Sub-Inhibitory Doses of Individual Constituents of Essential Oils Can Select for Staphylococcus aureus Resistant Mutants
Abstract
1. Introduction
2. Results
2.1. Isolation of S. aureus USA300 Derivative Strains with Increased Resistance to ICs
2.2. Effect of ICs on Mutation Frequency during Bacterial Growth
2.3. Evaluation of Derivative Strains against Lethal Treatments
2.4. Genomic Sequencing of WT and Derivative Strains
3. Discussion
4. Materials and Methods
4.1. Microorganisms and Growth Conditions
4.2. Determination of Minimum Inhibitory Concentration (MIC)
4.3. Isolation of Derivative Strains by Applying Sub-Inhibitory Doses of ICs During Bacterial Growth
4.4. Mutagenesis Frequency Evaluation
4.5. Evaluation of Increased Bacterial Tolerance
4.5.1. Lethal ICs Treatments
4.5.2. Lethal Heat treatments
4.5.3. Lethal PEF Treatments
4.6. Counting of Viable and Sublethally Injured Cells
4.7. Genome Sequencing and SNP Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kohanski, M.A.; DePristo, M.A.; Collins, J.J. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 2010, 37, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Foster, P.L. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 373–397. [Google Scholar] [CrossRef] [PubMed]
- de Souza, E.L.; da Cruz Almeida, E.T.; de Sousa Guedes, J.P. The potential of the incorporation of essential oils and their individual constituents to improve microbial safety in juices: A review. Compr. Rev. Food Sci. 2016, 15, 753–772. [Google Scholar] [CrossRef]
- Chueca, B.; Pagán, R.; García-Gonzalo, D. Oxygenated monoterpenes citral and carvacrol cause oxidative damage in Escherichia coli without the involvement of tricarboxylic acid cycle and Fenton reaction. Int. J. Food Microbiol. 2014, 189, 126–131. [Google Scholar] [CrossRef]
- Chueca, B.; Pagán, R.; García-Gonzalo, D. Differential mechanism of Escherichia coli inactivation by (+)-limonene as a function of cell physiological state and drug’s concentration. PLoS ONE 2014, 9, e94072. [Google Scholar] [CrossRef]
- Ait-Ouazzou, A.; Espina, L.; Gelaw, T.K.; de Lamo-Castellvi, S.; Pagan, R.; Garcia-Gonzalo, D. New insights in mechanisms of bacterial inactivation by carvacrol. J. Appl. Microbiol. 2013, 114, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Burt, S. Essential oils: Their antibacterial properties and potential applications in foods—a review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef] [PubMed]
- Leite de Souza, E. The effects of sublethal doses of essential oils and their constituents on antimicrobial susceptibility and antibiotic resistance among food-related bacteria: A review. Trends Food Sci. Technol. 2016, 56, 1–12. [Google Scholar] [CrossRef]
- Chueca, B.; Berdejo, D.; Gomes-Neto, N.J.; Pagan, R.; Garcia-Gonzalo, D. Emergence of hyper-resistant Escherichia coli MG1655 derivative strains after applying sub-inhibitory doses of individual constituents of essential oils. Front. Microbiol. 2016, 7, 273. [Google Scholar] [CrossRef] [PubMed]
- Chueca, B.; Renzoni, A.; Berdejo, D.; Pagan, R.; Kelley, W.L.; Garcia-Gonzalo, D. Whole-genome sequencing and genetic analysis reveal novel stress responses to individual constituents of essential oils in Escherichia coli. Appl. Environ. Microbiol. 2018, 84, e02538. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, A.; Laureti, L.; Crussard, S.; Abida, H.; Rodriguez-Rojas, A.; Blazquez, J.; Baharoglu, Z.; Mazel, D.; Darfeuille, F.; Vogel, J.; et al. β-lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 2013, 4, 1610. [Google Scholar] [CrossRef] [PubMed]
