Abstract
In relation to anti-inflammatory agents from medicinal plants, we have isolated three compounds from Atractylodes macrocephala; 1, 2-[(2E)-3,7-dimethyl-2,6-octadienyl]-6-methyl-2, 5-cyclohexadiene-1, 4-dione; 2, 1-acetoxy-tetradeca-6E,12E-diene-8, 10-diyne-3-ol; 3, 1,3-diacetoxy-tetradeca-6E, 12E-diene-8, 10-diyne. Compounds 1–3 showed concentration-dependent inhibitory effects on production of nitric oxide (NO) and prostaglandin E2 (PGE2) in lipopolysaccharide (LPS)-activated RAW 264.7 macrophages. Western blotting and RT-PCR analyses demonstrated that compounds 1–3 suppressed the protein and mRNA levels of inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2). Furthermore, compounds 1–3 inhibited transcriptional activity of nuclear factor-κB (NF-κB) and nuclear translocation of NF-κB in LPS-activated RAW 264.7 cells. The most active compound among them, compound 1, could reduce the mRNA levels of pro-inflammatory cytokines (IL-1β, IL-6 and TNF-α) and suppress the phosphorylation of MAPK including p38, JNK, and ERK1/2. Taken together, these results suggest that compounds 1–3 from A. macrocephala can be therapeutic candidates to treat inflammatory diseases.
1. Introduction
Atractylodes macrocephala, is a perennial herb that has been used as a traditional medicine in Korea, China, and Japan for thousands of years to treat gastrointestinal dysfunction. Examples may include loss of appetite, abdominal distention, and diarrhea [1,2]. The A. macrocephala has been reported to possess diverse biological activities, including improved functional defects in digestive system, as well as anti-tumor, anti-inflammatory, anti-aging, anti-oxidative, and anti-bacterial activities [3,4,5,6,7]. A. macrocephala have been reported to contain sesquiterpenoids, polyacetylenes, phenylpropanoids, flavonoids, and polysaccharides [8]. Some sesquiterpenoids such as atractylenolide I~III exhibited inhibitory effect on LPS-induced NO production in macrophages and neuroprotective effects in the Parkinson’s disease model [8,9,10,11]. Moreover, polyacetylenes including atractylodemayne A, E, F, G, and 14-acetoxy-12-senecioyloxytetradeca-2E,8E,10E-trien-4,6-diyn-1-ol showed anti-inflammatory potential against LPS-induced NO production and carrageenan-induced paw edema without their detailed action mechanisms [8,12,13].
Inflammation is a complicated physiological response to various immune stimuli such as infection and tissue injury, which is regulated by diverse inflammatory mediators. Although highly regulated inflammatory response is beneficial for body functions, immoderate and deregulated inflammation can cause tissue damages or chronic diseases such as cancer, diabetes, obesity, and Parkinson’s diseases [14,15,16].
Macrophage, major immune effector cell, plays an essential role as a responder to inflammation. Macrophage can be stimulated by external pathogen such as lipopolysaccharide (LPS). The activated macrophage contributes and accelerates immune response to induce expression of pro-inflammatory cytokines and inflammatory mediators including nitric oxide (NO) and prostaglandin E2 (PGE2) [17]. NO is synthesized by nitric oxide synthases (NOS) such as neuronal NOS (nNOS), endothelial NOS (eNOS), and inducible NOS (iNOS). Large amounts of NO are mostly released by iNOS which can be highly expressed in chronic inflammatory disorders [18]. Alternatively, PGE2 is synthesized by cyclooxygenase 1 (COX-1) or cyclooxygenase 2 (COX-2). PGE2 is abundantly produced by upregulated COX-2 in inflammatory state, which leads to pain and inflammation [19]. Therefore, inhibition of inflammatory responses in activated macrophages can be a preventive strategy to diminish excessive inflammation.
During searching for anti-inflammatory principles from A. macrocephala, we isolated one quinone compound and two polyacetylenes, which have not been reported for anti-inflammatory activity. In these studies, we evaluated the anti-inflammatory effect of three active constituents from A. macrocephala in LPS-activated RAW 264.7 macrophage cells and investigated their underlying action mechanisms.
2. Results and Discussion
Nitric oxide (NO), a free radical molecule, plays an important role in physiological activities in vascular and neuronal functions in normal conditions. Under inflammatory responses, activated macrophages release high level of NO which causes deleterious and severe inflammation [20]. PGE2, a lipid mediator synthesized from arachidonic acid, is abundantly produced in inflammatory reaction. They potentiate inflammation through eliciting vasodilation and increase of local blood flow [21]. Therefore, reducing the NO and PGE2 levels in LPS-activated macrophage cells is reasonable approach to suppress inflammatory response.
We purified compounds 1, 2, and 3 from A. macrocephala as inflammatory modulators in LPS-activated macrophages by the activity-guided purification process. Their structures were elucidated by the analysis of mass and nuclear magnetic resonance (NMR) spectroscopic data [22,23]. Compound 1 has quinone moiety, while compounds 2 and 3 have acetylene moiety (Figure 1A).
