Long-Chain Polyisoprenoids Are Synthesized by AtCPT1 in Arabidopsis thaliana
Abstract
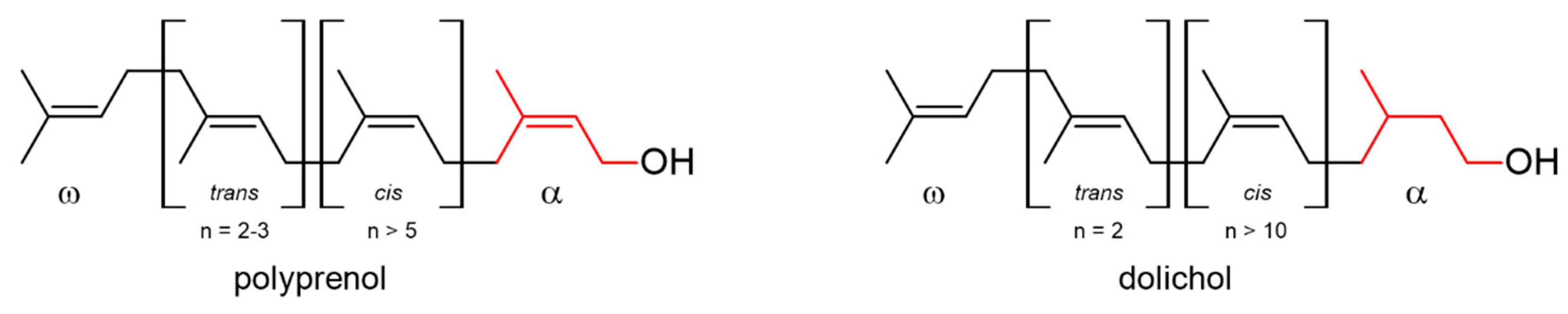
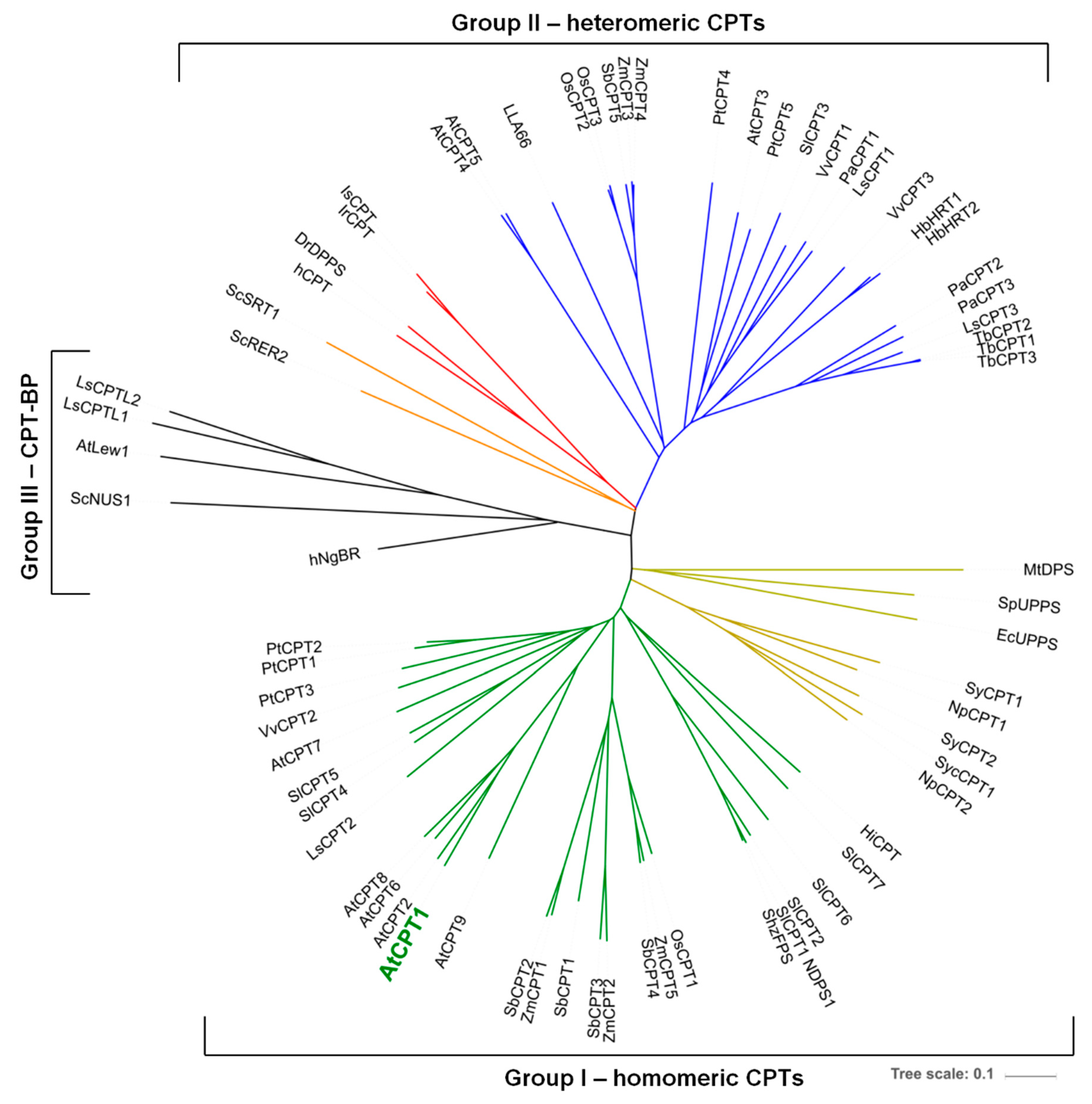
1. Introduction
2. Results and Discussion
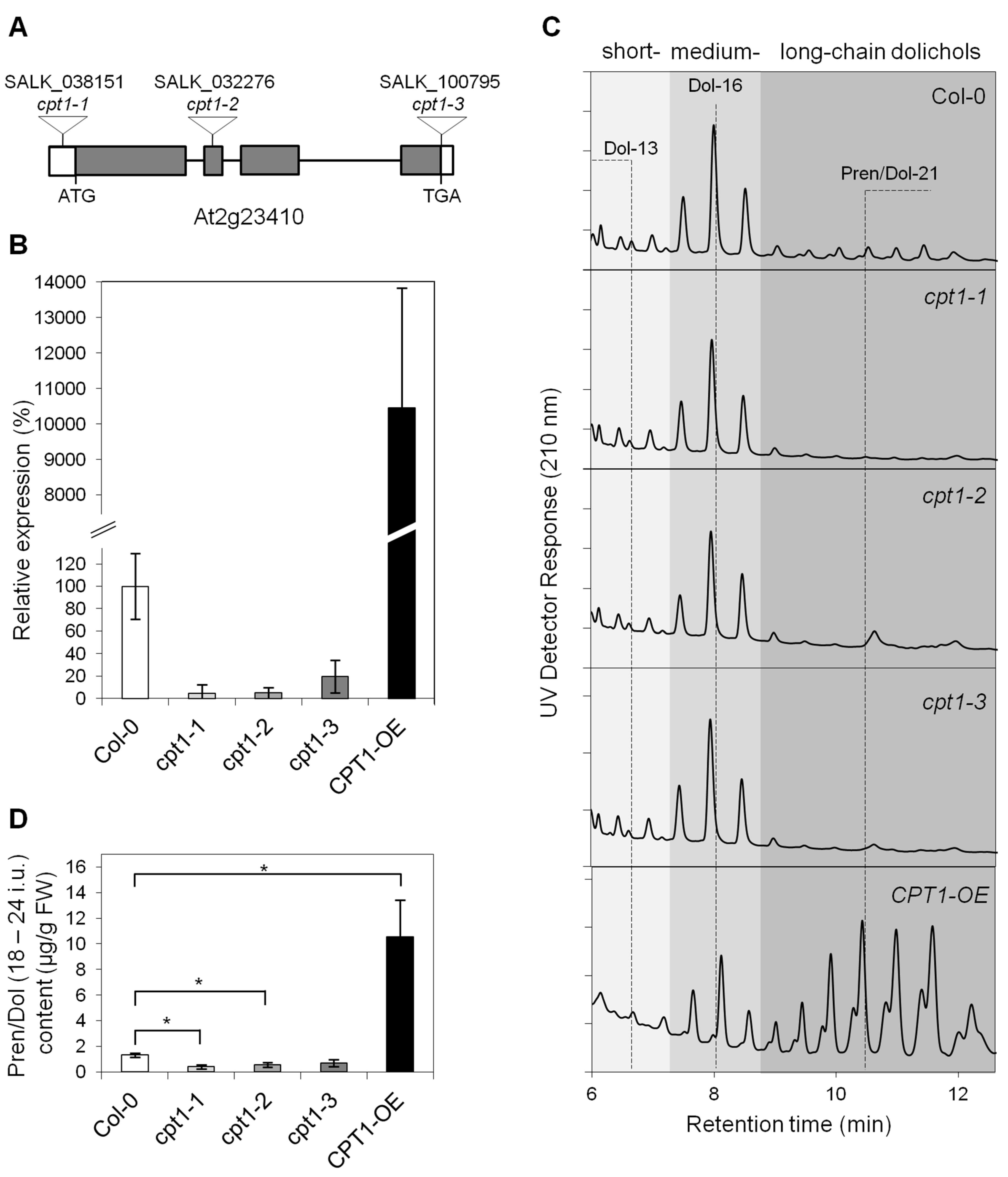