- Mackey, B.M. Injured bacteria. In The Microbiological Safety and Quality of Food; Lund, M., Baird-Par-ker, T.C., Gould, G.W., Eds.; Aspen Publisher: Gaithersburg, MD, USA, 2000; Volume I, pp. 315–341. [Google Scholar]
- Yap, P.S.X.; Yiap, B.C.; Ping, H.C.; Lim, S.H.E. Essential oils, a new horizon in combating bacterial antibiotic resistance. Open. Microbiol. J. 2014, 8, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Chávez-González, M.L.; Rodríguez-Herrera, R.; Aguilar, C.N. Essential oils: A natural alternative to combat antibiotics resistance. In Antibiotic Resistance; Rai, M., Ed.; Elsevier Academic Press: London Wall, London, UK, 2016; pp. 227–237. [Google Scholar]
- Silva da Luz, I.; Gomes Neto, N.J.; Tavares, A.G.; Nunes, P.C.; Magnani, M.; de Souza, E.L. Lack of induction of direct protection or cross-protection in Staphylococcus aureus by sublethal concentrations of Origanum vulgare L. essential oil and carvacrol in a meat-based medium. Arch. Microbiol. 2013, 195, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Apolonio, J.; Faleiro, M.L.; Miguel, M.G.; Neto, L. No induction of antimicrobial resistance in Staphylococcus aureus and Listeria monocytogenes during continuous exposure to eugenol and citral. FEMS Microbiol. Lett. 2014, 354, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Tavares, A.G.; Monte, D.F.; Albuquerque Ados, R.; Sampaio, F.C.; Magnani, M.; Siqueira Junior, J.P.; Souza, E.L. Habituation of enterotoxigenic Staphylococcus aureus to Origanum vulgare L. essential oil does not induce direct-tolerance and cross-tolerance to salts and organic acids. Braz. J. Microbiol. 2015, 46, 835–840. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Hammer, K.A.; Carson, C.F.; Riley, T.V. Frequencies of resistance to Melaleuca alternifolia (tea tree) oil and rifampicin in Staphylococcus aureus, Staphylococcus epidermidis and Enterococcus faecalis. Int. J. Antimicrob. Agents 2008, 32, 170–173. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef]
- Brewer, M.S. Natural antioxidants: Sources, compounds, mechanisms of action, and potential applications. Compr. Rev. Food Sci. Food Saf. 2011, 10, 221–247. [Google Scholar] [CrossRef]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Fridman, O.; Goldberg, A.; Ronin, I.; Shoresh, N.; Balaban, N.Q. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 2014, 513, 418–421. [Google Scholar] [CrossRef] [PubMed]
- Aiemsaard, J.; Aiumlamai, S.; Aromdee, C.; Taweechaisupapong, S.; Khunkitti, W. The effect of lemongrass oil and its major components on clinical isolate mastitis pathogens and their mechanisms of action on Staphylococcus aureus DMST 4745. Res. Vet. Sci. 2011, 91, e31–e37. [Google Scholar] [CrossRef] [PubMed]
- Espina, L.; Berdejo, D.; Alfonso, P.; García-Gonzalo, D.; Pagán, R. Potential use of carvacrol and citral to inactivate biofilm cells and eliminate biofouling. Food Control 2017, 82, 256–265. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Corry, J.E.; Miles, C.A. Heat resistance and mechanism of heat inactivation in thermophilic campylobacters. Appl. Environ. Microbiol. 2006, 72, 908–913. [Google Scholar] [CrossRef]
- Mackey, B.M.; Miles, C.A.; Parsons, S.E.; Seymour, D.A. Thermal denaturation of whole cells and cell components of Escherichia coli examined by differential scanning calorimetry. J. Gen. Microbiol. 1991, 137, 2361–2374. [Google Scholar] [CrossRef] [PubMed]
- Mañas, P.; Pagán, R. Microbial inactivation by new technologies of food preservation. J. Appl. Microbiol. 2005, 98, 1387–1399. [Google Scholar] [CrossRef] [PubMed]