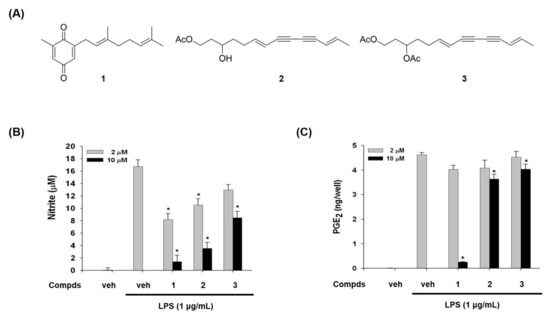
Figure 1.
Chemical structures of compounds 1–3 from A. macrocephala (A) and effects of compounds 1–3 on lipopolysaccharide (LPS)-induced nitric oxide (NO) and prostaglandin E2 (PGE2)production (B,C). (B) NO released from cells was assessed as nitrite form in culture supernatant. The amount of nitrite in culture medium was measured by using the Griess reagents, as described in materials and methods. (C) The levels of PGE2 in culture medium were determined by enzyme immunoassay method. Veh means vehicle. The values are expressed as the mean ± S.D. of three individual experiments. * p < 0.01 indicate significant differences from the LPS alone.
Compounds 1–3 inhibited LPS-induced NO production in a concentration-dependent manner, whereas LPS treatment dramatically increased NO in RAW 264.7 macrophage cells (Figure 1B). The IC50 values of compounds 1, 2, and 3 were 3.7, 21.1, and 60.4 μM, respectively. We have also examined the effects of compounds 1–3 on LPS-induced PGE2 production. As shown in Figure 1C, compound 1 suppressed PGE2 production, while the other two compounds 2 and 3 showed marginal suppression (Figure 1C). Especially IC50 for PGE2 of the compound 1 was 5.26 μM. These findings guided us to examine the effect of compounds 1–3 on expression levels of iNOS and COX-2 which produce NO and PGE2 as key mediators of inflammation.
To examine whether the suppressed production of NO and PGE2 by compounds 1–3 were related with modulation iNOS and COX-2 expressions, western blot and RT-PCR analyses were carried out. As shown in Figure 2A, compounds 1–3 (10 μM) attenuated the LPS-induced iNOS protein levels in RAW 264.7 cells whereas LPS treatment showed enhanced protein level of iNOS. Compounds 1–3 also suppressed protein levels of COX-2 compared with LPS treatment. Moreover, compounds 1–3 down-regulated the iNOS and COX-2 mRNA levels in LPS-stimulated RAW 264.7 cells (Figure 2B). Compound 1 among three compounds showed the most potent inhibitory effect on iNOS and COX-2 expressions. The suppressive effects of compounds 1–3 on iNOS and COX-2 expressions were in parallel with the NO and PGE2 levels. These results suggest that compounds 1–3 suppressed the production of NO and PGE2 inflammatory mediators by inhibiting expressions of iNOS and COX-2 protein and mRNA in LPS-induced macrophage cell system. Thus, we hypothesized that a transcription factor for iNOS and COX-2 gene expression would be affected by compounds 1–3.
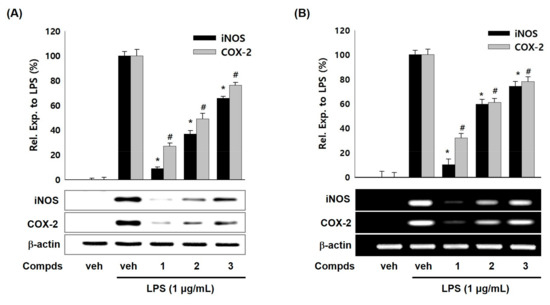
Figure 2.
Effects of compounds 1–3 on the expression of LPS-induced iNOS/COX-2 protein and mRNA in RAW 264.7 macrophages. (A) Cells were activated with LPS (1 μg/mL) in presence or absence of compounds 1–3 (10 μM) for 20 h. Cell lysates were prepared and the iNOS, COX-2, and β-actin protein levels were determined by Western blotting. (B) Cells were treated with compounds 1–3 (10 μM) and/or LPS (1 μg/mL) for 6 h. The mRNA levels for iNOS, COX-2, and β-actin were determined by RT-PCR. The relative intensity of iNOS/COX-2 to β-actin bands was measured by densitometry. Veh means vehicle. The values represented mean ± S.D. of three individual experiments. *, # p < 0.01 indicate significant difference (* iNOS, # COX-2) from LPS alone.
To disclose the action mode of compounds 1–3 for inhibition of transcriptional expression of iNOS and COX-2, we examined the effect of compounds 1–3 on activation of nuclear factor κB (NF-κB) which controls the expression of iNOS and COX-2 [24]. First, we assessed the secretory alkaline phosphatase (SEAP) activity by the transcriptional activation of NF-κB in T-RAW 264.7 cells with pNF-κB-SEAP-NPT reporter gene systems. As seen in Figure 3A, compounds 1–3 significantly repressed the SEAP activity, while LPS treatment markedly increased the SEAP activity. This result indicates that compounds 1–3 can suppress transcriptional activity of NF-κB.
