Repression of Acetaminophen-Induced Hepatotoxicity in HepG2 Cells by Polyphenolic Compounds from Lauridia tetragona (L.f.) R.H. Archer
Abstract
1. Introduction
2. Results
2.1. Polyphenolic Contents
2.2. Antioxidant Assay
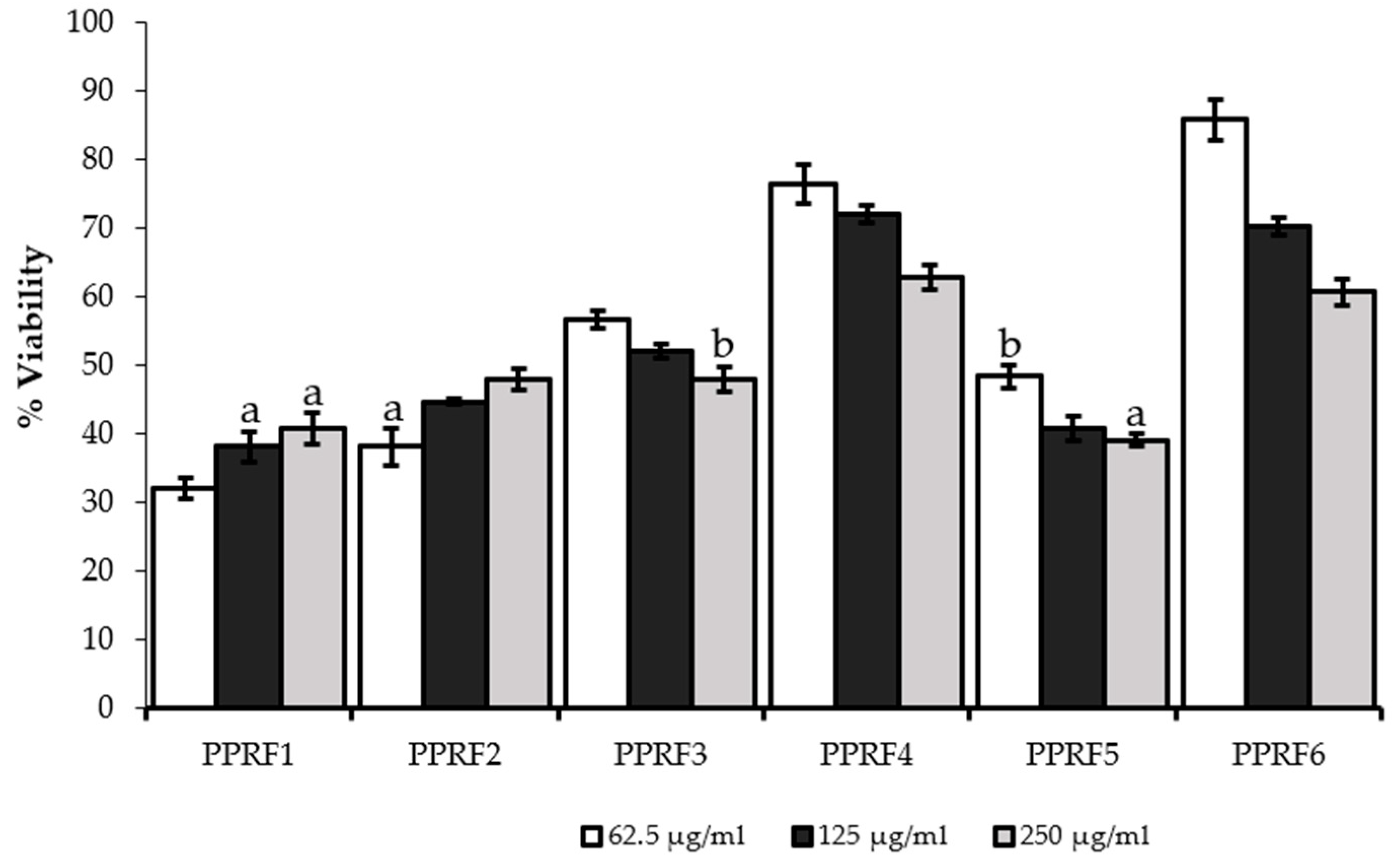
2.3. Cell Viability
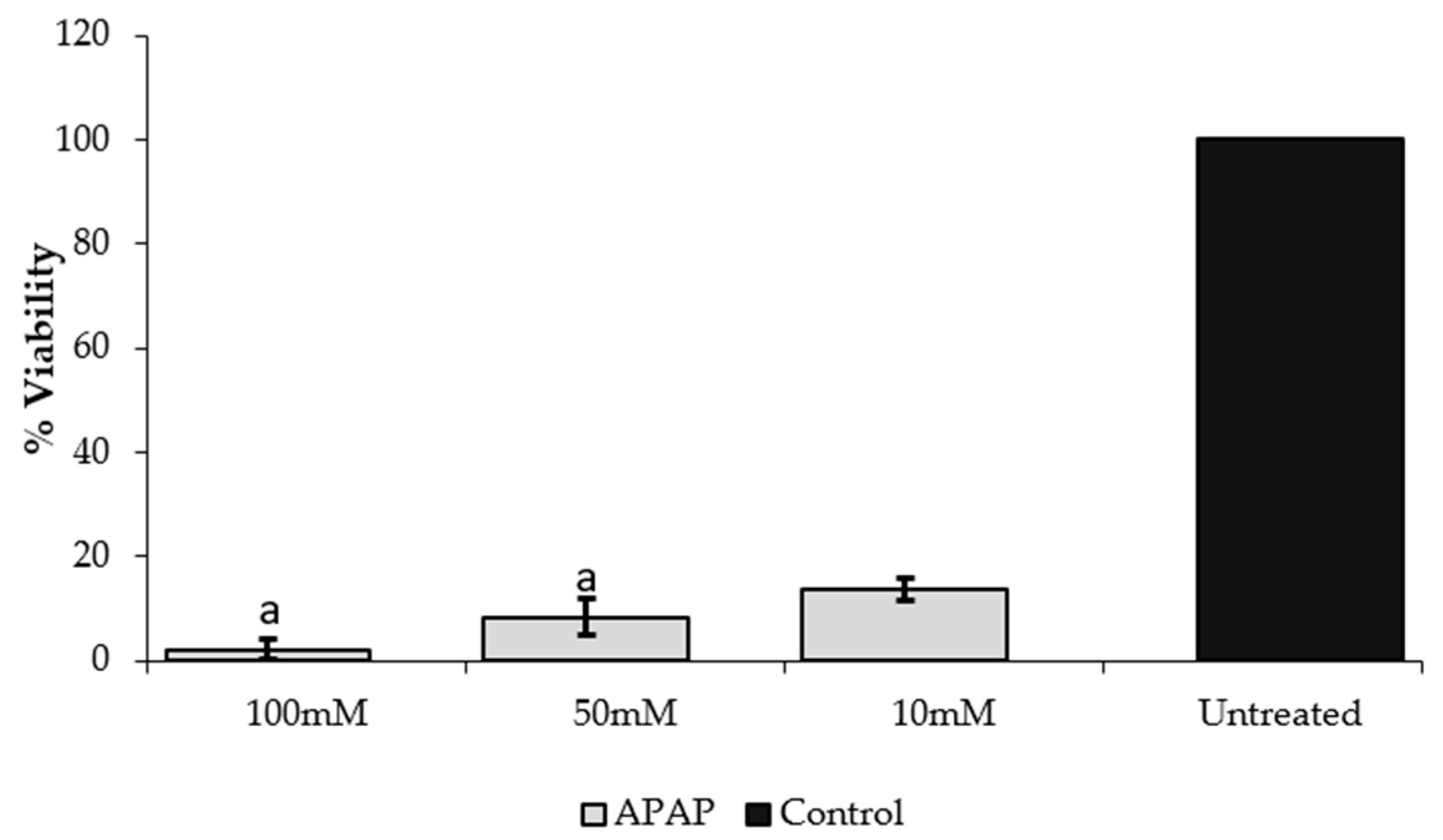
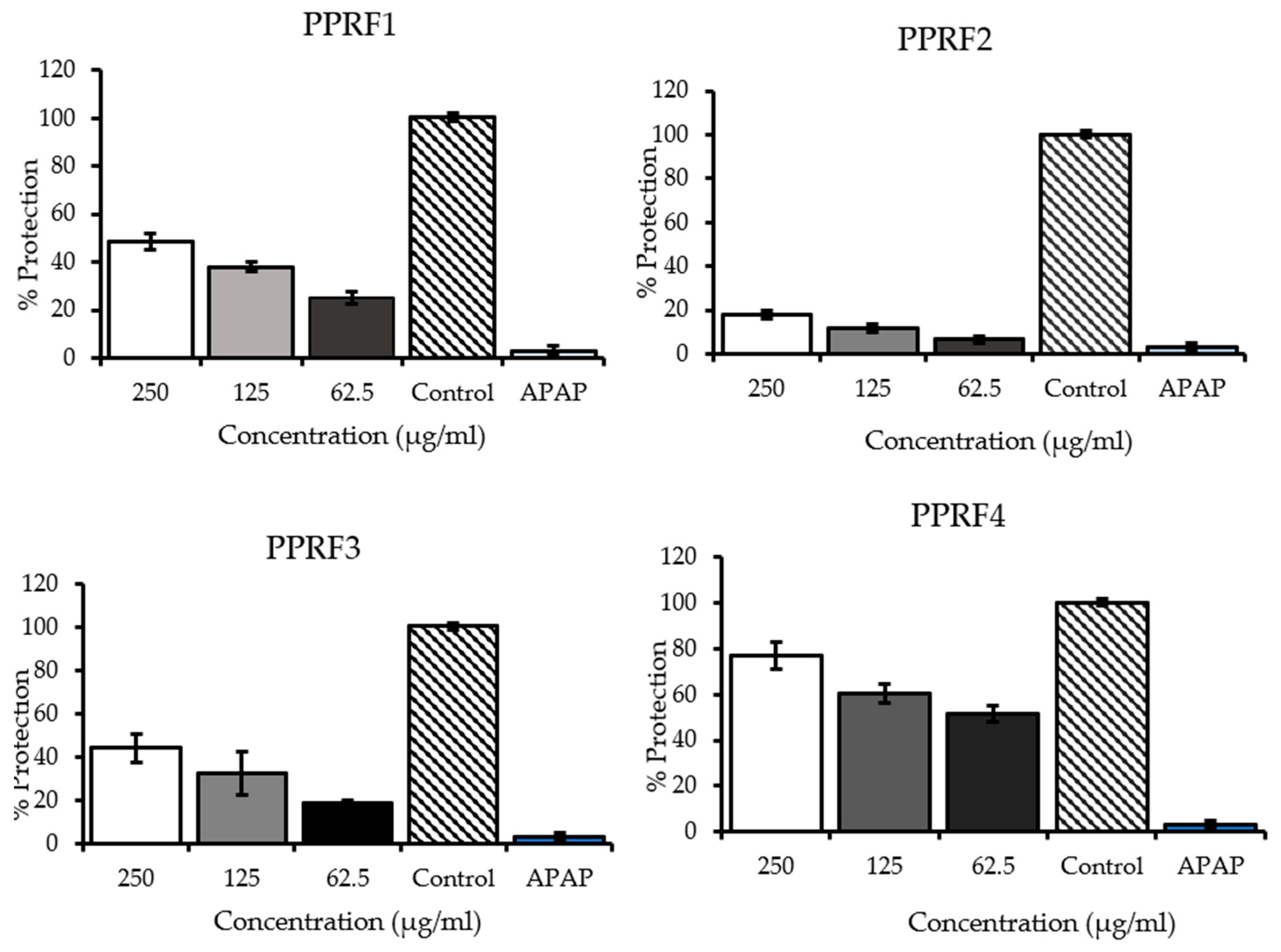
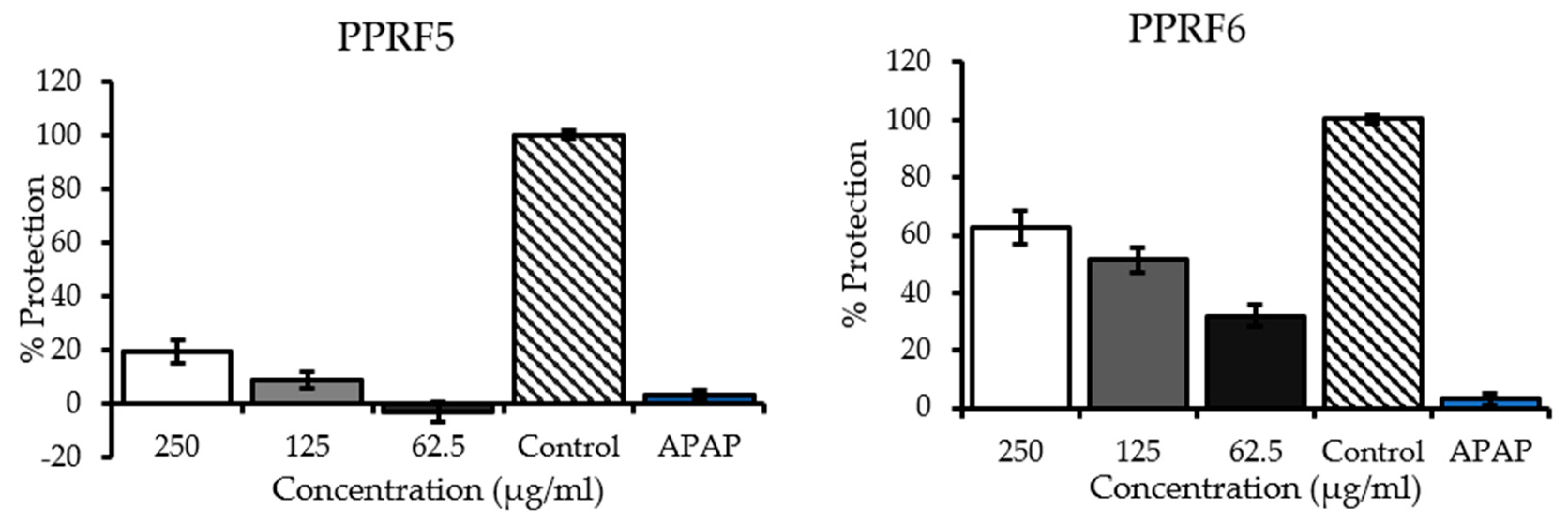
2.4. Inhibition of Acetaminophen-Induced Hepatotoxicity
2.5. Determination of Enzymatic Activities
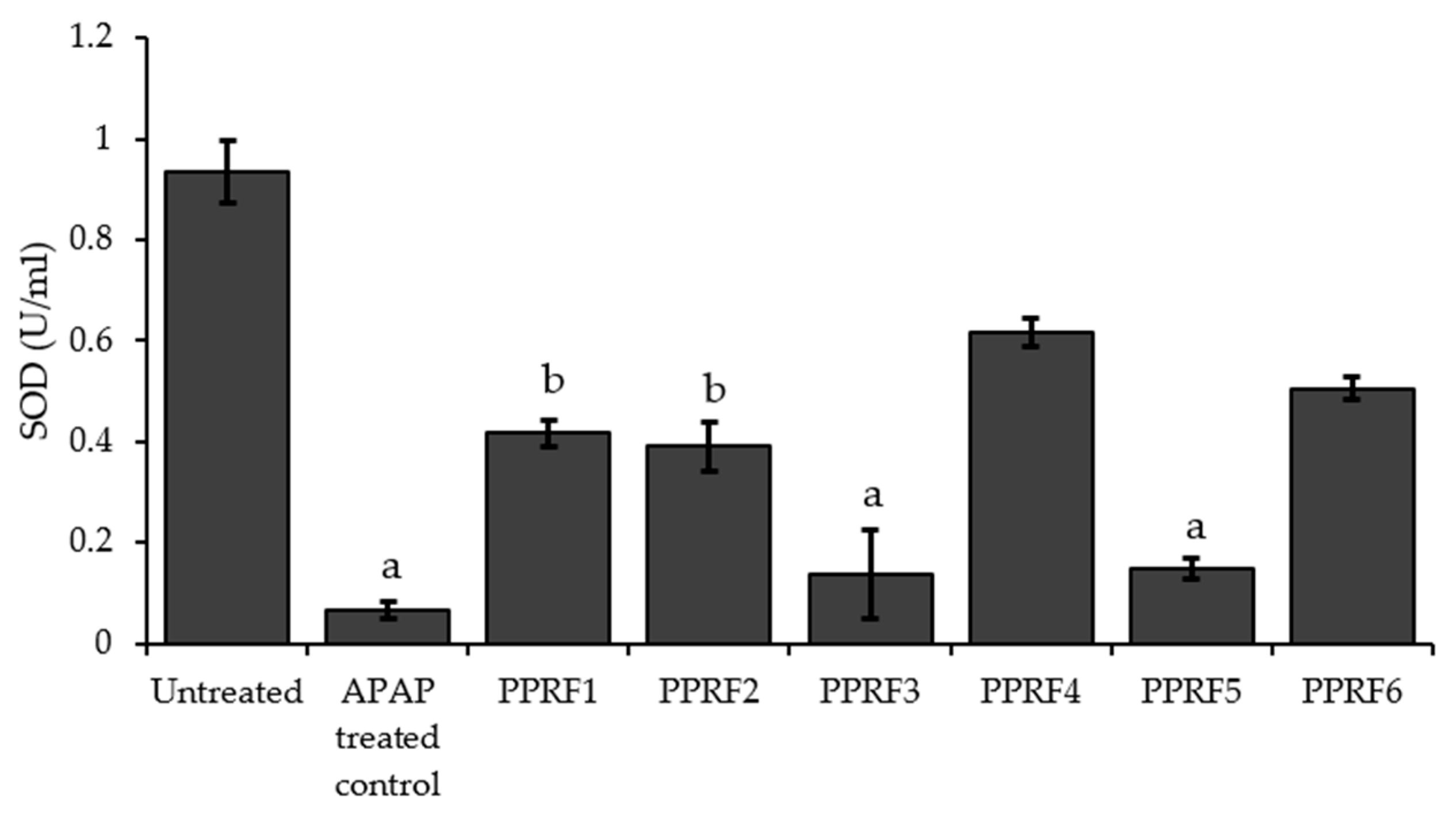
2.6. Superoxide Dismutase (SOD) Activity
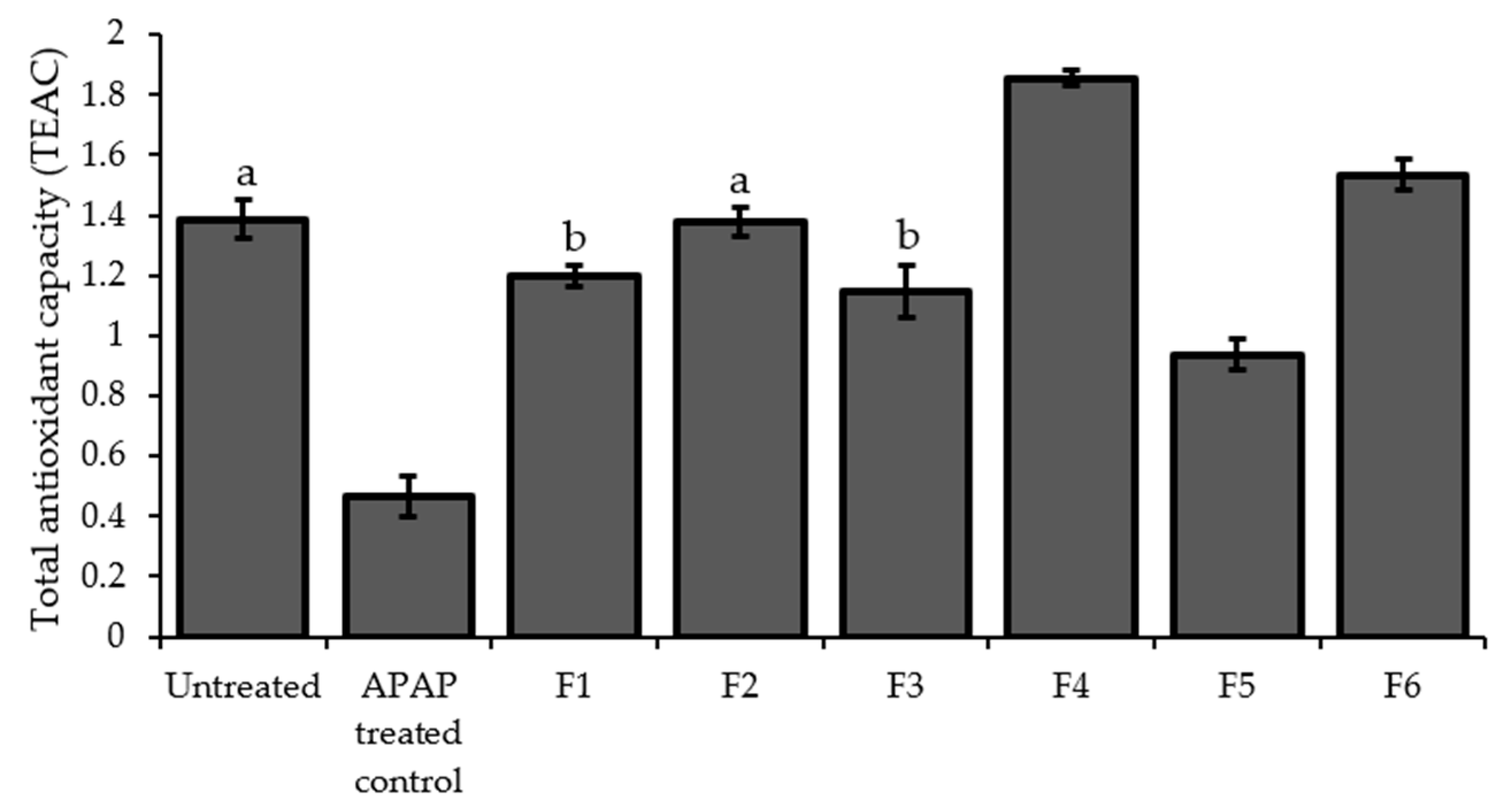
2.7. Total Antioxidant Capacity (TAOxC)
2.8. Reduced Glutathione (GSH) Content
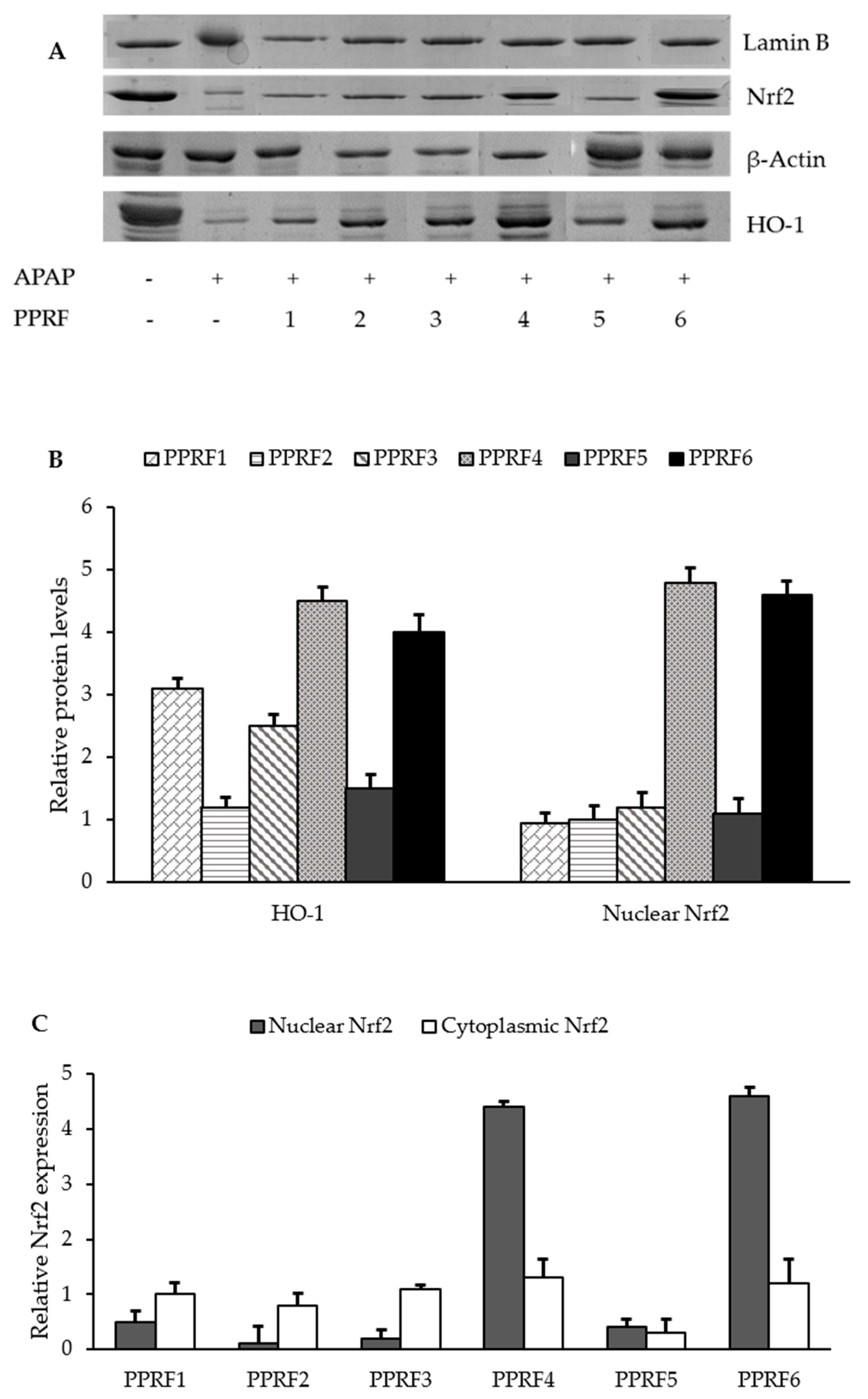
2.9. HO-1 and Nrf2 were Involved in the Protective Activity of PPRFs against APAP-Induced Hepatotoxicity
2.10. Compounds Identified in the PPRFs
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Plant Material
4.2.1. Fractionation
4.2.2. Determination of Total Phenolic Content
4.3. Antioxidant Assay
4.3.1. 2,2-diphenyl-1-picrylhydrazyl (DPPH) Radical Scavenging Assay
4.3.2. 2,2′-Azino-bis (3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) Radical Scavenging Assay
4.3.3. Ferric Reducing/Antioxidant Power (FRAP) Assay
4.4. Cell Culture
Cell Viability Assay
4.5. Inhibition of Acetaminophen-Induced Hepatotoxicity
4.6. Determination of Enzymatic Activities/ Biochemical Parameters of Liver Injury
4.7. Measurement of Superoxide Dismutase (SOD) Activity
4.8. Measurement of Antioxidant Capacity (TAOxC)
4.9. Measurement of Reduced Glutathione (GSH) Content
4.10. Protein Extraction
4.11. Western Blot Analysis
4.12. Ultra-High Performance Liquid Chromatography Mass Spectrometry Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hiraganahalli, B.D.; Chinampudur, V.C.; Dethe, S.; Mundkinajeddu, D.; Pandre, M.K.; Balachandran, J.; Agarwal, A. Hepatoprotective and antioxidant activity of standardized herbal extracts. Pharmacogn. Mag. 2012, 8, 116. [Google Scholar] [CrossRef] [PubMed]