- Chueca, B.; Pagán, R.; García-Gonzalo, D. Transcriptomic analysis of Escherichia coli MG1655 cells exposed to pulsed electric fields. Innov. Food Sci. Emerg. Technol. 2015, 29, 78–86. [Google Scholar] [CrossRef]
- Garcia, D.; Manas, P.; Gomez, N.; Raso, J.; Pagan, R. Biosynthetic requirements for the repair of sublethal membrane damage in Escherichia coli cells after pulsed electric fields. J. Appl. Microbiol. 2006, 100, 428–435. [Google Scholar] [CrossRef]
- Espina, L.; Gelaw, T.K.; de Lamo-Castellvi, S.; Pagan, R.; Garcia-Gonzalo, D. Mechanism of bacterial inactivation by (+)-limonene and its potential use in food preservation combined processes. PLoS ONE 2013, 8, e56769. [Google Scholar] [CrossRef]
- Somolinos, M.; Garcia, D.; Condon, S.; Mackey, B.; Pagan, R. Inactivation of Escherichia coli by citral. J. Appl. Microbiol. 2010, 108, 1928–1939. [Google Scholar] [CrossRef]
- Lewis, K.; Shan, Y. Why tolerance invites resistance. Science 2017, 355, 796. [Google Scholar] [CrossRef] [PubMed]
- Levin-Reisman, I.; Ronin, I.; Gefen, O.; Braniss, I.; Shoresh, N.; Balaban, N.Q. Antibiotic tolerance facilitates the evolution of resistance. Science 2017, 355, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Mehlan, H.; Bernhardt, J.; Hennig, A.; Michalik, S.; Surmann, K.; Pané-Farré, J.; Giese, A.; Weiss, S.; Backert, L.; et al. AureoWiki—The repository of the Staphylococcus aureus research and annotation community. Int. J. Med. Microbiol. 2017, 308, 558–568. [Google Scholar] [CrossRef] [PubMed]
- Chueca, B.; Perez-Saez, E.; Pagan, R.; Garcia-Gonzalo, D. Global transcriptional response of Escherichia coli MG1655 cells exposed to the oxygenated monoterpenes citral and carvacrol. Int. J. Food Microbiol. 2017, 257, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Nazzaro, F.; Fratianni, F.; De Martino, L.; Coppola, R.; De Feo, V. Effect of essential oils on pathogenic bacteria. Pharmaceuticals 2013, 6, 1451–1474. [Google Scholar] [CrossRef] [PubMed]
- Horsburgh, M.J.; Foster, T.J.; Barth, P.T.; Coggins, J.R. Chorismate synthase from Staphylococcus aureus. Microbiology 1996, 142, 2943–2950. [Google Scholar] [CrossRef] [PubMed]
- Foulongne, V.; Walravens, K.; Bourg, G.; Boschiroli, M.L.; Godfroid, J.; Ramuz, M.; O’Callaghan, D. Aromatic compound-dependent Brucella suis is attenuated in both cultured cells and mouse models. Infect. Immun. 2001, 69, 547–550. [Google Scholar] [CrossRef]
- Proctor, R.A.; von Eiff, C.; Kahl, B.C.; Becker, K.; McNamara, P.; Herrmann, M.; Peters, G. Small colony variants: A pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 2006, 4, 295–305. [Google Scholar] [CrossRef]
- Wakeman, C.A.; Hammer, N.D.; Stauff, D.L.; Attia, A.S.; Anzaldi, L.L.; Dikalov, S.I.; Calcutt, M.W.; Skaar, E.P. Menaquinone biosynthesis potentiates haem toxicity in Staphylococcus aureus. Mol. Microbiol. 2012, 86, 1376–1392. [Google Scholar] [CrossRef]
- Pitchandi, P.; Hopper, W.; Rao, R. Comprehensive database of chorismate synthase enzyme from shikimate pathway in pathogenic bacteria. BMC Pharmacol. Toxicol. 2013, 14, 29. [Google Scholar] [CrossRef]
- Sebkova, A.; Karasova, D.; Crhanova, M.; Budinska, E.; Rychlik, I. aro mutations in Salmonella enterica cause defects in cell wall and outer membrane integrity. J. Bacteriol. 2008, 190, 3155–3160. [Google Scholar] [CrossRef]