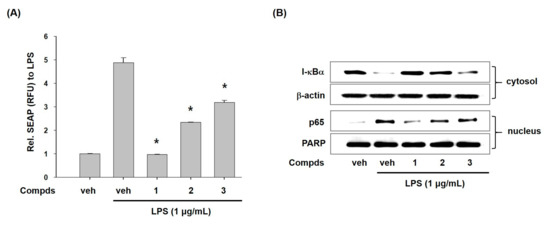
Figure 3.
Effects of compounds 1–3 on LPS-induced NF-κB activation. (A) Effects of compounds 1–3 on LPS-induced nuclear factor-κB (NF-κB) transcriptional activation in T-RAW cells (the stably transfected RAW 264.7 cells with pNF-κB-SEAP-NPT. The pNF-κB-SEAP-NPT plasmid that permits expression of the secretory alkaline phosphatase (SEAP) reporter gene in response to the NF-κB activity and contains the neomycin phosphotransferase (NPT) gene for geneticin resistance.). T-RAW cells were treated with compounds 1–3 (10 μM) for 2 h prior to stimulation of LPS for 16 h. The transcriptional activity was assessed by measuring SEAP activity and then expressed as relative fluorescence unit (RFU). (B) Effects of compounds 1–3 on I-κB-α degradation and p65 translocation to nucleus in LPS-stimulated RAW 264.7 macrophages. Cells were pre-treated with compounds 1–3 (10 μM) for 30 min prior to LPS treatment for 20 min. Cytoplasmic and nuclear extracts were prepared for western blotting of I-κB-α and p65, respectively. β-Actin and poly ADP-ribose polymerase (PARP) were used as loading controls. Veh means vehicle. The values are presented as mean ± S.D. of three individual experiments. *p < 0.01 indicate significant differences from the LPS alone.
Next, we assessed the effects of compounds 1–3 on LPS-induced degradation of I-κBα. NF-κB is a dimeric transcription factor complexed with p50 and p65 subunits. In normal conditions, NF-κB exists in cytoplasm as an inactive form (p50/p65 dimer) by physically binding with inhibitor-κB (I-κB) [25]. However, I-κB is phosphorylated, ubiquitinated, and quickly degraded to discharge p50/p65 in inflammatory situations. Released p50/p65 moves to nucleus and initiates the expression of inflammatory genes such as iNOS and COX-2 [26]. Here, we observed the effects of compounds 1–3 on degradation of I-κBα and translocation of p65 subunit in LPS-stimulated macrophages. As shown in Figure 3B, LPS treatment showed very low level of I-κBα protein whereas compounds 1–3 recovered level of I-κBα protein. Furthermore, treatment of cells with compounds 1–3 decreased the level of p65 in nucleus. These results indicated that compounds 1–3 inhibited NF-κB activation by modulating degradation of I-κBα and nuclear translocation of p65. These findings proposed that anti-inflammatory potential of compounds 1–3 could be partially associated with their inhibition of NF-κB.
NF-κB has been known to regulate a large array of genes involved in inflammatory responses. In addition, NF-κB is required for an induction of large numbers of inflammatory genes, including those encoding IL-1, IL-6 and TNF-α as well as iNOS and COX-2 [27]. Thus, we investigated the effect of compound 1 on LPS-induced pro-inflammatory cytokines such as IL-1, IL-6 and TNF-α, because compound 1 is the strongest inhibitor of NF-κB among the three compounds. As shown in Figure 4, compound 1 concentration-dependently reduced mRNA expressions of IL-1, IL-6 and TNF-α in LPS-stimulated RAW 264.7 macrophage cells. These suppressive effects of compound 1 on pro-inflammatory cytokines came from inhibition of I-κB degradation and NF-κB nuclear translocation.
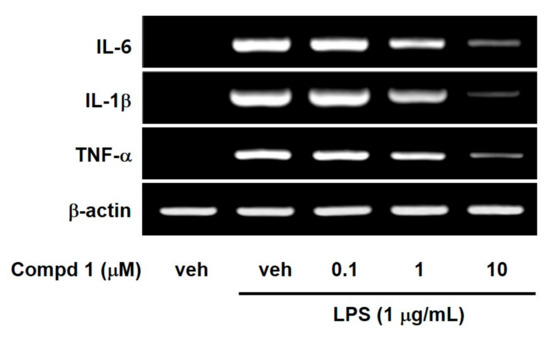
Figure 4.
Effects of compound 1 on the expression of IL-6, IL-1β and TNF-α mRNA in LPS-activated macrophages. RAW 264.7 cells were treated for 6 h with compound 1 at indicated concentrations during LPS (1 μg/mL) activation. The mRNA levels of IL-6, IL-1β, and TNF-α were determined by RT-PCR. Veh means vehicle. Images are the representative results of three separate experiments.