- De Andrade, K.; Moura, F.; dos Santos, J.; de Araújo, O.; de Farias Santos, J.; Goulart, M. Oxidative Stress and Inflammation in Hepatic Diseases: Therapeutic Possibilities of N-Acetylcysteine. Int. J. Mol. Sci. 2015, 16, 30269–30308. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-Q.; Chen, P.; Su, W.-W.; Wang, Y.-G.; Wu, H.; Peng, W.; Li, P.-B.; Huang, Z.-Q.; Chen, P.; Su, W.-W.; et al. Antioxidant activity and hepatoprotective potential of quercetin 7-rhamnoside in vitro and in vivo. Molecules 2018, 23, 1188. [Google Scholar] [CrossRef] [PubMed]
- Ved, A.; Gupta, A.; Rawat, A.K.S. Antioxidant and hepatoprotective potential of phenol-rich fraction of Juniperus communis Linn. leaves. Pharmacogn. Mag. 2017, 13, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Prescott, L.F. Paracetamol, alcohol and the liver. Br. J. Clin. Pharmacol. 2000, 49, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Dear, J.W.; Antoine, D.J.; Park, B.K. Where are we now with paracetamol? BMJ 2015, 351, h3705. [Google Scholar] [CrossRef]
- McCrae, J.C.; Morrison, E.E.; MacIntyre, I.M.; Dear, J.W.; Webb, D.J. Long-term adverse effects of paracetamol—A review. Br. J. Clin. Pharmacol. 2018, 84, 2218–2230. [Google Scholar] [CrossRef]
- Mazaleuskaya, L.L.; Sangkuhl, K.; Thorn, C.F.; FitzGerald, G.A.; Altman, R.B.; Klein, T.E. PharmGKB summary: Pathways of acetaminophen metabolism at the therapeutic versus toxic doses. Pharmacogenet. Genomics 2015, 25, 416–426. [Google Scholar] [CrossRef]
- Hodgman, M.J.; Garrard, A.R. A review of acetaminophen poisoning. Crit. Care Clin. 2012, 28, 499–516. [Google Scholar] [CrossRef]
- Odeyemi, S.; Bradley, G. Medicinal plants used for the traditional management of diabetes in the Eastern Cape, South Africa: Pharmacology and toxicology. Molecules 2018, 23, 2759. [Google Scholar] [CrossRef]
- De Vynck, J.C.; Van Wyk, B.-E.; Cowling, R.M. Indigenous edible plant use by contemporary Khoe-San descendants of South Africa’s Cape South Coast. South. Afr. J. Bot. 2016, 102, 60–69. [Google Scholar] [CrossRef]
- Oyedemi, S.; Bradley, G.; Afolayan, A.J. Ethnobotanical survey of medicinal plants used for the management of diabetes mellitus in the Nkonkobe municipality of South Africa. J. Med. Plants Res. 2009, 3, 1040–1044. [Google Scholar]
