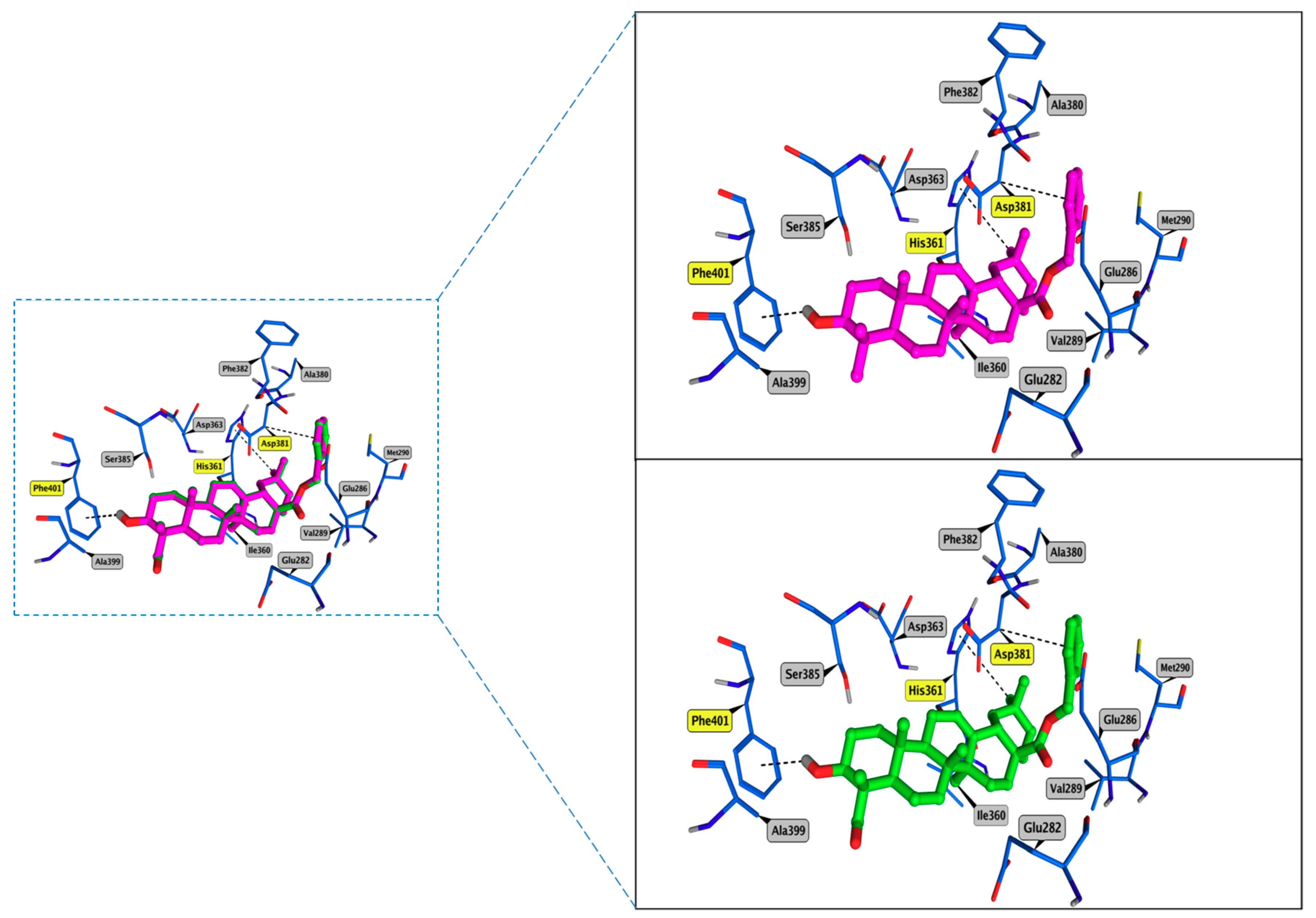
3.4. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fitzmaurice, C.; Akinyemiju, T.F.; Al Lami, F.H.; Alam, T.; Alizadeh-Navaei, R.; Allen, C.; Alsharif, U.; Alvis-Guzman, N.; Amini, E.; Anderson, B.O.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016 A Systematic Analysis for the Global Burden of Disease Study. Jama Oncol. 2018, 4, 1553–1568. [Google Scholar] [PubMed]
- Szczepański, T.; van der Velden, V.H.; van Dongen, J.J. Classification systems for acute and chronic leukaemias. Best Pract. Res. Clin. Haematol. 2013, 16, 561–582. [Google Scholar] [CrossRef]
- Clarkson, B.; Strife, A.; Wisniewski, D.; Lambek, C.L.; Liu, C. Chronic myelogenous leukemia as a paradigm of early cancer and possible curative strategies. Leukemia 2003, 17, 1211–1262. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.J.; Liu, Y.F.; Xu, L.Z.; Long, Z.J.; Huang, D.; Yang, Y.; Liu, B.; Feng, J.X.; Pan, Y.J.; Yan, J.S.; et al. The Philadelphia chromosome in leukemogenesis. Chin. J. Cancer 2016, 35. [Google Scholar] [CrossRef] [PubMed]
- Ernst, T.; Obstfelder, E.; Hochhaus, A. Chronic myeloid leukemia. Onkologe 2018, 24, 427–441. [Google Scholar] [CrossRef]
- Schiffer, C.A. Diagnosis and treatment of chronic myeloid leukemia. In Neoplastic Diseases of the Blood, 6th ed.; Wiernik, P.H., Dutcher, J.P., Gertz, M.A., Eds.; Springer: New York, NY, USA, 2018; Volume 20, p. 64. [Google Scholar]
- De Braekeleer, E.; Douet-Guilbert, N.; Rowe, D.; Bown, N.; Morel, F.; Berthou, C.; Ferec, C.; De Braekeleer, M. ABL1 fusion genes in hematological malignancies: A review. Eur. J. Haematol. 2011, 86, 361–371. [Google Scholar] [CrossRef]
- Iqbal, Z. A comprehensive analysis of breakpoint cluster region-abelson fusion oncogene splice variants in chronic myeloid leukemia and their correlation with disease biology. Indian, J. Hum. Genet. 2014, 20, 64–68. [Google Scholar] [CrossRef]
- Ross, T.S.; Mgbemena, V.E. Re-evaluating the role of BCR/ABL in chronic myelogenous leukemia. Mol. Cell Oncol. 2014, 1, e963450. [Google Scholar] [CrossRef]
- Vlahovic, G.; Crawford, J. Activation of tyrosine kinases in cancer. Oncologist 2003, 8, 531–538. [Google Scholar] [CrossRef]
- Lemmon, M.A.; Schlessinger, J. Cell Signaling by Receptor Tyrosine Kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef]