Mitogen-activated protein kinases (MAPKs) including p38, JNK, and ERK1/2 can regulate gene expression, cellular growth, and inflammatory response [28,29]. MAPKs can induce inflammatory cytokines in response to various inflammatory stimuli including LPS and TNF-α [30,31]. MAPKs signaling are positively associated with NF-κB activation [32,33]. To examine the effect of compound 1 on MAPKs signaling, we assessed the phosphorylation levels of MAPKs in LPS-activated macrophage cells. We observed the elevated phosphorylation of p38, JNK, and ERK1/2 by LPS treatment (Figure 5). However, compound 1 reduced the p38, JNK, and ERK1/2 phosphorylation in a concentration-dependent way with no alteration in total p38, JNK, and ERK1/2 levels. These results suggested that compound 1 could attenuate LPS-induced inflammation by suppression of MAPKs signaling pathway and NF-κB activation.
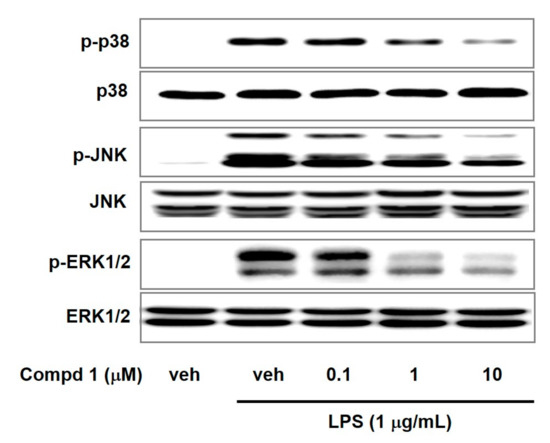
Figure 5.
Effects of compound 1 on phosphorylation of p38, JNK, and ERK1/2 in LPS-activated macrophages. RAW 264.7 cells were pretreated with compound 1 (0.1, 1, and 10 μM) for 30 min and incubated further with LPS (1 μg/mL) for 15 min. The protein levels of ERK, JNK and p38 in cell lysate were determined by western blotting. Veh means vehicle. Images are representative results of three independent experiments.
3. Materials and Methods
3.1. General Experimental Procedures
Mass spectra were obtained on a JEOL JMS-AX505WA mass spectrometer. NMR spectra were determined on a Varian UNITY INOVA 400 NMR spectrometer. Column chromatography was carried out over silica gel (40–60 μm, Merck, Kenilworth, NJ, USA). Fractions from column chromatography were monitored by thin layer chromatography (TLC) (silica gel 60 F254 and RP-C18 F254S, Merck) under UV light or by heating after spraying 10% H2SO4 in CH3OH (v/v).
3.2. Plant Materials, Extraction and Isolation
The dried rhizomes of A. macrocephala were purchased from the Kyungdong Herbal Market in Seoul, Korea. The air-dried material (10 kg) was extracted with 20 L of EtOAc three times. The EtOAc extract (310 g) was subjected to silica gel column chromatography eluting with n-hexane: EtOAc gradient system (100:1 to 1:2) to give 14 fractions. Fraction 9 (13.2 g), which suppressed NO production in LPS-stimulated RAW 264.7 cells, was further chromatographed on silica gel with n-hexane: acetone (30:1 to 1:2) to afford seven sub-fractions. By the activity-guided purification process, sub-fraction 9-6 (620 mg) was further purified by a silica gel column using n-hexane: EtOAc (30:1 to 1:2) to afford compound 1 (17 mg). Sub-fraction 9-4 (1.4 g) was re-chromatographed on silica gel with n-hexane: EtOAc (50:1 to 1:2) to yield compound 3 (53 mg). Fraction 12 (15.1 g) was chromatographed on silica gel with n-hexane: EtOAc gradient system (50:1 to 1:2) to afford 10 sub-fractions. Sub-fraction 12-6 (3.3 g) was further purified by a silica gel column with n-hexane: acetone (20:1 to 1:3) to yield compound 2 (45 mg). The structures of compounds 1–3 were identified as 2-[(2E)-3,7-dimethyl-2,6-octadienyl]-6-methyl-2,5-cyclohexadiene-1,4-dione (1), 1-acetoxy-tetradeca-6E,12E-diene-8,10-diyne-3-ol (2), and 1,3-diacetoxy-tetradeca-6E,12E-diene-8, 10-diyne (3) (Figure 1A) by spectroscopic analysis and comparison with the previously reported data [22,23]. Previously, compound 1 was reported from A. lancea [22] and A. macrocephala [34], and compounds 2 and 3 from A. koreana [23] and A. chinensis [35].