- Abosede Wintola, O.; Jide Afolayan, A. Toxicological evaluation of aqueous extract of Lauridia tetragona (L.F.) R.H. Archer leaf in wistar rats. Int. J. Pharmacol. 2018, 14, 239–247. [Google Scholar] [CrossRef]
- Dash, D.K.; Yeligar, V.C.; Nayak, S.S.; Ghosh, T.; Rajalingam, D.; Sengupta, P.; Maiti, B.C.; Maity, T.K. Evaluation of hepatoprotective and antioxidant activity of Ichnocarpus frutescens (Linn.) R.Br. on paracetamol-induced hepatotoxicity in rats. Trop. J. Pharm. Res. 2007, 6, 755–765. [Google Scholar] [CrossRef]
- Holden, P.R.; James, N.H.; Brooks, A.N.; Roberts, R.A.; Kimber, I.; Pennie, W.D. Identification of a possible association between carbon tetrachloride-induced hepatotoxicity and interleukin-8 expression. J. Biochem. Mol. Toxicol. 2000, 14, 283–290. [Google Scholar] [CrossRef]
- Harries, H.; Fletcher, S.; Duggan, C.; Baker, V. The use of genomics technology to investigate gene expression changes in cultured human liver cells. Toxicol. Vitr. 2001, 15, 399–405. [Google Scholar] [CrossRef]
- Torres-González, L.; Muñoz-Espinosa, L.E.; Rivas-Estilla, A.M.; Trujillo-Murillo, K.; Salazar-Aranda, R.; Waksman, N.; Torres, D.; Cordero-Pérez, P. Protective effect of four Mexican plants against CCl 4-induced damage on the Huh7 human hepatoma cell line. Ann. Hepatol. 2016, 10, 73–79. [Google Scholar]
- González, L.T.; Minsky, N.W.; Espinosa, L.E.M.; Aranda, R.S.; Meseguer, J.P.; Pérez, P.C. In vitro assessment of hepatoprotective agents against damage induced by acetaminophen and CCl4. BMC Complement. Altern. Med. 2017, 17, 39. [Google Scholar] [CrossRef]
- Madrigal-Santillán, E.; Madrigal-Bujaidar, E.; Álvarez-González, I.; Sumaya-Martínez, M.T.; Gutiérrez-Salinas, J.; Bautista, M.; Morales-González, Á.; García-Luna y González-Rubio, M.; Aguilar-Faisal, J.L.; Morales-González, J.A. Review of natural products with hepatoprotective effects. World J. Gastroenterol. 2014, 20, 14787–14804. [Google Scholar] [CrossRef]
- Kepekçi, R.A.; Polat, S.; Çelik, A.; Bayat, N.; Saygideger, S.D. Protective effect of Spirulina platensis enriched in phenolic compounds against hepatotoxicity induced by CCl4. Food Chem. 2013, 141, 1972–1979. [Google Scholar] [CrossRef]
- Cardile, A.P.; Mbuy, G.K.N. Anti-herpes virus activity of silibinin, the primary active component of Silybum marianum. J. Herb. Med. 2013, 3, 132–136. [Google Scholar] [CrossRef]