- Seong, W.J.; Kim, J.H.; Kwon, H.J. Comparison of complete rpoB gene sequence typing and multi-locus sequence typing for phylogenetic analysis of Staphylococcus aureus. J. Gen. Appl. Microbiol. 2013, 59, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Shan, W.; Ma, X.; Chang, W.; Zhou, X.; Lu, H.; Dai, Y. Molecular characterization of rifampicin-resistant Staphylococcus aureus isolates in a Chinese teaching hospital from Anhui, China. BMC Microbiol. 2012, 12, 240. [Google Scholar] [CrossRef]
- Aubry-Damon, H.; Soussy, C.J.; Courvalin, P. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1998, 42, 2590–2594. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Cui, L.; Katayama, Y.; Kozue, K.; Hiramatsu, K. Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J. Clin. Microbiol. 2011, 49, 2680–2684. [Google Scholar] [CrossRef] [PubMed]
- Kreiswirth, B.; Kornblum, J.; Arbeit, R.D.; Eisner, W.; Maslow, J.N.; McGeer, A.; Low, D.E.; Novick, R.P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science 1993, 259, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Kluytmans, J.A.J.W. Methicillin-resistant Staphylococcus aureus in food products: Cause for concern or case for complacency? Clin. Microbiol. Infect. 2010, 16, 11–15. [Google Scholar] [CrossRef]
- Patel, B.A.; Cockerill, F.R.; Bradford, P.A.; Eliopoulos, G.M.; Hindler, J.A.; Jenkins, S.G. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. In M07-A9 Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically Approved Standard, 10th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015; Volume 35, number 2. [Google Scholar]
- Rota, C.; Carraminana, J.J.; Burillo, J.; Herrera, A. In vitro antimicrobial activity of essential oils from aromatic plants against selected foodborne pathogens. J. Food Prot. 2004, 67, 1252–1256. [Google Scholar] [CrossRef]
- Rosche, W.A.; Foster, P.L. Determining mutation rates in bacterial populations. Methods (San Diego, Calif.) 2000, 20, 4–17. [Google Scholar] [CrossRef]
- Saldaña, G.; Cebrian, G.; Abenoza, M.; Sanchez-Gimeno, C.; Alvarez, I.; Raso, J. Assessing the efficacy of PEF treatments for improving polyphenol extraction during red wine vinifications. Innov. Food Sci. Emerg. Technol. 2017, 39, 179–187. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Proc, G.P.D. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

 ,
, ) and its derivative strains; CAR (
) and its derivative strains; CAR (  ,
, ) using 200 μL/L carvacrol (A), CIT (
) using 200 μL/L carvacrol (A), CIT ( ,
, ) using 2000 μL/L citral (B), and OXLIM (
) using 2000 μL/L citral (B), and OXLIM ( ,
, ) using 2000 μL/L (+)-limonene oxide (C) treatment, at pH 4.0 (black symbols) and pH 7.0 (white symbols). All treatments were performed at room temperature except (+)-limonene oxide, which were carried out at 37 °C. Data are means ± standard deviations (error bars) obtained from at least three independents experiments.
) using 2000 μL/L (+)-limonene oxide (C) treatment, at pH 4.0 (black symbols) and pH 7.0 (white symbols). All treatments were performed at room temperature except (+)-limonene oxide, which were carried out at 37 °C. Data are means ± standard deviations (error bars) obtained from at least three independents experiments.
 ,
, ) and its derivative strains; CAR (
) and its derivative strains; CAR (  ,
, ) using 200 μL/L carvacrol (A), CIT (
) using 200 μL/L carvacrol (A), CIT ( ,
, ) using 2000 μL/L citral (B), and OXLIM (
) using 2000 μL/L citral (B), and OXLIM ( ,
, ) using 2000 μL/L (+)-limonene oxide (C) treatment, at pH 4.0 (black symbols) and pH 7.0 (white symbols). All treatments were performed at room temperature except (+)-limonene oxide, which were carried out at 37 °C. Data are means ± standard deviations (error bars) obtained from at least three independents experiments.