2-[(2E)-3,7-Dimethy-2,6-octadienyl]-6-methyl-2,5-cyclohexadiene-1,4-dione (1) yellow- brownish oil, HREIMS m/z 258.1646 (calculated for C17H22O2, 258.1620), 1H-NMR (CDCl3, 400 MHz): δ 6.55 (1H, dq, J = 2.7, 1.6 Hz, H-5), 6.47 (1H, dt, J = 2.7, 1.6 Hz, H-3), 5.15 (1H, t sext, J = 7.3, 1.2 Hz, H-2′), 5.08 (1H, t sept, J = 6.8, 1.2 Hz, H-6′), 3.13(2H, br d, J = 7.3 Hz, H-1′), 2.08 (2H, m, H-5′), 2.06 (5H, m, H-4′ and H-7), 1.70 (3H, br s, H-9′),1.62 (3H, br s, H-10′), 1.60 (3H, br s, H-8′); 13C-NMR (CDCl3, 100 MHz): δ 188.0 (C-1 and C-4), 148.5 (C-3), 145.9 (C-5), 140.0 (C-3′), 133.1 (C-6), 132.2 (C-2), 131.8 (C-7′), 123.9 (C-6′), 118.0 (C-2′), 39.6 (C-4′), 27.5 (C-1′), 26.4 (C-5′), 25.7 (C-9′), 17.7 (C-8′), 16.0 (C-10′), 16.0 (C-7).
1-Acetoxy-tetradeca-6E,12E-diene-8,10-diyne-3-ol (2) yellowish oil, HREIMS m/z 260.1448 (calculated for C16H20O3, 260.1413), 1H-NMR (CDCl3, 400 MHz): δ 6.33 (1H, dq, J = 15.0, 7.0 Hz, H-13), 6.27 (1H, dt, J = 15.0, 7.5 Hz, H-6), 5.58 (1H, dd, J = 15.0, 2.0 Hz, H-12), 5.55 (1H, dd, J = 15.0, 2.0 Hz, H-7), 4.37 (1H, m, H-1a), 4.11 (1H, m, H-1b), 3.7 (1H, m, H-3), 2.28 (2H, m, H-5), 2.07 (3H, s, Acetyl) 1.82 (3H, dd, J = 7.0, 2.0 Hz, H-14), 1.78 (1H, m, H-2a), 1.68 (1H, m, H-2b), 1.58 (2H, m, H-4); 13C-NMR (CDCl3, 100 MHz): δ 171.6 (aceetyl, C=O), 147.4 (C-6), 143.4 (C-13), 109.8 (C-12), 109.2 (C-7), 79.9 (C-11), 79.4 (C-8), 72.9 (C-10), 72.2 (C-9), 67.7 (C-3), 61.5 (C-1), 36.4 (C-2), 36.0 (C-4), 29.4 (C-5), 20.9 (acetyl, CH3), 18.9 (C-14).
1,3-Diacetoxy-tetradeca-6E,12E-diene-8,10-diyne (3) yellowish oil, HRFABMS m/z [M + H]+ 303.1468 (calculated for C18H22O4, 303.1596), 1H-NMR (CDCl3, 400 MHz): δ 6.33 (1H, dq, J = 15.0, 7.0 Hz, H-13), 6.25 (1H, dt, J = 15.0, 7.0 Hz, H-6), 5.57 (2H, br d, J = 15.0 Hz, H-7 and H-12), 5.00 (1H, tt, J = 7.0, 6.5 Hz, H-3), 4.10 (2H, t, J = 6.5 Hz, H-1), 2.09 (2H, br dt, J = 7.0, 7.0 Hz, H-5), 2.06 (6H, s, Acetyl), 1.89 (2H, dt, J = 6.5, 6.5 Hz, H-2), 1.84 (3H, dd, J = 7.0, 15.0 Hz, H-14) 1.70 (2H, m, H-4); 13C-NMR (CDCl3, 100 MHz): δ 170.9 (acetyl, C=O), 170.5 (acetyl, C=O), 146.5 (C-6), 143.5 (C-13), 109.8 (C-12), 109.5 (C-7), 80.0 (C-11), 79.2 (C-8), 73.1 (C-10), 72.3 (C-9), 70.3 (C-3), 60.6 (C-1), 33.0 (C-2), 33.0 (C-4), 29.1 (C-5), 21.0 (acetyl, CH3), 20.9 (acetyl, CH3), 18.9 (C-14).
3.3. Cell Culture
RAW 264.7 murine macrophage cells (ATCC, Rockville, MD, USA) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin (Life technologies, Frederick, MD, USA) at 37 °C in 5% CO2 in a humidified atmosphere.
3.4. Nitrite Assay
RAW 264.7 cells were stimulated with LPS (1 μg/mL) in the absence or presence of compounds for 20 h. NO released from cells was assessed by detecting nitrite in culture supernatant. Aliquots (100 μL) of culture media were incubated with 150 μL of Griess reagent (1% sulfanilamide, 0.1% naphthylethylene diamine in 2.5% phosphoric acid solution) at room temperature for 10 min. Absorbance at 540 nm was read by using a microplate reader (Molecular Devices, CA, USA). The concentration of NO was determined by the sodium nitrite standard curve.
3.5. PGE2 Assay
To observe the effects of compounds on COX-2, cells were seeded with aspirin (500 µM) to inactivate the COX-1. After 2 h, cells were washed with fresh media three times and incubated with LPS (1 µg/mL) in presence of compounds for 20 h. The PGE2 levels in culture media were determined using enzyme immunoassay kit (Cayman Chemical, Ann Arbor, MI) according to the manufacturer’s instruction. In brief, 50 μL of supernatant of the culture medium and 50 μL PGE2 tracer were put into the PGE2 EIA plate and incubated for 18 h at room temperature. The wells were washed with 10 mM phosphate buffer (pH 7.4) containing 0.05% Tween 20. Then 200 μL of Ellman’s reagent was added to the well and incubated in the dark. Following the developing step, the absorbance was read at 405 nm by a microplate reader. A standard curve was prepared simultaneously with PGE2 standard ranging from 0.06 to 6 pg/mL.