- McGill, M.R.; Williams, C.D.; Xie, Y.; Ramachandran, A.; Jaeschke, H. Acetaminophen-induced liver injury in rats and mice: Comparison of protein adducts, mitochondrial dysfunction, and oxidative stress in the mechanism of toxicity. Toxicol. Appl. Pharmacol. 2012, 264, 387–394. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.R.; Schaffer, L.F.; Busanello, A.; Barbosa, C.P.; Peroza, L.R.; de Freitas, C.M.; Krum, B.N.; Bressan, G.N.; Boligon, A.A.; Athayde, M.L.; et al. Silymarin has antioxidant potential and changes the activity of Na+/K+-ATPase and monoamine oxidase in vitro. Ind. Crops Prod. 2015, 70, 347–355. [Google Scholar] [CrossRef]
- Gažák, R.; Purchartová, K.; Marhol, P.; Živná, L.; Sedmera, P.; Valentová, K.; Kato, N.; Matsumura, H.; Kaihatsu, K.; Křen, V. Antioxidant and antiviral activities of silybin fatty acid conjugates. Eur. J. Med. Chem. 2010, 45, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Salomone, F.; Galvano, F.; Li Volti, G. Molecular bases underlying the hepatoprotective effects of coffee. Nutrients 2017, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Atmaca, M.; Bilgin, H.M.; Obay, B.D.; Diken, H.; Kelle, M.; Kale, E. The hepatoprotective effect of coumarin and coumarin derivates on carbon tetrachloride-induced hepatic injury by antioxidative activities in rats. J. Physiol. Biochem. 2011, 67, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Wang, F.; Duan, M.; Cao, L.; Zhang, Y.; Yao, X.; Tang, J. Coumarin analogues from the Citrus grandis (L.) osbeck and their hepatoprotective activity. J. Agric. Food Chem. 2019, 67, 1937–1947. [Google Scholar] [CrossRef]
- Li, C.; Li, L.; Yang, C.; Zhong, Y.; Wu, D.; Shi, L.; Chen, L.; Li, Y. Hepatoprotective effects of Methyl ferulic acid on alcohol-induced liver oxidative injury in mice by inhibiting the NOX4/ROS-MAPK pathway. Biochem. Biophys. Res. Commun. 2017, 493, 277–285. [Google Scholar] [CrossRef]
- Rush, G.F.; Gorski, J.R.; Ripple, M.G.; Sowinski, J.; Bugelski, P.; Hewitt, W.R. Organic hydroperoxide-induced lipid peroxidation and cell death in isolated hepatocytes. Toxicol. Appl. Pharmacol. 1985, 78, 473–483. [Google Scholar] [CrossRef]