) using 2000 μL/L (+)-limonene oxide (C) treatment, at pH 4.0 (black symbols) and pH 7.0 (white symbols). All treatments were performed at room temperature except (+)-limonene oxide, which were carried out at 37 °C. Data are means ± standard deviations (error bars) obtained from at least three independents experiments.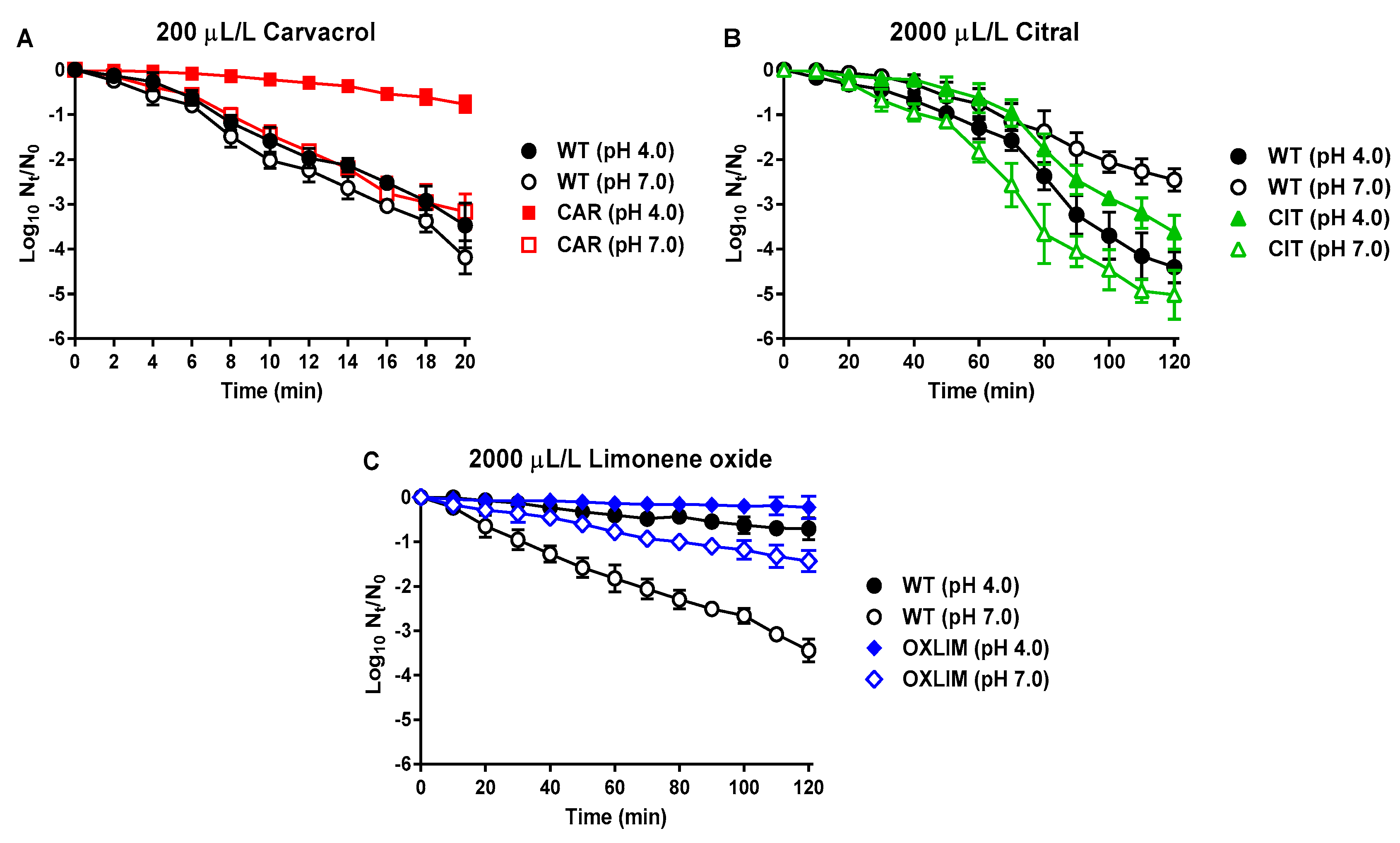
 ,
, ) and its derivative strains: CAR (
) and its derivative strains: CAR ( ,
, ), CIT (
), CIT ( ,
, ), and OXLIM (
), and OXLIM ( ,
, ) using heat treatments (60 °C) at pH 4.0 (A, black symbols) and pH 7.0 (B, white symbols). Data are means ± standard deviations (error bars) obtained from at least three independents experiments.
) using heat treatments (60 °C) at pH 4.0 (A, black symbols) and pH 7.0 (B, white symbols). Data are means ± standard deviations (error bars) obtained from at least three independents experiments.
 ,
, ) and its derivative strains: CAR (
) and its derivative strains: CAR ( ,
, ), CIT (
), CIT ( ,
, ), and OXLIM (
), and OXLIM ( ,
, ) using heat treatments (60 °C) at pH 4.0 (A, black symbols) and pH 7.0 (B, white symbols). Data are means ± standard deviations (error bars) obtained from at least three independents experiments.