- Du, Z.F.; Lovly, C.M. Mechanisms of receptor tyrosine kinase activation in cancer. Mol Cancer 2018, 17. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Iqbal, N. Imatinib: A breakthrough of targeted therapy in cancer. Chemother. Res. Prac. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Sacha, T. Imatinib in chronic myeloid leukemia: An overview. Mediterr. J. Hematol. Infect. Dis. 2014, 6, e2014007. [Google Scholar] [CrossRef] [PubMed]
- Pophali, P.A.; Patnaik, M.M. The Role of New Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia. Cancer J. 2016, 22, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gutierrez, J.V.; Herrera, P.; Abalo, L.L.; Rey, M.D.; Calbacho, M.; Ramos, L.; Ramos, P.; Montalban, C.; Lopez-Jimenez, J. Impact of Second-Generation Tyrosine Kinase Inhibitors As Second Line Treatment for Patients with Chronic Myeloid Leukemia. Blood 2011, 118, 1615. [Google Scholar]
- Wehrle, J.; Pahl, H.L.; von Bubnoff, N. Ponatinib: A Third-Generation Inhibitor for the Treatment of CML. Recent Results Canc. 2014, 201, 99–107. [Google Scholar]
- Jabbour, E.; Kantarjian, H.; Cortes, J. Use of Second- and Third-Generation Tyrosine Kinase Inhibitors in the Treatment of Chronic Myeloid Leukemia: An Evolving Treatment Paradigm. Cl. Lymph. Myelom. Leuk. 2015, 15, 323–334. [Google Scholar] [CrossRef]
- Rossari, F.; Minutolo, F.; Orciuolo, E. Past, present, and future of Bcr-Abl inhibitors: From chemical development to clinical efficacy. J. Hematol. Oncol. 2018, 11. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Bba-Gen. Subjects 2013, 1830, 3670–3695. [Google Scholar] [CrossRef]
- Nasr, H.; Abdel-Aziz, M.S.; Radwan, M.O.; Shaaban, M. Bioactive Secondary Metabolites from Terrestrial Streptomyces baarnensis MH4. Brit. J. Pharm. Res. 2015, 5, 72–81. [Google Scholar] [CrossRef]
- Bishayee, A.; Sethi, G. Bioactive natural products in cancer prevention and therapy: Progress and promise. Semin. Cancer Biol. 2016, 40–41, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Meng, X.; Li, Y.; Zhao, C.N.; Tang, G.Y.; Li, H.B. Antibacterial and Antifungal Activities of Spices. Int. J. Mol. Sci. 2017, 18, 1283. [Google Scholar] [CrossRef]
- Aung, T.N.; Qu, Z.P.; Kortschak, R.D.; Adelson, D.L. Understanding the Effectiveness of Natural Compound Mixtures in Cancer through Their Molecular Mode of Action. Int. J. Mol. Sci. 2017, 18, 656. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.; El-Alfy, A.T.; Ezel, K.; Radwan, M.O.; Shilabin, A.G.; Kochanowska-Karamyan, A.J.; Abd-Alla, H.I.; Otsuka, M.; Hamann, M.T. Marine Inspired 2-(5-Halo-1H-indol-3-yl)-N, N-dimethylethanamines as Modulators of Serotonin Receptors: An Example Illustrating the Power of Bromine as Part of the Uniquely Marine Chemical Space. Mar. Drugs 2017, 15, 248. [Google Scholar] [CrossRef]