2.1. AtCPT1 Synthesizes the Family of Long-Chain Dolichols in Planta
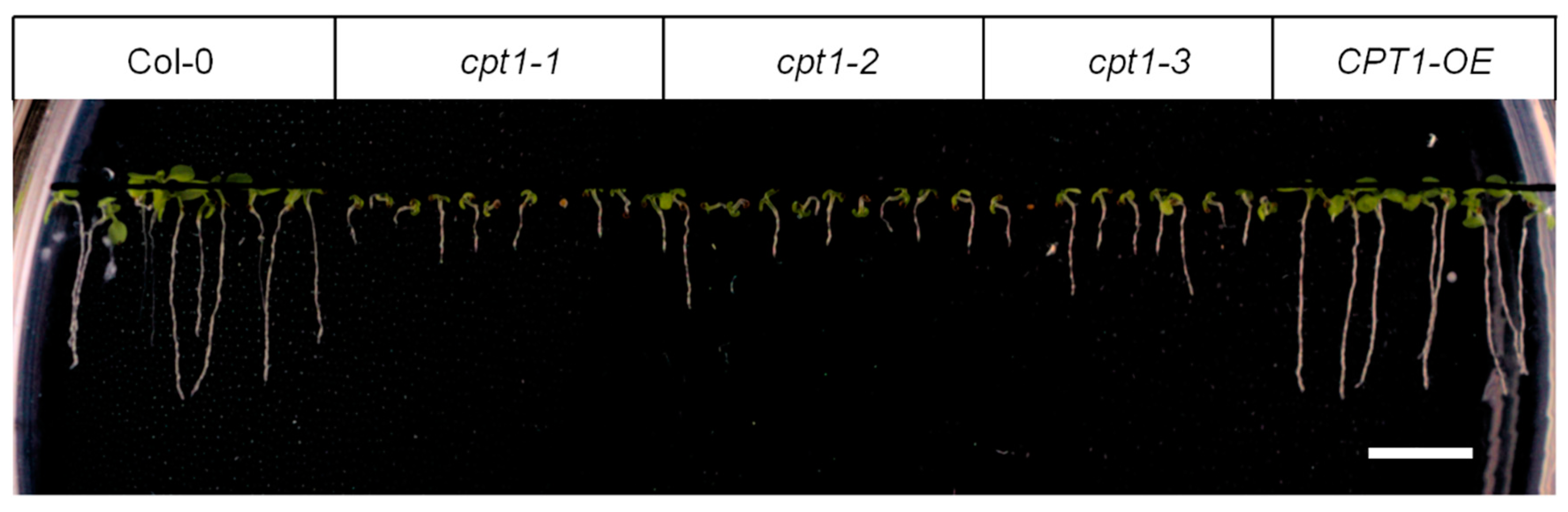
2.2. Shortage of Long-Chain Dolichols Leads to the Growth Defects
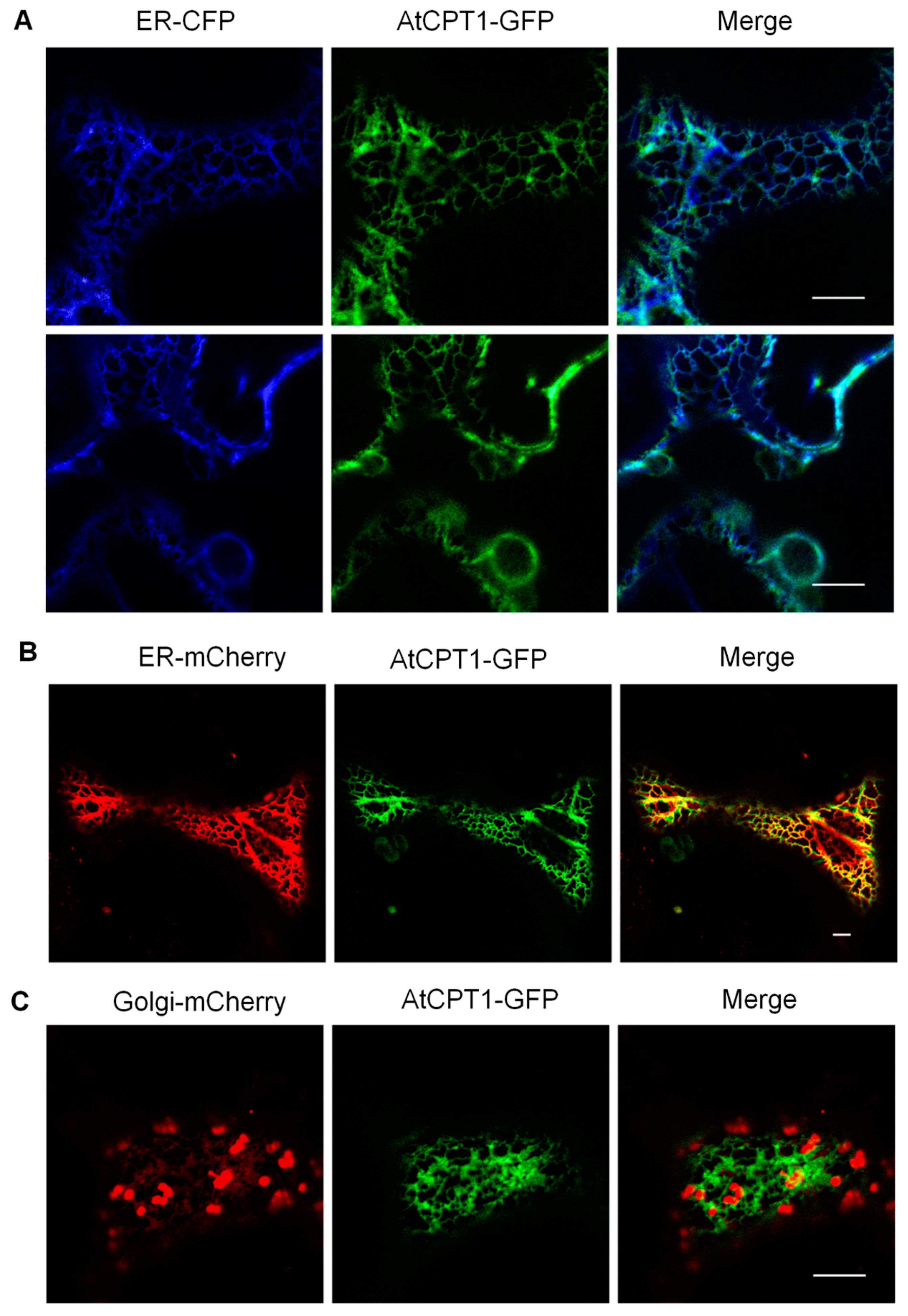
2.3. AtCPT1 Is Localized to the ER
2.4. AtCPT1 Complements the Function of Yeast Rer2 cis-Prenyltransferase
3. Materials and Methods
3.1. Chemicals
3.2. Plant Materials and Growth Conditions
3.3. Yeast Materials
- SS328 (wild type, MATα ade2-101 ura3-52his3Δ 200lys2-801) and
- YG932 (rer2Δ mutant, MATα rer2Δ::kanMX4 ade2-101 ura3-52his3Δ 200 lys-801) were kind gifts of Dr. C.J. Waechter (University of Kentucky College of Medicine, Lexington, KY, USA).