) using heat treatments (60 °C) at pH 4.0 (A, black symbols) and pH 7.0 (B, white symbols). Data are means ± standard deviations (error bars) obtained from at least three independents experiments.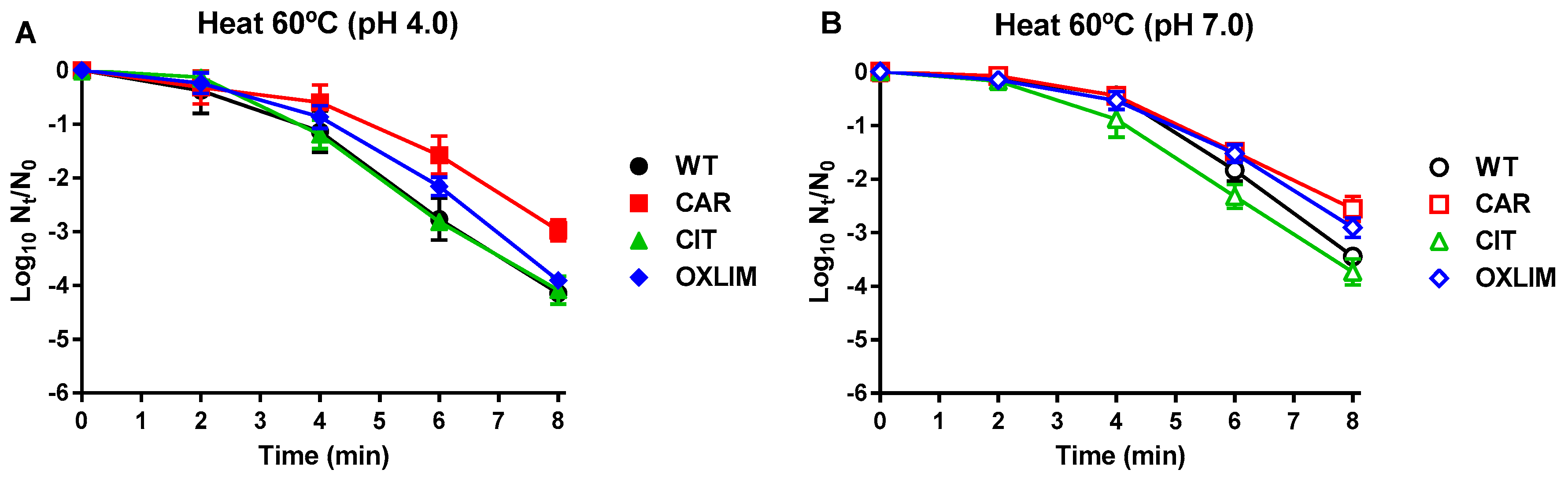
 ,
, ) and its derivative strains: CAR (
) and its derivative strains: CAR ( ,
, ), CIT (
), CIT ( ,
, ), and OXLIM (
), and OXLIM ( ,
, ) using pulsed electric fields treatments (PEF: 25 kV/cm, 1 Hz, 3 µs/pulse) at pH 4.0 (A, black symbols) and pH 7.0 (B, white symbols). Data are means ± standard deviations (error bars) obtained from at least three independents experiments.
) using pulsed electric fields treatments (PEF: 25 kV/cm, 1 Hz, 3 µs/pulse) at pH 4.0 (A, black symbols) and pH 7.0 (B, white symbols). Data are means ± standard deviations (error bars) obtained from at least three independents experiments.
 ,
, ) and its derivative strains: CAR (
) and its derivative strains: CAR ( ,
, ), CIT (
), CIT ( ,
, ), and OXLIM (
), and OXLIM ( ,
, ) using pulsed electric fields treatments (PEF: 25 kV/cm, 1 Hz, 3 µs/pulse) at pH 4.0 (A, black symbols) and pH 7.0 (B, white symbols). Data are means ± standard deviations (error bars) obtained from at least three independents experiments.
