A New Strategy for Rapidly Screening Natural Inhibitors Targeting the PCSK9/LDLR Interaction In Vitro
Abstract
1. Introduction
2. Results and Discussions
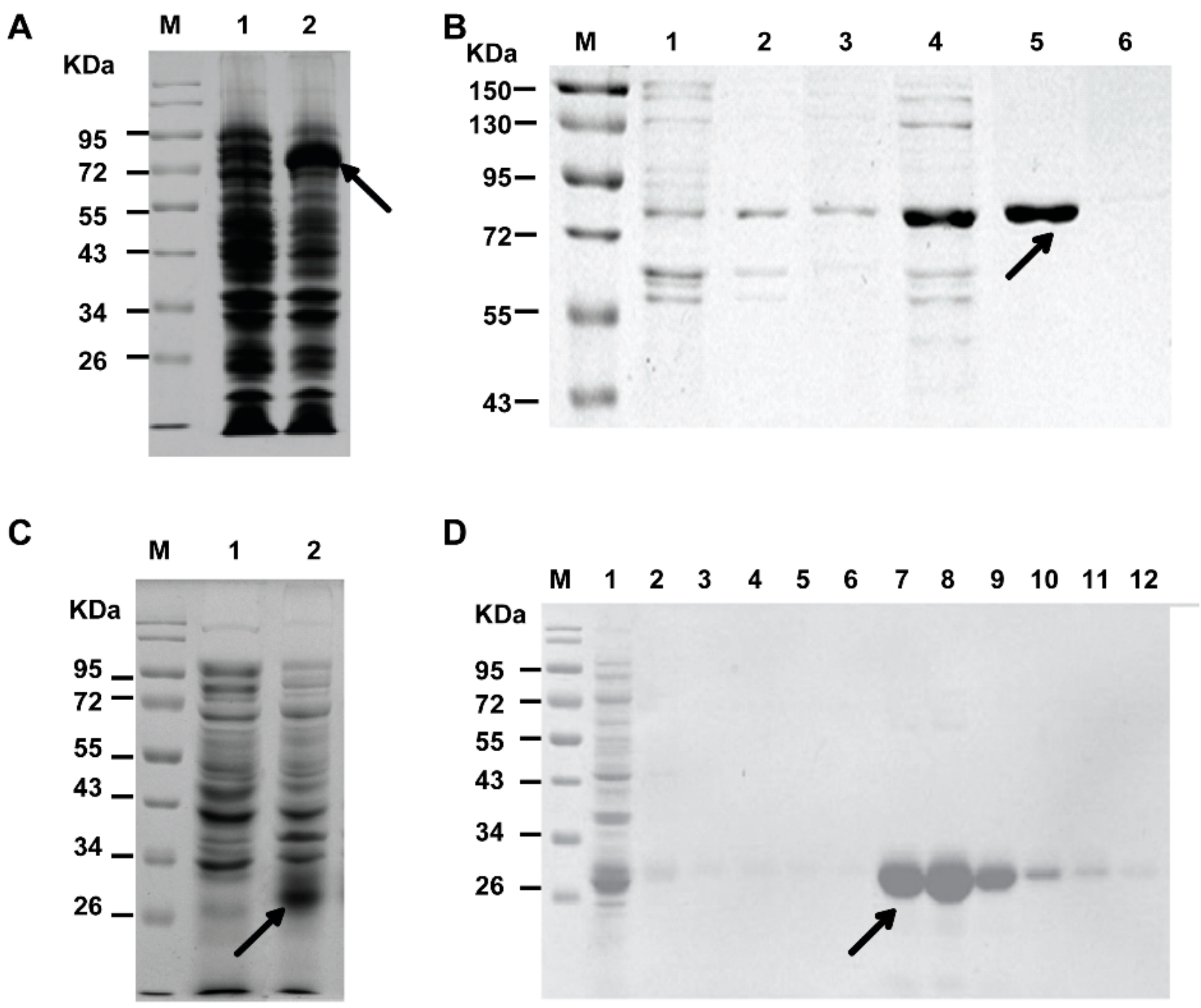
2.1. Expression and Purification of His-PCSK9 and GST-EGF-A
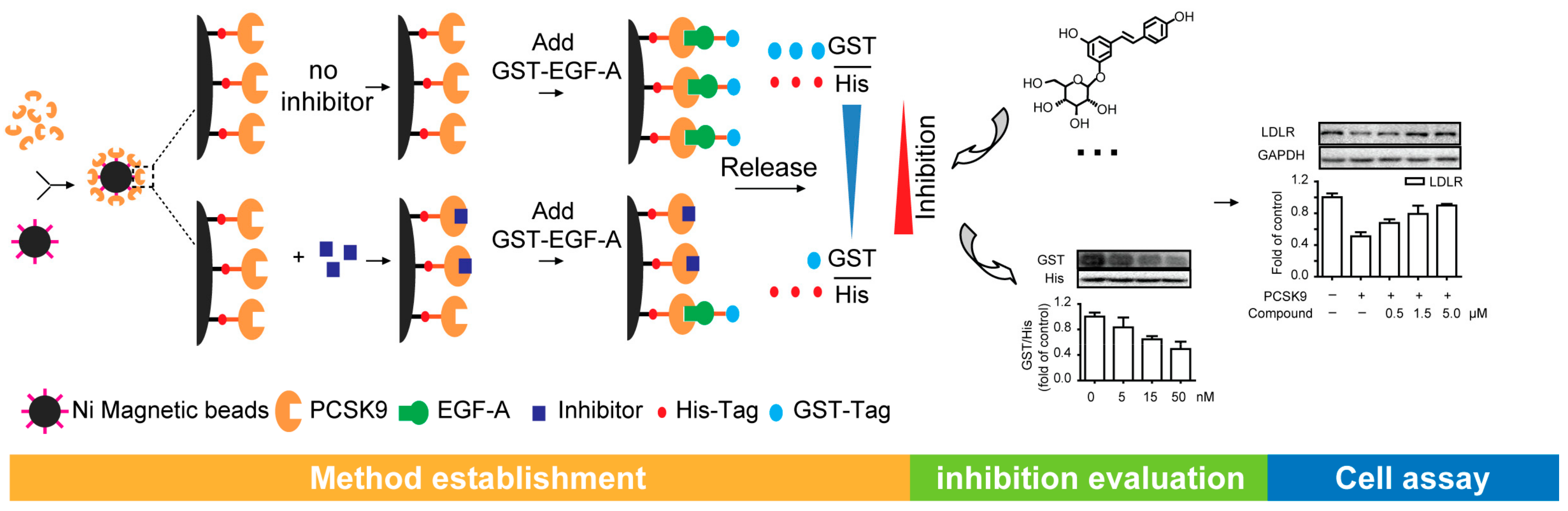
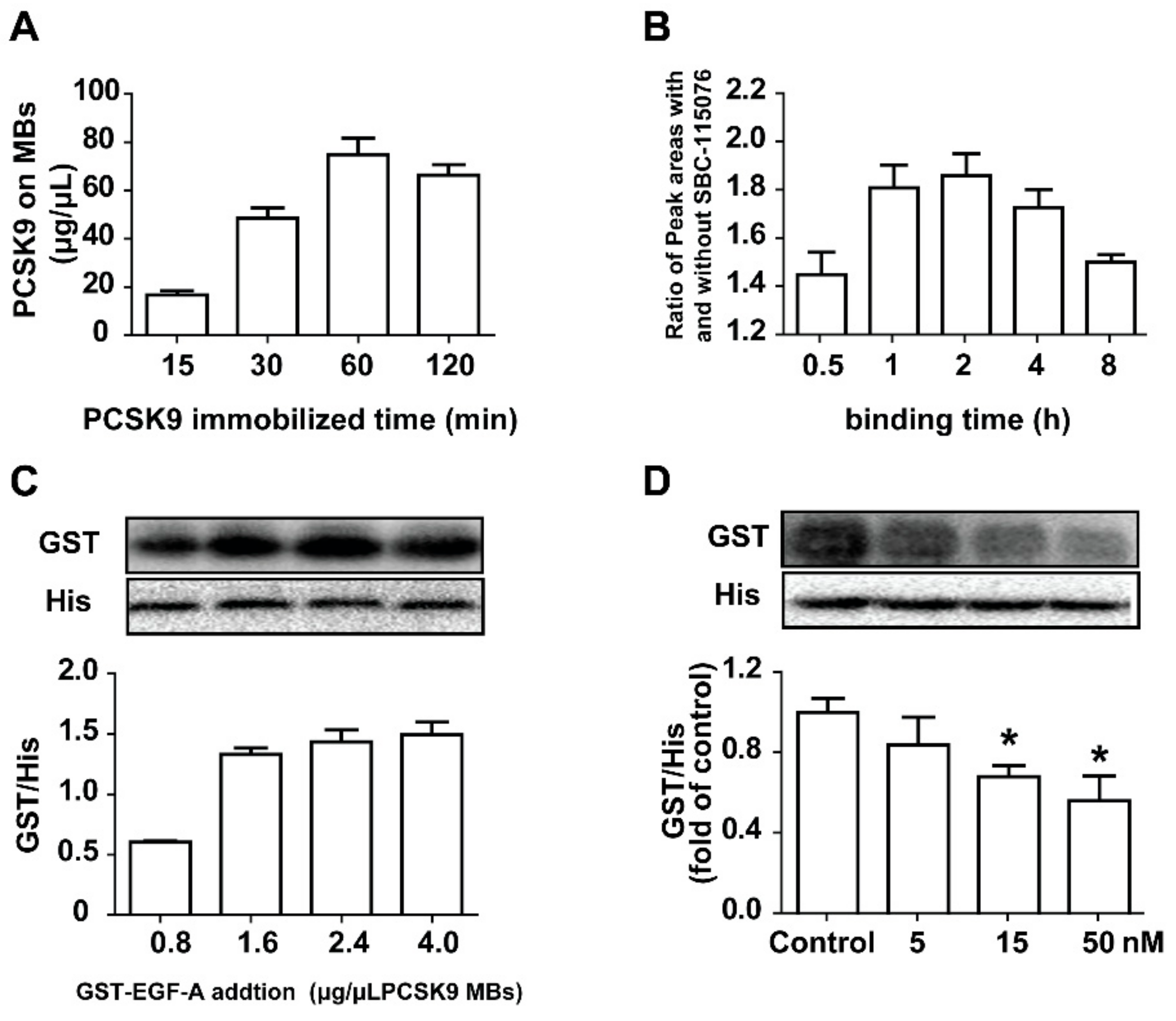
2.2. Establishment of the Method for Evaluating the Inhibitory Activities on PCSK9/LDLR Interaction
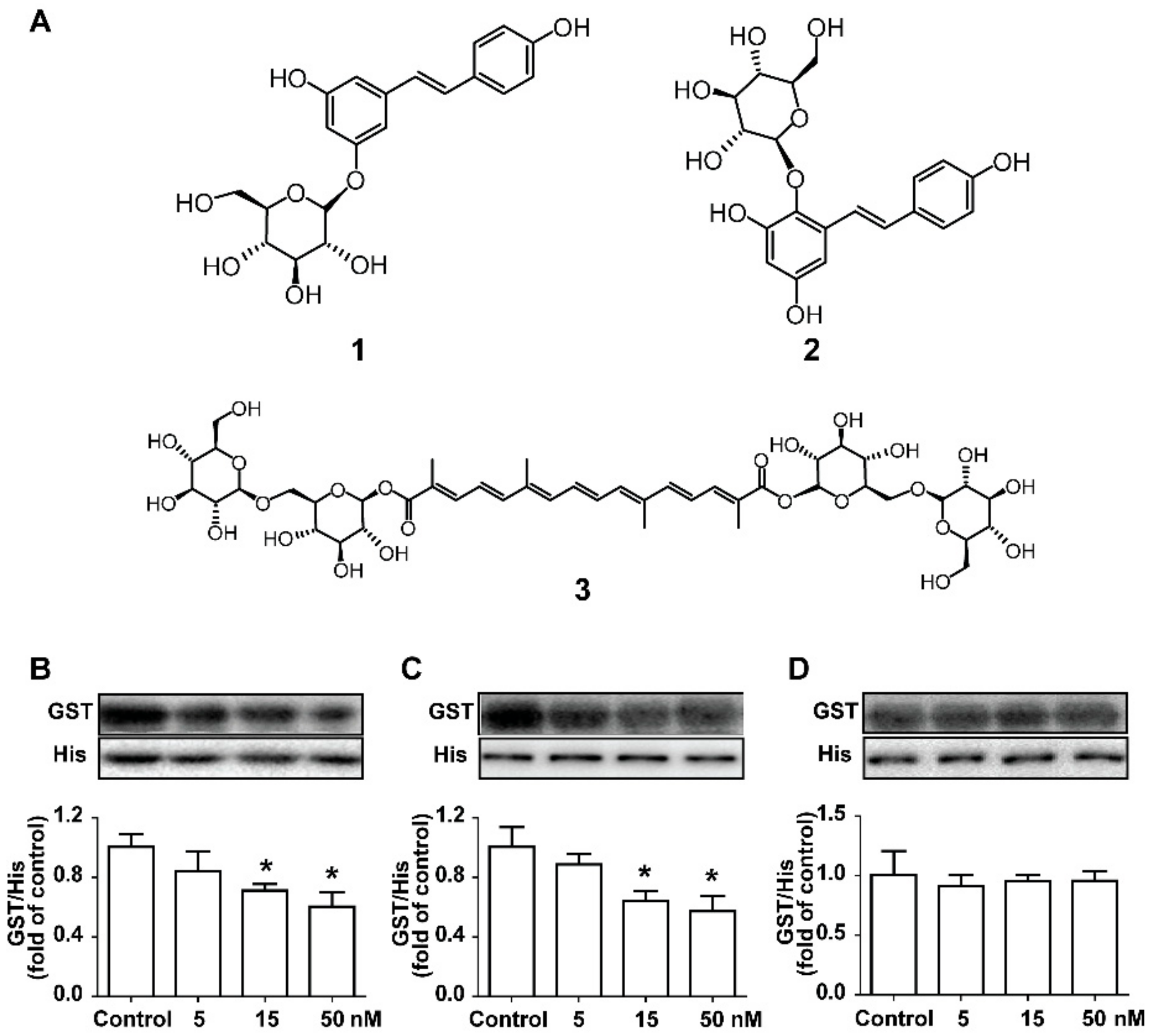
2.3. Screening the Potential Natural Products Interrupting the PCSK9/LDLR Interaction
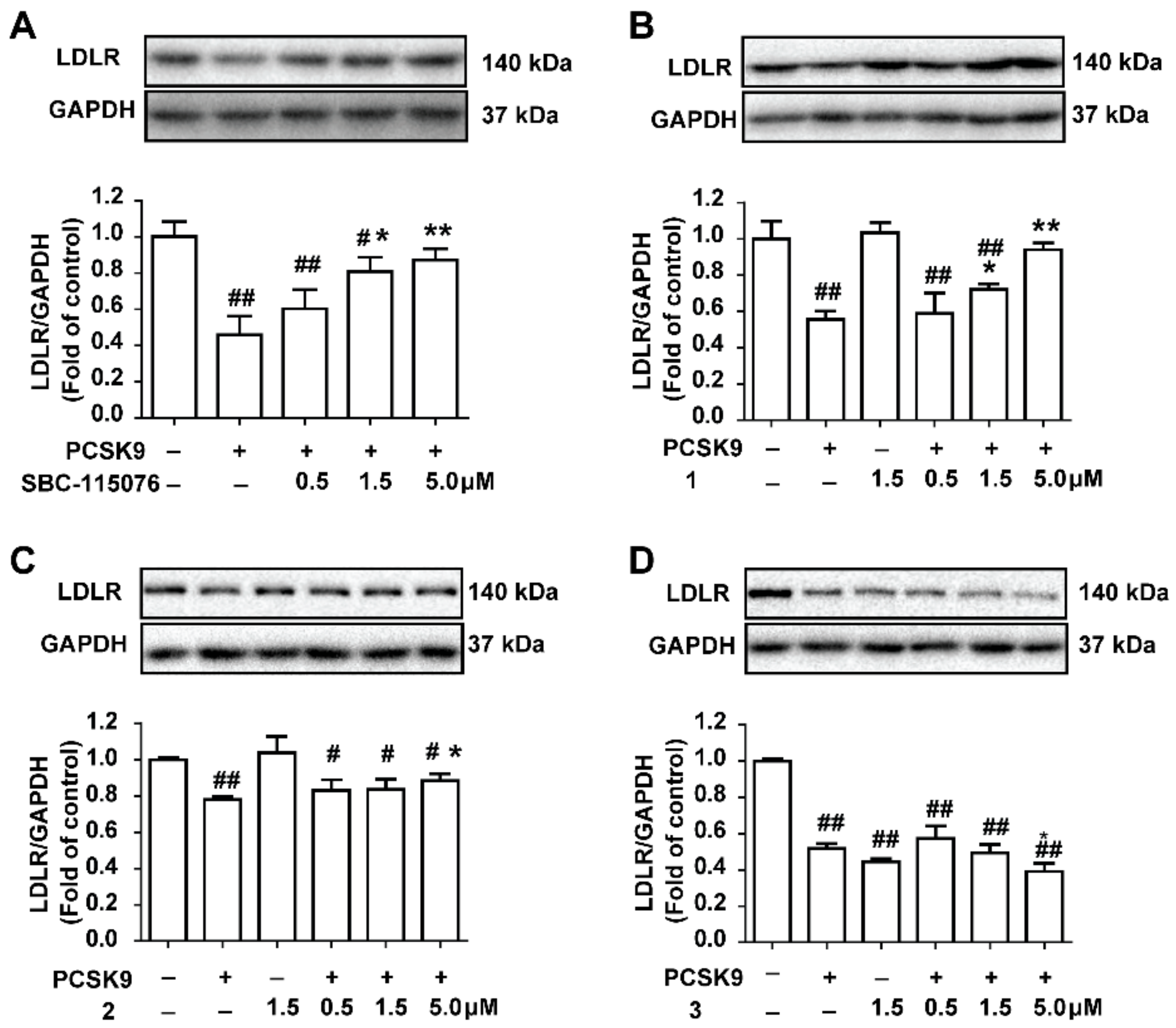
2.4. The Potential Natural Inhibitors Prevent PCSK9-Mediated LDLR Degradation in HepG2 Cells
3. Materials and Methods
3.1. Apparatus, Chemicals and Reagents
3.2. Construction, Expression, and Purification of His-PCSK9 Protein
3.3. Construction, Expression, and Purification of GST-EGF-A Protein
3.4. Interaction between PCSK9 and EGF-A by Fishing with Magnetic Beads
3.5. Evaluation of Inhibitory Activities on PCSK9/LDLR Interaction
3.6. HPLC Analysis for SBC-115076
3.7. Cell Culture and Measurement of LDLR in HepG2 Cells
3.8. Western Blot Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Myler, R.K.; Ryan, C.; Dunlap, R.; Shaw, R.E.; Bashour, T.T.; Cumberland, D.C.; Mooney, M.R. Dyslipoproteinemias in atherosclerosis, thrombosis and restenosis after coronary angioplasty. J. Invasive Cardiol. 1995, 7, 33–46. [Google Scholar] [PubMed]