- Armentano, M.F.; Bisaccia, F.; Miglionico, R.; Russo, D.; Nolfi, N.; Carmosino, M.; Andrade, P.B.; Valentão, P.; Sissokho Diop, M.; Milella, L. Antioxidant and proapoptotic activities of Sclerocarya birrea [(A. Rich.) Hochst.] methanolic root extract on the hepatocellular carcinoma cell line HepG2. BioMed Res. Int. 2015, 2015, 561589. [Google Scholar] [CrossRef]
- Odeyemi, S.; Afolayan, A.; Bradley, G. In vitro anti-inflammatory and free radical scavenging activities of crude saponins extracted from Albuca bracteata Jacq. Bulb. Afr. J. Tradit. Complement. Altern. Med. 2015, 12, 34–40. [Google Scholar] [CrossRef]
- Odeyemi, S.; Afolayan, A.; Bradley, G. Phytochemical analysis and anti-oxidant activities of Albuca bracteata Jacq. and Albuca setosa Jacq bulb extracts used for the management of diabetes in the Eastern Cape, South Africa. Asian Pac. J. Trop. Biomed. 2017, 7, 577–584. [Google Scholar] [CrossRef]
- Thaipong, K.; Boonprakob, U.; Crosby, K.; Cisneros-Zevallos, L.; Hawkins Byrne, D. Comparison of ABTS, DPPH, FRAP, and ORAC assays for estimating antioxidant activity from guava fruit extracts. J. Food Compos. Anal. 2006, 19, 669–675. [Google Scholar] [CrossRef]
- Odeyemi, S.W.; Afolayan, A.J. Identification of antidiabetic compounds from polyphenolic-rich fractions of Bulbine abyssinica A. rich leaves. Pharmacognosy Res. 2018, 10, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-U.; Bae, E.-A.; Kim, D.-H. Hepatoprotective effects of irisolidone on tert-butyl hyperoxide-induced liver injury. Biol. Pharm. Bull. 2005, 28, 531–533. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kinjo, J.; Hirakawa, T.; Tsuchihashi, R.; Nagao, T.; Okawa, M.; Nohara, T.; Okabe, H. Hepatoprotective Constituents in Plants 14. Effects of Soyasapogenol B, Sophoradiol, and their glucuronides on the cytotoxicity of tert-butyl hydroperoxide to HepG2 cells. Biol. Pharm. Bull. 2003, 26, 1357–1360. [Google Scholar] [CrossRef] [PubMed][Green Version]
Sample Availability: Samples of the PPRFs 1–6 are available from the authors. |


| Samples | Phenolic Content |
|---|---|
| µg QE/mg of sample | |
| PPRF1 | 20.51 ± 2.9 |
| PPRF2 | 16.38 ± 1.08 |
| PPRF3 | 26.47 ± 1.00 |