) using pulsed electric fields treatments (PEF: 25 kV/cm, 1 Hz, 3 µs/pulse) at pH 4.0 (A, black symbols) and pH 7.0 (B, white symbols). Data are means ± standard deviations (error bars) obtained from at least three independents experiments.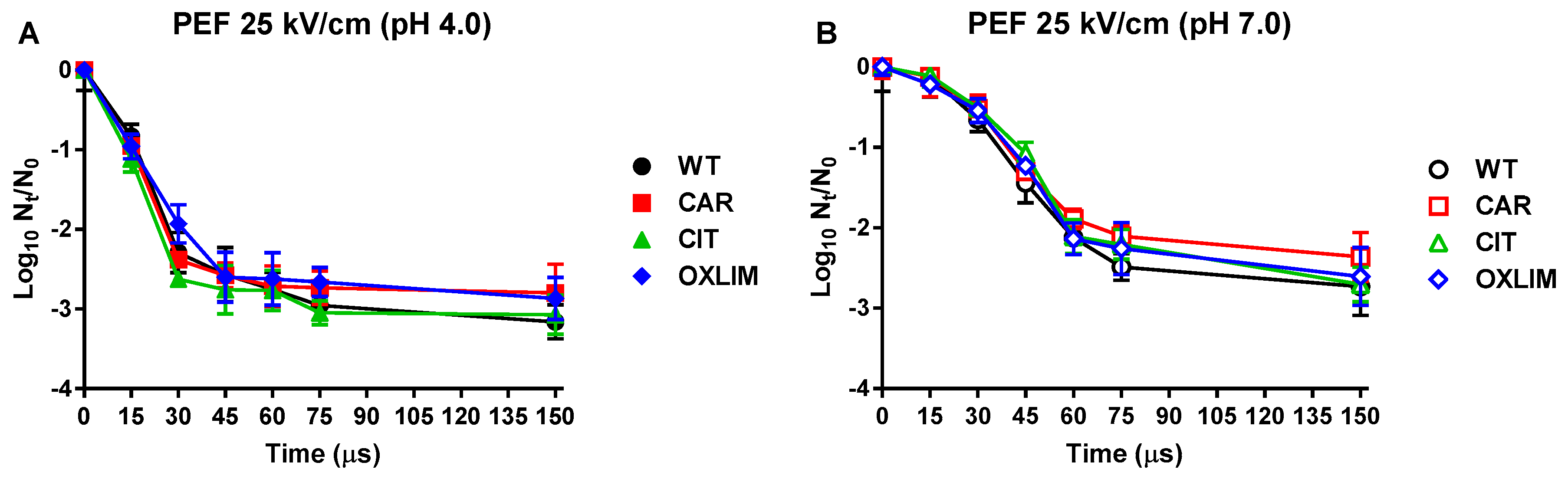
 ,
, ) and CAR derivative strain (
) and CAR derivative strain ( ,
, ) using carvacrol (200 μL/L; A) and heat treatments (60 °C; B) at pH 4.0. The treated samples were recovered in TSBYE without (black symbols) and with 14% of NaCl (semi-transparent symbols), used as selective agent to detect sublethal damage. Data are means ± standard deviations (error bars) obtained from at least three independents experiments. Discontinuous line shows the detection limit.
) using carvacrol (200 μL/L; A) and heat treatments (60 °C; B) at pH 4.0. The treated samples were recovered in TSBYE without (black symbols) and with 14% of NaCl (semi-transparent symbols), used as selective agent to detect sublethal damage. Data are means ± standard deviations (error bars) obtained from at least three independents experiments. Discontinuous line shows the detection limit.
 ,
, ) and CAR derivative strain (
) and CAR derivative strain ( ,
, ) using carvacrol (200 μL/L; A) and heat treatments (60 °C; B) at pH 4.0. The treated samples were recovered in TSBYE without (black symbols) and with 14% of NaCl (semi-transparent symbols), used as selective agent to detect sublethal damage. Data are means ± standard deviations (error bars) obtained from at least three independents experiments. Discontinuous line shows the detection limit.
) using carvacrol (200 μL/L; A) and heat treatments (60 °C; B) at pH 4.0. The treated samples were recovered in TSBYE without (black symbols) and with 14% of NaCl (semi-transparent symbols), used as selective agent to detect sublethal damage. Data are means ± standard deviations (error bars) obtained from at least three independents experiments. Discontinuous line shows the detection limit.