- Scharenberg, F.; Zidorn, C. Genuine and Sequestered Natural Products from the Genus Orobanche (Orobanchaceae, Lamiales). Molecules 2018, 23, 2821. [Google Scholar] [CrossRef]
- Dzubak, P.; Hajduch, M.; Vydra, D.; Hustova, A.; Kvasnica, M.; Biedermann, D.; Markova, L.; Urban, M.; Sarek, J. Pharmacological activities of natural triterpenoids and their therapeutic implications. Nat. Prod. Rep. 2006, 23, 394–411. [Google Scholar] [CrossRef] [PubMed]
- Laszczyk, M.N. Pentacyclic Triterpenes of the Lupane, Oleanane and Ursane Group as Tools in Cancer Therapy. Planta. Med. 2009, 75, 1549–1560. [Google Scholar] [CrossRef]
- Lu, Z.Z.; Jin, Y.L.; Qiu, L.; Lai, Y.R.; Pan, J.X. Celastrol, a novel HSP90 inhibitor, depletes Bcr-Abl and induces apoptosis in imatinib-resistant chronic myelogenous leukemia cells harboring T315I mutation. Cancer Lett. 2010, 290, 182–191. [Google Scholar] [CrossRef]
- Radwan, M.O.; Ismail, M.A.H.; El-Mekkawy, S.; Ismail, N.S.M.; Hanna, A.G. Synthesis and biological activity of new 18 beta-glycyrrhetinic acid derivatives. Arab. J. Chem. 2016, 9, 390–399. [Google Scholar] [CrossRef]
- Wang, R.; Zheng, Q.X.; Wang, W.; Feng, L.; Li, H.J.; Huai, Q.Y. Design and Synthesis of New Anticancer Glycyrrhetinic Acids and Oleanolic Acids. Biol. Pharm. Bull. 2017, 40, 703–710. [Google Scholar] [CrossRef]
- Ciftci, H. Effects of glycyrrhetic acid on human chronic myelogenous leukemia cells. Turk. J. Pharm. Sci. in press. [CrossRef]
- Ozturk, S.E.; Babahan, I.; Ozmen, A. Synthesis, characterization and in vitro anti-neoplastic activity of gypsogenin derivatives. Bioorg. Chem. 2014, 53, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Emirdag-Ozturk, S.; Karayildirim, T.; Capaci-Karagoz, A.; Alankus-Caliskan, O.; Ozmen, A.; Poyrazoglu-Coban, E. Synthesis, antimicrobial and cytotoxic activities, and structure-activity relationships of gypsogenin derivatives against human cancer cells. Eur. J. Med. Chem. 2014, 82, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Mu, Y.; Wang, F.; Song, L.; Sun, J.; Liu, Y.; Sun, J. Synthesis of gypsogenin derivatives with capabilities to arrest cell cycle and induce apoptosis in human cancer cells. R Soc. Open Sci. 2018, 5, 171510. [Google Scholar] [CrossRef] [PubMed]