| PPRF4 | 22.13 ± 2.09 |
| PPRF5 | 15.62 ± 1.93 |
| PPRF6 | 16.35 ± 2.79 |
| Samples | DPPH IC50 (µmol AAE/mL) | ABTS IC50 (µmol AAE/mL) | FRAP IC50 (µg AAE/mL) |
|---|---|---|---|
| PPRF1 | 130.459 ± 24.9 | 310.459 ± 24.9 | 1761.49 ± 22.4 |
| PPRF2 | 212.95 ± 26.57 | 302.95 ± 26.57 | 1676.44 ± 70.75 |
| PPRF3 | 124.153 ± 12.39 | 251.51 ± 12.39 | 1262.13 ± 32.94 |
| PPRF4 | 187.23 ± 20.69 | 296.23 ± 20.69 | 4423.06 ± 26.06 |
| PPRF5 | 99.59 ± 13.92 | 258.11 ± 8.32 | 2235. 63 ± 21.21 |
| PPRF6 | 137.77 ± 19.66 | 351.11 ± 20.31 | 2220.89 ± 29.62 |
| Samples | IC50 |
|---|---|
| μg/mL | |
| PPRF1 | 251.935 ± 6.7 |
| PPRF2 | 788.14 ± 4.2 |
| PPRF3 | 287.893 ± 2.11 |
| PPRF4 | 50.243 ± 8.03 |
| PPRF5 | 510.35 ± 3.4 |
| PPRF6 | 154.59 ± 1.9 |
| Samples | APAP | Dose | AST | ALT | LDH |
|---|---|---|---|---|---|
| (µg/mL) | (mU/mL) | (mU/mL) | |||
| PPRF1 | + | 500 | 32.60 ± 3.1 # | 29.06 ± 3.6 | 39.82 ± 4.2 # |
| + | 250 | 39.80 ± 9.3 | 33.39 ± 3.3 | 44.94 ± 2.5 # | |
| + | 125 | 44.15± 3.9 | 35.10 ± 3.5 | 49.82 ± 2.2 # | |
| PPRF2 | + | 500 | 34.24 ± 4.3 # | 23.36 ± 3.2 # | 34.67 ± 2.5 # |
| + | 250 | 41.10 ± 2.1 | 29.49 ± 2.1 | 38.62 ± 5.2 # | |
| + | 125 | 46.35± 4.2 | 33.49 ± 3.7 | 40.01 ± 0.2 # | |
| PPRF3 | + | 500 | 26.22 ± 3.6 # | 20.26 ± 2.2 # | 26.01 ± 3.2 # |
| + | 250 | 31.40 ± 1.3 # | 25.15 ± 1.5 # | 27.57 ± 8.1 # | |
| + | 125 | 36.33± 4.6 | 29.17 ± 5.2 | 31.11 ± 4.2 # | |
| PPRF4 | + | 500 | 12.98 ± 4.3 # | 9.26 ± 4.2 # | 27.88 ± 3.1 # |
| + | 250 | 19.78 ± 2.1 # | 15.48 ± 2.2 # | 23.8 ± 4.1 # | |
| + | 125 | 25.25± 3.2 # | 19.96 ± 2.6 # | 17.18 ± 1.6 # | |
| PPRF5 | + | 500 | 31.44 ± 7.3 # | 27.26 ± 2.4 | 32.19 ± 2.4 # |
| + | 250 | 36.51 ± 3.9 | 31.42 ± 4.4 | 35.84 ± 4.2 # | |
| + | 125 | 41.20 ± 9.7 | 36.33 ± 4.7 | 39.61 ± 5.3 # | |
| PPRF6 | + | 500 | 18.34 ± 4.4 # | 25.43 ± 4.9 | 33.05 ± 0.8 # |
| + | 250 | 22.41 ± 4.3 # | 31.02 ± 6.2 | 29.23 ± 3.4 # | |
| + | 125 | 35.11 ± 3.6 # | 38.10 ± 1.5 | 22.11 ± 2.3 # | |
| Control | - | - | 5.20 ± 1.2 | 3.26 ± 1.7 | 0.89 ± 2.3 |
| APAP-treated control | + | 100 mM | 46.50 ± 6.4 * | 35.05 ± 5.3 * | 65.95 ± 7.3 * |
| Compound Number | Major Compounds | Fraction(s) | tR (min) | Formula | Measured Mass (m/z) | Positive ESI Mode | ||