3.6. Western Blot Analysis
RAW 264.7 cells (5 × 105 cells/60 mm dish) were treated with or without test compounds in presence of LPS (1 μg/mL) for 20 h. Cells were lysed gently with cell lysis buffer (Cell Signaling Technologies, Beverly, MA, USA). Cell lysates were centrifuged at 4 °C and supernatant were subjected to the quantitation of protein concentrations by the Bradford method. For preparation of cytosol and nuclear extracts, cells were treated with test compounds for 30 min prior to the stimulation with 1 μg/mL LPS. Following 15 min treatment of LPS, cells were collected by using NE-PER nuclear and cytoplasmic extraction reagents according to the manufacturer’s instructions (Pierce Biotechnology, Rockford, IL, USA). The protein lysates were then subjected to SDS-PAGE and transferred onto PVDF membranes. After blocking with 5% non-fat milk for 1 h, membranes were incubated with the primary antibody overnight at 4 °C. Antibodies against iNOS (BD Biosciences, Franklin Lakes, NJ, USA), COX-2 (Cayman Chemical Company, Ann Arbor, MI, USA), I-κB-α, p65 (Santa Cruz Biotechnology, Rockford, IL, USA), ERK1/2, phospho-ERK1/2, p38, phospho-p38, JNK, phospho-JNK, and β-actin (Cell Signaling Technology, Beverly, MA, USA) were used for immunoblot analysis. After incubation with the secondary antibody for 1 h at room temperature, proteins were detected by VersaDoc 3000 (Bio-Rad, Hercules, CA, USA) with ECL reagents (GE Health Care Life Sciences, Marborugh, MA, USA).
3.7. Reverse Transcription and Polymerase Chain Reaction (RT-PCR) Analysis
Cells (1.8 × 106 cells/60mm dish) were stimulated for 6 h with LPS (1 μg/mL) in the presence or absence of test compounds. Total RNA was extracted by TRIzol (Life technologies, Frederick, MD, USA) according to the manufacturer’s instructions. RNA was reverse-transcribed into cDNA using reverse-transcriptase (Life technologies, Frederick, MD, USA) and random-hexamer (Cosmo, Seoul, Korea). The cDNA amplification was performed by using a recombinant Taq polymerase (Promega, Madison, WI, USA) and primers for iNOS, COX-2, IL-6, IL-1β, TNF-α, and β-actin. The amplified PCR products were separated on 2% agarose gels and stained with ethidium bromide.
3.8. Measurement of NF-κB Transcriptional Activity
NF-κB transcriptional activity was determined by using the stably transfected RAW 264.7 cells with pNF-κB-SEAP-NPT (T-RAW 264.7 cells) as described previously with some modifications [36,37]. T-RAW 264.7 cells were kindly gifted by Professor Yeong Shik Kim, Seoul National University, Korea. T-RAW 264.7 cells were plated on a 24 well plate overnight. Test compounds were added to cells 2 h before the treatment with LPS (1 µg/mL). After 16 h incubation, aliquots of culture medium were heated at 65 °C for 6 min and then the activity of SEAP (secretory alkaline phosphatase) was assessed. The transcriptional activity was expressed as relative fluorescence unit (RFU).
3.9. Statistical Analysis
All values were expressed as mean ± S.D. of three experiments. Statistical analysis was carried out with Student’s t-test. A p-value of <0.05 was considered as significantly different.
4. Conclusions
In the present study, we explored the anti-inflammatory potential of compounds 1–3 from Atractylodes macrocephala. Compounds 1–3 reduce NO and PGE2 production, and also suppress the iNOS and COX-2 expression in LPS-stimulated RAW 264.7 cells. The underlying mechanism proved that compounds 1–3 suppressed NF-κB through inhibiting I-κB degradation and the NF-κB nuclear accumulation. Moreover, compound 1, the most potent among three components, decreased levels of pro-inflammatory cytokines including IL-6, IL-1β, and TNF-α, and also suppressed MAPKs phosphorylation in LPS-activated RAW 264.7 cells. Taken together, compounds 1–3 from A. macrocephala can be therapeutic candidates to treat inflammatory diseases.
Author Contributions
D.J. and G.-z.D. performed the experiments and analyzed data; H.J.L. analyzed the data and prepared the manuscript; J.-H.R. designed the whole experiments, analyzed the data, reviewed and edited the manuscript.