- He, X.X.; Zhang, R.; Zuo, P.Y.; Liu, Y.W.; Zha, X.N.; Shan, S.S.; Liu, C.Y. The efficacy advantage of evolocumab (AMG 145) dosed at 140 mg every 2 weeks versus 420 mg every 4 weeks in patients with hypercholesterolemia: Evidence from a meta-analysis. Eur. J. Intern. Med. 2017, 38, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, D.; Witztum, J.L. Inhibition of PCSK9: A powerful weapon for achieving ideal LDL cholesterol levels. Proc. Natl. Acad. Sci. USA 2009, 106, 9546–9547. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tumanut, C.; Gavigan, J.A.; Huang, W.J.; Hampton, E.N.; Tumanut, R.; Suen, K.F.; Trauger, J.W.; Spraggon, G.; Lesley, S.A.; et al. Secreted PCSK9 promotes LDL receptor degradation independently of proteolytic activity. Biochem. J. 2007, 406, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Lagace, T.A.; Curtis, D.E.; Garuti, R.; McNutt, M.C.; Park, S.W.; Prather, H.B.; Anderson, N.N.; Ho, Y.K.; Hammer, R.E.; Horton, J.D. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Investig. 2006, 116, 2995–3005. [Google Scholar] [CrossRef] [PubMed]