| WT | CAR | CIT | OXLIM | |
|---|---|---|---|---|
| Carvacrol | 100 | 150 | 150 | 150 |
| Citral | 150 | 150 | 250 | 200 |
| (+)-Limonene oxide | 750 | 750 | 1000 | 1500 |
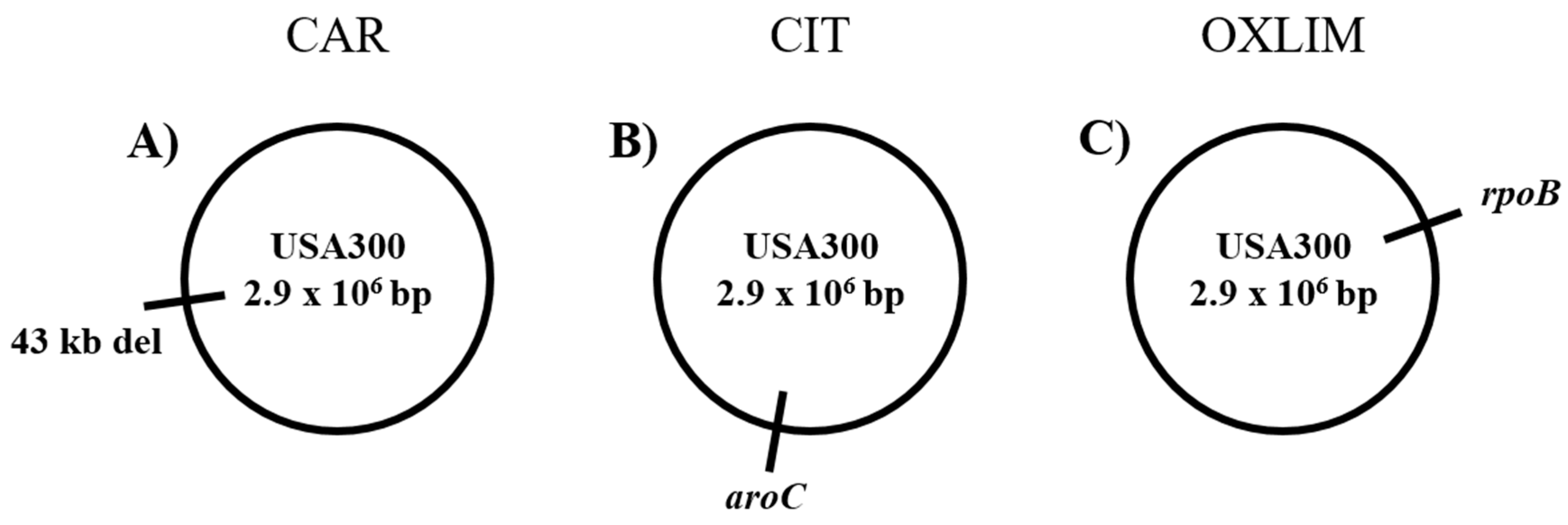
| Genome Position | Strain | Mutation | Gene | Locus tag | Description | Change |
|---|---|---|---|---|---|---|
| 2,084,675–2,127,700 | CAR | Large deletion | Δ (hlb1–int3) | SAUSA300_RS10505-SAUSA300_RS10835 | Deletion of 65 genes | - |
| 1,524,394 | CIT | SNP (A→T) | aroC | SAUSA300_RS07395 | Chorismate synthase | N187K |
| 588,138 | OXLIM | SNP (C→T) | rpoB | SAUSA300_RS02820 | DNA-directed RNA polymerase subunit beta | A862V |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berdejo, D.; Chueca, B.; Pagán, E.; Renzoni, A.; Kelley, W.L.; Pagán, R.; Garcia-Gonzalo, D. Sub-Inhibitory Doses of Individual Constituents of Essential Oils Can Select for Staphylococcus aureus Resistant Mutants. Molecules 2019, 24, 170. https://doi.org/10.3390/molecules24010170
Berdejo D, Chueca B, Pagán E, Renzoni A, Kelley WL, Pagán R, Garcia-Gonzalo D. Sub-Inhibitory Doses of Individual Constituents of Essential Oils Can Select for Staphylococcus aureus Resistant Mutants. Molecules. 2019; 24(1):170. https://doi.org/10.3390/molecules24010170
Chicago/Turabian StyleBerdejo, Daniel, Beatriz Chueca, Elisa Pagán, Adriana Renzoni, William L. Kelley, Rafael Pagán, and Diego Garcia-Gonzalo. 2019. "Sub-Inhibitory Doses of Individual Constituents of Essential Oils Can Select for Staphylococcus aureus Resistant Mutants" Molecules 24, no. 1: 170. https://doi.org/10.3390/molecules24010170
APA StyleBerdejo, D., Chueca, B., Pagán, E., Renzoni, A., Kelley, W. L., Pagán, R., & Garcia-Gonzalo, D. (2019). Sub-Inhibitory Doses of Individual Constituents of Essential Oils Can Select for Staphylococcus aureus Resistant Mutants. Molecules, 24(1), 170. https://doi.org/10.3390/molecules24010170