Funding
This study was supported by the National Research Foundation of Korea (NRF). Grant funded by the Korean Government (MSIP) (No. 2011-0030074 and No. 2018R1D1A3B07050361).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kim, J.H.; Doh, E.J.; Lee, G. Evaluation of Medicinal Categorization of Atractylodes japonica Koidz. by Using Internal Transcribed Spacer Sequencing Analysis and HPLC Fingerprinting Combined with Statistical Tools. Evid. Based Complement. Alternat. Med. 2016, 2016, 2926819. [Google Scholar] [CrossRef] [PubMed]
- Li, L.S.; Qu, R.Y.; Wang, W.; Guo, H. Significance of changes of gastrointestinal peptides in blood and ileum of experimental spleen deficiency rats. World J. Gastroenterol. 2003, 9, 553–556. [Google Scholar] [CrossRef]
- Wang, R.; Zhou, G.; Wang, M.; Peng, Y.; Li, X. The Metabolism of Polysaccharide from Atractylodes macrocephala Koidz and Its Effect on Intestinal Microflora. Evid. Based Complement. Alternat. Med. 2014, 2014, 926381. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, F.; Ye, N.; Huang, C.; Yuan, X. Atractylodes macrocephala polysaccharides induces mitochondrial-mediated apoptosis in glioma C6 cells. Int. J. Biol. Macromol. 2014, 66, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Duan, H.; He, L. Inhibitory effect of atractylenolide I on angiogenesis in chronic inflammation in vivo and in vitro. Eur. J. Pharmacol. 2009, 612, 143–152. [Google Scholar] [CrossRef]
- Li, X.; Lin, J.; Han, W.; Mai, W.; Wang, L.; Li, Q.; Lin, M.; Bai, M.; Zhang, L.; Chen, D. Antioxidant ability and mechanism of rhizoma Atractylodes macrocephala. Molecules 2012, 17, 13457–13472. [Google Scholar] [CrossRef]
- Shu, Y.T.; Kao, K.T.; Weng, C.S. In vitro antibacterial and cytotoxic activities of plasma-modified polyethylene terephthalate nonwoven dressing with aqueous extract of Rhizome Atractylodes macrocephala. Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 77, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Zhang, Q.L.; Hua, J.W.; Cheng, W.L.; Qin, L.P. The traditional uses, phytochemistry, and pharmacology of Atractylodes macrocephala Koidz.: A review. J. Ethnopharmacol. 2018, 226, 143–167. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, C.; Sun, T.M.; Ran, X.K.; Kang, T.G.; Dou, D.Q. Two new compounds from Atractylodes macrocephala with neuroprotective activity. J. Asian Nat. Prod. Res. 2017, 19, 35–41. [Google Scholar] [CrossRef]
- Hoang, L.S.; Tran, M.H.; Lee, J.S.; Ngo, Q.M.; Woo, M.H.; Min, B.S. Inflammatory inhibitory activity of sesquiterpenoids from Atractylodes macrocephala rhizomes. Chem. Pharm. Bull. 2016, 64, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Q.; He, L.C.; Jin, J.Q. Atractylenolide I and atractylenolide III inhibit lipopolysaccharide-induced TNF-alpha and NO production in macrophages. Phytother. Res. 2007, 21, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.M.; Yang, X.W. Bioactivity-guided isolation of polyacetylenes with inhibitory activity against NO production in LPS-activated RAW264.7 macrophages from the rhizomes of Atractylodes macrocephala. J. Ethnopharmacol. 2014, 151, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Q.; He, L.C.; Dong, H.Y.; Jin, J.Q. Screening for the anti-inflammatory activity of fractions and compounds from Atractylodes macrocephala koidz. J. Ethnopharmacol. 2007, 114, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Murata, M. Inflammation and cancer. Environ. Health Prev. Med. 2018, 23, 50. [Google Scholar] [CrossRef] [PubMed]
- Bastard, J.P.; Jaachi, M.; Laqathu, C.; Kim, M.J.; Caron, M.; Vidal, H.; Capeau, J.; Feve, B. Recent advances in the relationship between obesity, inflammation, and insulin resistance. Eur. Cytokine Netw. 2006, 17, 4–12. [Google Scholar] [PubMed]
- De Virgilio, A.; Greco, A.; Fabbrini, G.; Inqhilleri, M.; Rizzo, M.I.; Gallo, A.; Conte, M.; Rosato, C.; Ciniqlio Appiani, M.; de Vincentiis, M. Parkinson’s disease: Autoimmunity and neuroinflammation. Autoimmun. Rev. 2016, 15, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Hamidzadeh, K.; Christensen, S.M.; Dalby, E.; Chandrasekaran, P.; Mosser, D.M. Macrophages and the Recovery from Acute and Chronic Inflammation. Annu. Rev. Physiol. 2017, 79, 567–592. [Google Scholar] [CrossRef]