- Abifadel, M.; Elbitar, S.; El Khoury, P.; Ghaleb, Y.; Chemaly, M.; Moussalli, M.L.; Rabes, J.P.; Varret, M.; Boileau, C. Living the PCSK9 adventure: From the identification of a new gene in familial hypercholesterolemia towards a potential new class of anticholesterol drugs. Curr. Atheroscler. Rep. 2014, 16, 439. [Google Scholar] [CrossRef] [PubMed]
- Yadav, K.; Sharma, M.; Ferdinand, K.C. Proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitors: Present perspectives and future horizons. Nutr. Metab. Cardiovasc. Dis. NMCD 2016, 26, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Gusarova, V.; Stahl, N.; Gurnett-Bander, A.; Kyratsous, C.A. Alirocumab, a Therapeutic Human Antibody to PCSK9, Does Not Affect CD81 Levels or Hepatitis C Virus Entry and Replication into Hepatocytes. PLoS ONE 2016, 11, e0154498. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Eigenbrot, C.; Zhou, L.; Shia, S.; Li, W.; Quan, C.; Tom, J.; Moran, P.; Di Lello, P.; Skelton, N.J.; et al. Identification of a small peptide that inhibits PCSK9 protein binding to the low density lipoprotein receptor. J. Boil. Chem. 2014, 289, 942–955. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ye, J.; Li, J.; Chen, C.; Huang, J.; Liu, P.; Huang, H. Polydatin ameliorates lipid and glucose metabolism in type 2 diabetes mellitus by downregulating proprotein convertase subtilisin/kexin type 9 (PCSK9). Cardiovasc. Diabetol. 