- YG938 (srt1Δ mutant, MATα Δ srt1::kanMX4 ade2-101 ura3-52 his3Δ200 lys2-801) was obtained from Dr. M. Aebi, (ETH, Zurich, Switzerland).
3.4. Preparation of AtCPT1 and LEW1 Expression Constructs
3.5. Generation of Arabidopsis Plants Overexpressing AtCPT1
3.6. Transient Expression of AtCPT1 in Nicotiana benthamiana
3.7. Generation of Yeast Overexpressing AtCPT1
3.8. Polyisoprenoid Extraction and Purification
3.9. HPLC/UV Analysis of Polyisoprenoids
3.10. Confocal Microscopy
3.11. Statistical Analysis
3.12. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Burda, P.; Aebi, M. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta (BBA) Gen. Subj. 1999, 1426, 239–257. [Google Scholar] [CrossRef]
- Pattison, R.J.; Amtmann, A. N-glycan production in the endoplasmic reticulum of plants. Trends Plant Sci. 2009, 14, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Nothaft, H.; Szymanski, C.M. Protein glycosylation in bacteria: Sweeter than ever. Nat. Rev. Genet. 2010, 8, 765–778. [Google Scholar] [CrossRef]
- Gutkowska, M.; Bieńkowski, T.; Hung, V.S.; Wanke, M.; Hertel, J.; Danikiewicz, W.; Swiezewska, E. Proteins are polyisoprenylated in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2004, 322, 998–1004. [Google Scholar] [CrossRef] [PubMed]
- Bajda, A.; Konopka-Postupolska, D.; Krzymowska, M.; Hennig, J.; Skorupinska-Tudek, K.; Surmacz, L.; Wójcik, J.; Matysiak, Z.; Chojnacki, T.; Skorzynska-Polit, E.; et al. Role of polyisoprenoids in tobacco resistance against biotic stresses. Physiol. Plant 2009, 135, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Swiezewska, E.; Danikiewicz, W. Polyisoprenoids: Structure, biosynthesis and function. Prog. Lipid Res. 2005, 44, 235–258. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, T.A.; Surowiecki, P.; Siekierska, H.; Kania, M.; Van Gelder, K.; Rea, K.A.; Virta, L.K.; Vatta, M.; Gawarecka, K.; Wojcik, J.; et al. Polyprenols Are Synthesized by a Plastidial cis-Prenyltransferase and Influence Photosynthetic Performance[OPEN]. Plant Cell 2017, 29, 1709–1725. [Google Scholar] [CrossRef]
- Akhtar, T.A.; Matsuba, Y.; Schauvinhold, I.; Yu, G.; Lees, H.A.; Klein, S.E.; Pichersky, E. The tomato cis-prenyltransferase gene family. Plant J. 2013, 73, 640–652. [Google Scholar] [CrossRef]
- Cantagrel, V.; Lefeber, D.J.; Ng, B.G.; Guan, Z.Q.; Silhavy, J.L.; Bielas, S.L.; Lehle, L.; Hombauer, H.; Adamowicz, M.; Swiezewska, E.; et al. SRD5A3 Is Required for Converting Polyprenol to Dolichol and Is Mutated in a Congenital Glycosylation Disorder. Cell 2010, 142, 203–217. [Google Scholar] [CrossRef]
- Jozwiak, A.; Gutkowska, M.; Gawarecka, K.; Surmacz, L.; Buczkowska, A.; Lichocka, M.; Nowakowska, J.; Swiezewska, E. POLYPRENOL REDUCTASE2 Deficiency Is Lethal in Arabidopsis Due to Male Sterility[OPEN]. Plant Cell 2015, 27, 3336–3353. [Google Scholar] [CrossRef]
- Harrison, K.D.; Park, E.J.; Gao, N.; Kuo, A.; Rush, J.S.; Waechter, C.J.; Lehrman, M.A.; Sessa, W.C. Nogo-B receptor is necessary for cellular dolichol biosynthesis and protein N-glycosylation. EMBO J. 2011, 30, 2490–2500. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Grabińska, K.A.; Guan, Z.; Stránecký, V.; Hartmannová, H.; Hodaňová, K.; Barešová, V.; Sovová, J.; Jozsef, L.; Ondrušková, N.; et al. Mutation of Nogo-B receptor, a subunit of cis-prenyltransferase, causes a congenital disorder of glycosylation. Cell Metab. 