- Ciftci, H.I.; Ozturk, S.E.; Ali, T.F.S.; Radwan, M.O.; Tateishi, H.; Koga, R.; Ocak, Z.; Can, M.; Otsuka, M.; Fujita, M. The First Pentacyclic Triterpenoid Gypsogenin Derivative Exhibiting Anti-ABL1 Kinase and Anti-chronic Myelogenous Leukemia Activities. Biol. Pharm. Bull. 2018, 41, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, J.; Gong, Y.; Liu, J.; Zhang, L.; Hua, W.; Sun, H. Synthesis and Biological Evaluation of Asiatic Acid Derivatives as Inhibitors of Glycogen Phosphorylases. Chem. Biodivers. 2009, 6, 864–874. [Google Scholar] [CrossRef] [PubMed]
- Wolfram, R.K.; Heller, L.; Csuk, R. Targeting Mitochondria: Esters of Rhodamine B with Triterpenoids Are Mitocanic Triggers of Apoptosis. Eur. J. Med. Chem. 2018, 152, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Hernández, D.; Demuner, A.J.; Barbosa, L.C.A.; Csuk, R.; Heller, L. Hederagenin as a Triterpene Template for the Development of New Antitumor Compounds. Eur. J. Med. Chem. 2015, 105, 57–62. [Google Scholar] [CrossRef]
- Bayrak, N.; Yildirim, H.; Tuyun, A.F.; Kara, E.M.; Celik, B.O.; Gupta, G.K.; Ciftci, H.I.; Fujita, M.; Otsuka, M.; Nasiri, H.R. Synthesis, Computational Study, and Evaluation of In Vitro Antimicrobial, Antibiofilm, and Anticancer Activities of New Sulfanyl Aminonaphthoquinone Derivatives. Lett Drug Des. Discov. 2017, 14, 647–661. [Google Scholar] [CrossRef]
- Ali, T.F.S.; Iwamaru, K.; Ciftci, H.I.; Koga, R.; Matsumoto, M.; Oba, Y.; Kurosaki, H.; Fujita, M.; Okamoto, Y.; Umezawa, K.; et al. Novel metal chelating molecules with anticancer activity. Striking effect of the imidazole substitution of the histidine-pyridine-histidine system. Bioorgan Med. Chem. 2015, 23, 5476–5482. [Google Scholar] [CrossRef]
- Tateishi, H.; Monde, K.; Anraku, K.; Koga, R.; Hayashi, Y.; Ciftci, H.I.; DeMirci, H.; Higashi, T.; Motoyama, K.; Arima, H.; et al. A clue to unprecedented strategy to HIV eradication: “Lock-in and apoptosis”. Sci. Rep-Uk 2017, 7, 8957. [Google Scholar] [CrossRef] [PubMed]
- Altintop, M.D.; Ciftci, H.I.; Radwan, M.O.; Sever, B.; Kaplancikli, Z.A.; Ali, T.F.S.; Koga, R.; Fujita, M.; Otsuka, M.; Ozdemir, A. Design, Synthesis, and Biological Evaluation of Novel 1,3,4-Thiadiazole Derivatives as Potential Antitumor Agents against Chronic Myelogenous Leukemia: Striking Effect of Nitrothiazole Moiety. Molecules 2018, 23, 59. [Google Scholar] [CrossRef] [PubMed]
- Ciftci, H.I.; Fujino, H.; Koga, R.; Yamamoto, M.; Kawamura, S.; Tateishi, H.; Iwatani, Y.; Otsuka, M.; Fujita, M. Mutational analysis of HIV-2 Vpx shows that proline residue 109 in the poly-proline motif regulates degradation of SAMHD1. Febs. Lett. 2015, 589, 1505–1514. [Google Scholar] [CrossRef] [PubMed]