2016, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Tai, M.H.; Chen, P.K.; Chen, P.Y.; Wu, M.J.; Ho, C.T.; Yen, J.H. Curcumin enhances cell-surface LDLR level and promotes LDL uptake through downregulation of PCSK9 gene expression in HepG2 cells. Mol. Nutr. Food Res. 2014, 58, 2133–2145. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.C.; Chen, P.Y.; Wu, M.J.; Tai, M.H.; Yen, J.H. Tanshinone IIA Modulates Low Density Lipoprotein Uptake via Down-Regulation of PCSK9 Gene Expression in HepG2 Cells. PLoS ONE 2016, 11, e0162414. [Google Scholar] [CrossRef] [PubMed]
- Nhoek, P.; Chae, H.S.; Masagalli, J.N.; Mailar, K.; Pel, P.; Kim, Y.M.; Choi, W.J.; Chin, Y.W. Discovery of Flavonoids from Scutellaria baicalensis with Inhibitory Activity Against PCSK 9 Expression: Isolation, Synthesis and Their Biological Evaluation. Molecules 2018, 23, 504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.W.; Lagace, T.A.; Garuti, R.; Zhao, Z.; McDonald, M.; Horton, J.D.; Cohen, J.C.; Hobbs, H.H. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Boil. Chem. 2007, 282, 18602–18612. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Pang, L.; Zhang, R.; Murgolo, N.J.; Lan, H.; Hedrick, J.A. PCSK9 binds to multiple receptors and can be functionally inhibited by an EGF-A peptide. Biochem. Biophys. Res. Commun. 