2014, 20, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Brasher, M.I.; Surmacz, L.; Leong, B.; Pitcher, J.; Swiezewska, E.; Pichersky, E.; Akhtar, T.A. A two-component enzyme complex is required for dolichol biosynthesis in tomato. Plant J. 2015, 82, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Chakrabarty, R.; Tran, H.T.; Kwon, E.J.; Kwon, M.; Nguyen, T.D.; Ro, D.K. A lettuce (Lactuca sativa) homolog of human Nogo-B receptor interacts with cis-prenyltransferase and is necessary for natural rubber biosynthesis. J. Biol. Chem. 2015, 290, 1898–1914. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Kwon, E.J.; Ro, D. cis-Prenyltransferase and Polymer Analysis from a Natural Rubber Perspective. Methods Enzymol. 2016, 576, 121–145. [Google Scholar] [PubMed]
- Nguyen, N.Q.; Lee, S.C.; Lee, O.R. cis-Prenyltransferase interacts with a Nogo-B receptor homolog for dolichol biosynthesis in Panax ginseng Meyer. J. Ginseng Res. 2017, 41, 403–410. [Google Scholar] [CrossRef]
- Lakusta, A.M.; Kwon, M.; Kwon, E.-J.G.; Stonebloom, S.; Scheller, H.V.; Ro, D.K. Molecular Studies of the Protein Complexes Involving Cis-Prenyltransferase in Guayule (Parthenium argentatum), an Alternative Rubber-Producing Plant. Front. Plant Sci. 2019, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- Asawatreratanakul, K.; Zhang, Y.-W.; Wititsuwannakul, D.; Wititsuwannakul, R.; Takahashi, S.; Rattanapittayaporn, A.; Koyama, T. Molecular cloning, expression and characterization of cDNA encoding cis-prenyltransferases from Hevea brasiliensis. A key factor participating in natural rubber biosynthesis. JBIC J. Biol. Inorg. Chem. 2003, 270, 4671–4680. [Google Scholar]
- Schmidt, T.; Lenders, M.; Hillebrand, A.; Van Deenen, N.; Munt, O.; Reichelt, R.; Eisenreich, W.; Fischer, R.; Prüfer, D.; Gronover, C.S. Characterization of rubber particles and rubber chain elongation in Taraxacum koksaghyz. BMC Biochem. 2010, 11, 11. [Google Scholar] [CrossRef]
- Liu, M.C.; Wang, B.J.; Huang, J.K.; Wang, C.S. Expression, Localization and Function of a cis-Prenyltransferase in the Tapetum and Microspores of Lily Anthers. Plant Cell Physiol. 2011, 52, 1487–1500. [Google Scholar] [CrossRef]
- Surmacz, L.; Swiezewska, E. Polyisoprenoids—Secondary metabolites or physiologically important superlipids? Biochem. Biophys. Res. Commun. 2011, 407, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Jozwiak, A.; Ples, M.; Skorupinska-Tudek, K.; Kania, M.; Dydak, M.; Danikiewicz, W.; Swiezewska, E. Sugar availability modulates polyisoprenoid and phytosterol profiles in Arabidopsis thaliana hairy root culture. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2013, 1831, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Kera, K.; Takahashi, S.; Sutoh, T.; Koyama, T.; Nakayama, T. Identification and characterization of acis,trans-mixed heptaprenyl diphosphate synthase fromArabidopsis thaliana. FEBS J. 2012, 279, 3813–3827. [Google Scholar] [CrossRef] [PubMed]
- Surmacz, L.; Plochocka, D.; Kania, M.; Danikiewicz, W.; Swiezewska, E. cis-Prenyltransferase AtCPT6 produces a family of very short-chain polyisoprenoids in planta. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2014, 1841, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Cunillera, N.; Arró, M.; Forés, O.; Manzano, D.; Ferrer, A. Characterization of dehydrodolichyl diphosphate synthase of Arabidopsis thaliana, a key enzyme in dolichol biosynthesis. FEBS Lett. 2000, 477, 170–174. [Google Scholar] [CrossRef]