- Koga, R.; Radwan, M.O.; Ejima, T.; Kanemaru, Y.; Tateishi, H.; Ali, T.F.S.; Ciftci, H.I.; Shibata, Y.; Taguchi, Y.; Inoue, J.; et al. A Dithiol Compound Binds to the Zinc Finger Protein TRAF6 and Suppresses Its Ubiquitination. ChemMedChem 2017, 12, 1935–1941. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Radwan, M.O.; Hamasaki, A.; Ejima, A.; Obata, E.; Koga, R.; Tateishi, H.; Okamoto, Y.; Fujita, M.; Nakao, M.; et al. A novel inhibitor of farnesyltransferase with a zinc site recognition moiety and a farnesyl group. Bioorg. Med. Chem. Lett. 2017, 27, 3862–3866. [Google Scholar] [CrossRef] [PubMed]
- Radwan, M.O.; Koga, R.; Hida, T.; Ejima, T.; Kanemaru, Y.; Tateishi, H.; Okamoto, Y.; Inoue, J.; Fujita, M.; Otsuka, M. Minimum structural requirements for inhibitors of the zinc finger protein TRAF6. Bioorg. Med Chem. Lett. 2019, 29, 2162–2167. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, N.; Radwan, M.O.; Amano, M.; Endo, S.; Fujii, E.; Hayashi, H.; Ueno, S.; Ueno, N.; Tatetsu, H.; Hata, H.; et al. Novel p97/VCP inhibitor induces endoplasmic reticulum stress and apoptosis in both bortezomib-sensitive and-resistant multiple myeloma cells. Cancer Sci. 2019, 00, 1–13. [Google Scholar] [CrossRef] [PubMed]
- El-Shaheny, R.; Fuchigami, T.; Yoshida, S.; Radwan, M.O.; Nakayama, M. Complementary HPLC, in Silico Toxicity, and Molecular Docking Studies for Investigation of the Potential Influences of Gastric Acidity and Nitrite Content on Paracetamol Safety. Microchem. J. 2019, 150, 104107. [Google Scholar] [CrossRef]
- Sever, B.; Altıntop, D.M.; Radwan, M.O.; Özdemir, A.; Otsuka, M.; Fujita, M.; Ciftci, H.I. Design, synthesis and biological evaluation of a new series of thiazolyl-pyrazolines as dual EGFR and HER2 inhibitors. Eur. J. Med. Chem. 2019, 182, 111648. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds (1c, PT1, PT2, PT3, PT4, PT5, PT6, GP1, GP2, GP3, GP4, GP5 and GP6) are available from the authors. |

| ID | IC50 Values (μM) a | SI b | ||
|---|---|---|---|---|
| K562 Cell Line | PBMC | |||
| Free Triterpenes | GP | 12.67 ± 2.15 | >100 | >7.89 |
| AA | 29.31 ± 3.42 | >100 | >3.41 | |
| BA | 52.27 ± 4.79 | >100 | >1.91 | |
| GA | 41.03 ± 5.34 | >100 | >2.43 | |
| HE | >100 | >100 | >1 | |
| OA | 13.46 ± 2.55 | >100 | >7.43 | |
| UA | 16.51 ± 1.13 | >100 | >6.06 | |
| Triterpenes benzyl esters | 1c | 8.94 ± 1.73 | >100 | >11.19 |
| PT1 | 4.10 ± 0.85 | 15.54 ± 1.08 | 3.79 | |
| PT2 | 8.49 ± 1.22 | 27.96 ± 2.74 | 3.29 | |
| PT3 | 69.50 ± 8.43 | >100 | >1.44 | |
| PT4 | 6.48 ± 1.27 | 23.08 ± 3.11 | 3.56 | |
| PT5 | 5.46 ± 1.51 | 50.21 ± 4.58 | 9.19 | |