2008, 375, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, L.; Kong-Beltran, M.; Li, W.; Moran, P.; Wang, J.; Quan, C.; Tom, J.; Kolumam, G.; Elliott, J.M.; et al. Calcium-independent inhibition of PCSK9 by affinity-improved variants of the LDL receptor EGF(A) domain. J. Mol. Boil. 2012, 422, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.J.; Lagace, T.A.; McNutt, M.C.; Horton, J.D.; Deisenhofer, J. Molecular basis for LDL receptor recognition by PCSK9. Proc. Natl. Acad. Sci. USA 2008, 105, 1820–1825. [Google Scholar] [CrossRef] [PubMed]
- Bottomley, M.J.; Cirillo, A.; Orsatti, L.; Ruggeri, L.; Fisher, T.S.; Santoro, J.C.; Cummings, R.T.; Cubbon, R.M.; Lo Surdo, P.; Calzetta, A.; et al. Structural and biochemical characterization of the wild type PCSK9-EGF(AB) complex and natural familial hypercholesterolemia mutants. J. Boil. Chem. 2009, 284, 1313–1323. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.M.; Adijiang, A.; Mah, M.; Zhang, D.W. Characterization of the role of EGF-A of low density lipoprotein receptor in PCSK9 binding. J. Lipid Res. 2013, 54, 3345–3357. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Sun, L.N.; Xing, W.W.; Huang, B.K.; Jia, M.; Wu, J.Z.; Zhang, H.; Qin, L.P. Lipid-lowering effects of polydatin from Polygonum cuspidatum in hyperlipidemic hamsters. Phytomed. Int. J. Phytother. Phytopharm. 