- Oh, S.K. Molecular Cloning, Expression, and Functional Analysis of a cis-Prenyltransferase from Arabidopsis thaliana IMPLICATIONS IN RUBBER BIOSYNTHESIS. J. Biol. Chem. 2000, 275, 18482–18488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ohyama, K.; Boudet, J.; Chen, Z.; Yang, J.; Zhang, M.; Muranaka, T.; Maurel, C.; Zhu, J.-K.; Gong, Z. Dolichol Biosynthesis and Its Effects on the Unfolded Protein Response and Abiotic Stress Resistance in Arabidopsis[W][OA]. Plant Cell 2008, 20, 1879–1898. [Google Scholar] [CrossRef]
- Szkopinska, A.; Swiezewska, E.; Rytka, J. Interplay between the cis-prenyltransferases and polyprenol reductase in the yeast Saccharomyces cerevisiae. Biochimie 2006, 88, 271–276. [Google Scholar] [CrossRef]
- Sato, M.; Sato, K.; Nishikawa, S.I.; Hirata, A.; Kato, J.I.; Nakano, A. The Yeast RER2 Gene, Identified by Endoplasmic Reticulum Protein Localization Mutations, Encodes cis -Prenyltransferase, a Key Enzyme in Dolichol Synthesis. Mol. Cell. Biol. 1999, 19, 471–483. [Google Scholar] [CrossRef]
- Sato, M.; Fujisaki, S.; Sato, K.; Nishimura, Y.; Nakano, A. Yeast Saccharomyces cerevisiae has two cis-prenyltransferases with different properties and localizations. Implication for their distinct physiological roles in dolichol synthesis. Genes Cells 2001, 6, 495–506. [Google Scholar] [CrossRef]
- Hoffmann, R.; Grabińska, K.; Guan, Z.; Sessa, W.C.; Neiman, A.M. Long-Chain Polyprenols Promote Spore Wall Formation in Saccharomyces cerevisiae. Genetics 2017, 207, 1371–1386. [Google Scholar] [CrossRef] [PubMed]
- Grabińska, K.A.; Edani, B.H.; Park, E.J.; Kraehling, J.R.; Sessa, W.C. A conserved C-terminal RXG motif in the NgBR subunit of cis-prenyltransferase is critical for prenyltransferase activity. J. Biol. Chem. 2017, 292, 17351–17361. [Google Scholar] [CrossRef] [PubMed]
- Gibeaut, D.; Thomine, S.; Lelievre, F.; Boufflet, M.; Guern, J.; Barbier-Brygoo, H. Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol. 1997, 115, 317–319. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Matsuoka, K.; Jinbo, T.; Kimura, T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Glazebrook, J. Arabidopsis: A Laboratory Manual; Cold Spring Harbor: New York, NY, USA, 2002; pp. 119–141. [Google Scholar]
- Surmacz, L.; Wójcik, J.; Kania, M.; Bentinger, M.; Danikiewicz, W.; Dallner, G.; Surowiecki, P.; Cmoch, P.; Swiezewska, E. Short-chain polyisoprenoids in the yeast Saccharomyces cerevisiae—New companions of the old guys. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2015, 1851, 1296–1303. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Surowiecki, P.; Onysk, A.; Manko, K.; Swiezewska, E.; Surmacz, L. Long-Chain Polyisoprenoids Are Synthesized by AtCPT1 in Arabidopsis thaliana. Molecules 2019, 24, 2789. https://doi.org/10.3390/molecules24152789
Surowiecki P, Onysk A, Manko K, Swiezewska E, Surmacz L. Long-Chain Polyisoprenoids Are Synthesized by AtCPT1 in Arabidopsis thaliana. Molecules. 2019; 24(15):2789. https://doi.org/10.3390/molecules24152789
Chicago/Turabian StyleSurowiecki, Przemyslaw, Agnieszka Onysk, Katarzyna Manko, Ewa Swiezewska, and Liliana Surmacz. 2019. "Long-Chain Polyisoprenoids Are Synthesized by AtCPT1 in Arabidopsis thaliana" Molecules 24, no. 15: 2789. https://doi.org/10.3390/molecules24152789
APA StyleSurowiecki, P., Onysk, A., Manko, K., Swiezewska, E., & Surmacz, L. (2019). Long-Chain Polyisoprenoids Are Synthesized by AtCPT1 in Arabidopsis thaliana. Molecules, 24(15), 2789. https://doi.org/10.3390/molecules24152789