| PT6 | 21.77 ± 1.24 | 43.31 ± 1.93 | 1.99 | |
| Gypsogenin substituted- benzyl esters | GP1 | 5.53 ± 1.78 | 25.05 ± 2.92 | 4.53 |
| GP2 | 4.78 ± 1.25 | 52.76 ± 3.82 | 11.04 | |
| GP3 | 4.58 ± 1.49 | 23.22 ± 3.08 | 5.07 | |
| GP4 | 5.13 ± 1.38 | 28.28 ± 2.25 | 5.51 | |
| GP5 | 3.19 ± 1.21 | 25.60 ± 2.18 | 8.03 | |
| Control | Imatinibc | 5.49 ± 1.81 | 28.66 ± 3.16 | 5.22 |
| IC50 Values (μM) | ||||||
|---|---|---|---|---|---|---|
| HL-60 | MT-2 | Jurkat | HeLa | MCF7 | A549 | |
| GP2 | 5.27 ± 0.42 | 36.82 ± 4.21 | 9.14 ± 0.71 | 35.28 ± 2.84 | 51.58 ± 4.75 | >100 |
| GP5 | 4.36 ± 0.28 | 7.26 ± 0.97 | 4.83 ± 0.35 | 5.66 ± 0.32 | 15.31 ± 2.11 | 24.55 ± 1.94 |
| PT5 | 15.71 ± 1.36 | 34.70 ± 3.85 | 13.85 ± 1.58 | 67.05 ± 4.19 | 53.91 ± 6.46 | >100 |
| Imatinib | 11.75 ± 1.08 | 12.96 ± 1.33 | 9.69 ± 0.63 | 28.68 ± 1.92 | 90.56 ± 5.81 | 95.07 ± 8.33 |
| IC50 Values (μM) | |||||
|---|---|---|---|---|---|
| 1c a | GP2 | GP5 | PT5 | Imatinib | |
| ABL1 | 8.71 ± 1.14 | 7.19 ± 0.65 | 6.16 ± 0.63 | 1.44 ± 0.12 | 0.21 ± 0.44 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
I. Ciftci, H.; O. Radwan, M.; E. Ozturk, S.; Ulusoy, N.G.; Sozer, E.; E. Ellakwa, D.; Ocak, Z.; Can, M.; F.S. Ali, T.; I. Abd-Alla, H.; et al. Design, Synthesis and Biological Evaluation of Pentacyclic Triterpene Derivatives: Optimization of Anti-ABL Kinase Activity. Molecules 2019, 24, 3535. https://doi.org/10.3390/molecules24193535
I. Ciftci H, O. Radwan M, E. Ozturk S, Ulusoy NG, Sozer E, E. Ellakwa D, Ocak Z, Can M, F.S. Ali T, I. Abd-Alla H, et al. Design, Synthesis and Biological Evaluation of Pentacyclic Triterpene Derivatives: Optimization of Anti-ABL Kinase Activity. Molecules. 2019; 24(19):3535. https://doi.org/10.3390/molecules24193535
Chicago/Turabian StyleI. Ciftci, Halil, Mohamed O. Radwan, Safiye E. Ozturk, N. Gokce Ulusoy, Ece Sozer, Doha E. Ellakwa, Zeynep Ocak, Mustafa Can, Taha F.S. Ali, Howaida I. Abd-Alla, and et al. 2019. "Design, Synthesis and Biological Evaluation of Pentacyclic Triterpene Derivatives: Optimization of Anti-ABL Kinase Activity" Molecules 24, no. 19: 3535. https://doi.org/10.3390/molecules24193535
APA StyleI. Ciftci, H., O. Radwan, M., E. Ozturk, S., Ulusoy, N. G., Sozer, E., E. Ellakwa, D., Ocak, Z., Can, M., F.S. Ali, T., I. Abd-Alla, H., Yayli, N., Tateishi, H., Otsuka, M., & Fujita, M. (2019). Design, Synthesis and Biological Evaluation of Pentacyclic Triterpene Derivatives: Optimization of Anti-ABL Kinase Activity. Molecules, 24(19), 3535. https://doi.org/10.3390/molecules24193535