|---|---|---|---|---|---|---|---|---|
| Adduct Ions | Mass Error (ppm) | MSn Fragment Ions | ||||||
| 1 | Diethyl phthalate | 1,2,3,4,5,6 | 0.2182 | C12H14O4 | 222.24 | 221.089 [M + H]+ | 0.5 | 65.039 [C5H4]+H+, 93.034 [C6H4O]+H+, 121.028 [C7H4O2]+H+ |
| 2 | Homovanillic acid | 6 | 0.8032 | C9H10O4 | 182.17 | 181.058 [M + H]+ | 1.2 | 110.036 [C6H4O2+H]+H+, 110.036 [C6H4O2+H]+H+, |
| 3 | Varenicline | 2,4,5,6 | 1.2265 | C13H13N3 | 212. 11 | 211.267 [M + H]+ | −0.7 | 77.039 [C6H4]+H+, 94.065 [C6H7N]+H+, 119.06 [C7H8N2-H]+, 195.092 [C13H11N2]+ |
| 4 | Pedaliin | 1,3 | 1.8622 | C22H22O12 | 479.23 | 478.112 [M + H]+ | 7.1 | 83.049 [C5H4O+2H]+H+, 83.049 [C5H4O+2H]+H+, 111.044 [C6H5O2+H]+H+, 111.044 [C6H5O2+H]+H+, 139.039 [C7H5O3+H]+H+, 139.039 [C7H5O3+H]+H+ |
| 5 | 3-Isopropylcatechol | 2,4,5,6 | 3.2675 | C9H12O2 | 153.09 | 152.084 | −0.4 | 67.05426 [C5H8-H]+ |
| 6 | 2-Methoxy-4-vinylphenol | 4,5,6 | 3.829 | C9H10O2 | 151.07 | 150.177 [M + H]+ | 0.6 | 103.054 [C8H6]+H+, 117.034 [C8H6O-H]+, 121.065 [C8H7O+H]+H+ |
| 7 | Caffeine | 4,6 | 4.169 | C8H10N4O2 | 195.08 | 194.19 [M + H]+ | −0.2 | 110.035 [C4H6N3O-2H]+, 110.035 [C4H6N3O-2H]+, 152.058 [C7H7N2O2]+H+, 152.058 [C7H7N2O2]+H+ |
| 8 | 3-tert-Butyl-5-methylcatechol | 4,6 | 4.2581 | C11H16O2 | 181. 12 | 180.247 [M + H]+ | −0.1 | 93.07 [C7H10-H]+, 121.065 [C8H11O-2H]+, 149.06 [C9H10O2-H]+ |
| 9 | Coumarin | 4,6 | 5.0014 | C9H6O2 | 147.619 | 146.037 [M + H]+ | 5.6 | 77.039 [C6H4]+H+, 103.053 [C8H6]+H+, 117.034 [C8H6O-H]+ |
| 10 | Gambiriin A2 | 1 | 5.193 | C30H28O12 | 581.1654 | 580.1581 [M + H]+ | −1.3 | 503.134 [C28H25O9-2H]+, 563.155 [C30H27O11]+ |
| 11 | Gentioflavine | 2,4,6 | 5.2539 | C10H11NO3 | 194.72 | 193.07393 [M + H]+ | −1.3 | 97.028 [C5H5O2]+, 112.039 [C5H6NO2]+, 127.039 [C6H5O3+H]+H+ |
| 12 | Herniarin | 2,4,5,6 | 6.0091 | C10H8O3 | 177.0546 | 176.047 [M + H]+ | −1.1 | 121.028 [C7H6O2-H]+, 149.023 [C8H6O3-H]+ |
| 13 | Ferulic acid | 2,4,5,6 | 6.242 | C10H10O4 | 195.18 | 194.0594 [M + H]+ | −0.1 | 77.0386 [C6H5]+, 117.034 [C8H6O-H]+, 121.065 [C8H6O+2H]+H+, 145.028 [C9H6O2-H]+, 149.06 [C9H9O2]+, 177.055 [C10H9O3]+ |
| 14 | 3-Aminophenol | 1,3 | 8.0365 | C6H7NO | 110.13 | 109.05279 [M + H]+ | 0.8 | 77.039 [C6H4]+H+ |