- Guzik, T.J.; Korbut, R.; Adamek-Guzik, T. Nitric oxide and superoxide in inflammation and immune regulation. J. Physiol. Pharmacol. 2003, 54, 469–487. [Google Scholar] [PubMed]
- Attiq, A.; Jalil, J.; Husain, K.; Ahmad, W. Raging the war against inflammation with natural products. Front. Pharmacol. 2018, 9, 976. [Google Scholar] [CrossRef]
- Lind, M.; Hayes, A.; Capmda, M.; Petrovic, D.; Rodrigo, L.; Kruzliak, P.; Zulli, A. Inducible nitric oxide synthase: Good or bad? Biomed. Pharmacother. 2017, 93, 370–375. [Google Scholar] [CrossRef]
- Kawahara, K.; Hohjoh, H.; Inazumi, T.; Tsuchiya, S.; Sugimoto, Y. Prostaglandin E2-induced inflammation: Relevance of prostaglandin E receptors. Biochim. Biophys. Acta 2015, 1851, 414–421. [Google Scholar] [CrossRef]
- Resch, M.; Steigel, A.; Chen, Z.L.; Bauer, R. 5-Lipoxygenase and cyclooxygenase-1 inhibitory active compounds from Atractylodes lancea. J. Nat. Prod. 1998, 61, 347–350. [Google Scholar] [CrossRef]
- Pachaly, P.; Lansing, A.; Neugebauer, M.; Sin, K.S. Acetylenes from Atractylis koreana. Planta Med. 1990, 56, 469–471. [Google Scholar] [CrossRef]
- Ahn, K.S.; Noh, E.J.; Zhao, H.S.; Jung, S.H.; Kang, S.S.; Kim, Y.S. Inhibition of inducible nitric oxide synthase and cyclooxygenase II by Platycodon grandiflorum saponins via suppression of nuclear factor-kappaB activation in RAW 264.7 cells. Life Sci. 2005, 76, 2315–2328. [Google Scholar] [CrossRef]
- Yamamoto, M.; Takeda, K. Role of nuclear IkappaB proteins in the regulation of host immune responses. J. Infect. Chemother. 2008, 14, 265–269. [Google Scholar] [CrossRef]
- Wan, F.; Lenardo, M.J. The nuclear signaling of NF-kappaB: Current knowledge, new insights, and future perspectives. Cell Res. 2010, 20, 24–33. [Google Scholar] [CrossRef]
- Liang, Y.; Zhou, Y.; Shen, P. NF-kappaB and its regulation on the immune system. Cell. Mol. Immunol. 2004, 1, 343–350. [Google Scholar]
- Chan, E.D.; Riches, D.W. IFN-gamma + LPS induction of iNOS is modulated by ERK, JNK/SAPK, and p38(mapk) in a mouse macrophage cell line. Am. J. Physiol. Cell Physiol. 2001, 280, C441–C450. [Google Scholar] [CrossRef]
- Chan, E.D.; Winston, B.W.; Uh, S.T.; Wynes, M.W.; Rose, D.M.; Riches, D.W. Evaluation of the role of mitogen-activated protein kinases in the expression of inducible nitric oxide synthase by IFN-gamma and TNF-alpha in mouse macrophages. J. Immunol. 1999, 162, 415–422. [Google Scholar]
- Kyriakis, J.M. Activation of the AP-1 transcription factor by inflammatory cytokines of the TNF family. Gene Expr. 1999, 7, 217–231. [Google Scholar]
- Mahtani, K.R.; Brook, M.; Dean, J.S.; Sully, G.; Saklatvala, J.; Clark, A.R. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 2001, 21, 6461–6469. [Google Scholar] [CrossRef]
- Karunakaran, S.; Ravindranath, V. Activation of p38 MAPK in the substantia nigra leads to nuclear translocation of NF-kappaB in MPTP-treated mice: Implication in Parkinson’s disease. J. Neurochem. 2009, 109, 1791–1799. [Google Scholar] [CrossRef]
- Lee, S.M.; Kim, E.J.; Suk, K.; Lee, W.H. Stimulation of Fas (CD95) induces production of pro-inflammatory mediators through ERK/JNK-dependent activation of NF-kappaB in THP-1 cells. Cell. Immunol. 2011, 271, 157–162. [Google Scholar] [CrossRef]
- Li, L.; Zhao, R.; Li, Y.; Wang, W.H. Antitumor activity of 2-[(2E)-3,7-dimethyl-2,6-octadienyl]-6-methyl-2,5-cyclohexadiene-1,4-dione isolated from the aerial part of Atractylodes macrocephala in hepatocellular carcinoma. Mol. Med. Rep. 2017, 16, 6299–6305. [Google Scholar] [CrossRef][Green Version]
- Meng, H.; Li, G.; Dai, R.; Ma, Y.; Zhang, K.; Zhang, C.; Li, X.; Wang, J. Chemical constituents of Atractylodes chinensis(DC.) Koidz. Biochem. Syst. Ecol. 2010, 38, 1220–1223. [Google Scholar] [CrossRef]
- Moon, K.Y.; Hahn, B.S.; Lee, J.; Kim, Y.S. A cell-based assay system for monitoring NF-kappaB activity in human HaCat transfectant cells. Anal. Biochem. 2001, 292, 17–21. [Google Scholar] [CrossRef]
- Ahn, K.S.; Aggarwal, B.B. Transcription factor NF-kappaB: A sensor for smoke and stress signals. Ann. N. Y. Acad. Sci. 2005, 1056, 218–233. [Google Scholar] [CrossRef]
Sample Availability: Not available. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).