| 15 | 4-[(2-tert-butoxy-2-oxo-ethyl)-(3-methoxy-3-oxo-propyl)amino]-3-nitro-benzenesulfonic acid | 1,3 | 8.7380 | C23H16O8 | 419.212 | 418.105 [M + H]+ | 3.5 | 315.065 [C12H15N2O6S]+, 315.065 [C12H15N2O6S]+, 330.088 [C13H17N2O6S]+H+, 401.101 [C16H22N2O8S-H]+ |
| 16 | 4-Piperidone | 1,3 | 9.0382 | C5H9NO | 99.13 | 100.068 [M + H]+ | 10.2 | 72.081 [C4H9N]+H+ |
| 17 | Coumarin 314 | 1,3 | 10.372 | C18H19NO4 | 314.1387 | 313.131 [M + H]+ | −1.3 | 77.0386 [C6H4]+H+, 93.07 [C7H6+2H]+H+, 103.054 [C8H6]+H+, 117.034 [C8H6O-H]+, 121.065 [C8H9O]+, 145.028 [C9H6O2-H]+, 149.06 [C9H9O2]+, 177.055 [C10H9O3]+ |
| 18 | Petunidin 3-O-glucoside | 1 | 11.1382 | C22H22O12 | 479.1184 | 478.111 [M + H]+ | 5.4 | 85.028 [C4H5O2]+, 302.042 [C15H9O7]+H+, 317.066 [C16H12O7]+H+ |
| 19 | Venlafaxine | 1,3 | 11.8120 | C17H28NO2 | 278.2115 | 277.204 [M + H]+ | −1.7 | 163.112 [C11H15O]+, 219.174 [C15H20O+2H]+H+, 222.149 [C13H19NO2]+H+, |
| 20 | Sterculic acid | 4,6 | 9.94 | C19H34O2 | 295.2632 | 294.256 [M + H]+ | −0.2 | 69.06992 [C5H10-H]+, 83.086 [C6H12-H]+, 97.101 [C7H14-H]+, 111.117 [C8H12+2H]+H+, 123.117 [C9H14]+H+, 135.117 [C10H16-H]+, 149.133 [C11H18-H]+, 163.148 [C12H20-H]+, 263.237 [C18H30O]+H+ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Odeyemi, S.; Dewar, J. Repression of Acetaminophen-Induced Hepatotoxicity in HepG2 Cells by Polyphenolic Compounds from Lauridia tetragona (L.f.) R.H. Archer. Molecules 2019, 24, 2118. https://doi.org/10.3390/molecules24112118
Odeyemi S, Dewar J. Repression of Acetaminophen-Induced Hepatotoxicity in HepG2 Cells by Polyphenolic Compounds from Lauridia tetragona (L.f.) R.H. Archer. Molecules. 2019; 24(11):2118. https://doi.org/10.3390/molecules24112118
Chicago/Turabian StyleOdeyemi, Samuel, and John Dewar. 2019. "Repression of Acetaminophen-Induced Hepatotoxicity in HepG2 Cells by Polyphenolic Compounds from Lauridia tetragona (L.f.) R.H. Archer" Molecules 24, no. 11: 2118. https://doi.org/10.3390/molecules24112118
APA StyleOdeyemi, S., & Dewar, J. (2019). Repression of Acetaminophen-Induced Hepatotoxicity in HepG2 Cells by Polyphenolic Compounds from Lauridia tetragona (L.f.) R.H. Archer. Molecules, 24(11), 2118. https://doi.org/10.3390/molecules24112118