2009, 16, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Lu, J.M.; Wang, Y.F.; Gu, W.; Zhao, R.H.; Yu, J. Prevention Mechanism of 2,3,5,4′-Tetrahydroxy-stilbene-2-O-beta-d-glucoside on Lipid Accumulation in Steatosis Hepatic L-02 Cell. Pharmacogn. Mag. 2017, 13, 245–253. [Google Scholar] [PubMed]
- Javandoost, A.; Afshari, A.; Nikbakht-Jam, I.; Khademi, M.; Eslami, S.; Nosrati, M.; Foroutan-Tanha, M.; Sahebkar, A.; Tavalaie, S.; Ghayour-Mobarhan, M.; et al. Effect of crocin, a carotenoid from saffron, on plasma cholesteryl ester transfer protein and lipid profile in subjects with metabolic syndrome: A double blind randomized clinical trial. ARYA Atheroscler. 2017, 13, 245–252. [Google Scholar] [PubMed]
- Benjannet, S.; Rhainds, D.; Essalmani, R.; Mayne, J.; Wickham, L.; Jin, W.; Asselin, M.C.; Hamelin, J.; Varret, M.; Allard, D.; et al. NARC-1/PCSK9 and its natural mutants: Zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Boil. Chem. 2004, 279, 48865–48875. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.N.; Breslow, J.L. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc. Natl. Acad. Sci. USA 2004, 101, 7100–7105. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Moon, Y.A.; Horton, J.D. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. J. Boil. Chem. 2004, 279, 50630–50638. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Lu, C.; Ryan, R.O. A two-step binding model of PCSK9 interaction with the low density lipoprotein receptor. J. Boil. Chem. 2011, 286, 5464–5470. [Google Scholar] [CrossRef] [PubMed]
- Miao, Q.; Shi, X.P.; Ye, M.X.; Zhang, J.; Miao, S.; Wang, S.W.; Li, B.; Jiang, X.X.; Zhang, S.; Hu, N.; et al. Polydatin attenuates hypoxic pulmonary hypertension and reverses remodeling through protein kinase C mechanisms. Int. J. Mol. Sci. 2012, 13, 7776–7787. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Lu, J.; Wang, Y.; Gu, W.; Yu, J.; Zhao, R. Naturally Occurring Stilbenoid TSG Reverses Non-Alcoholic Fatty Liver Diseases via Gut-Liver Axis. PLoS ONE 2015, 10, e0140346. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; He, Y.; Lin, P.; Li, Y.; Sun, R.; Gu, W.; Yu, J.; Zhao, R. In vitro effects of active components of Polygonum Multiflorum Radix on enzymes involved in the lipid metabolism. J. Ethnopharmacol. 2014, 153, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Meguid, S.; Abou-Gharbia, M.; Blass, B.; Childers, W.; Elshourbagy, N. Anti-Proprotein Convertase Subtilisin Kexin Type 9 (Anti-PCSK9) Compounds and Methods of Using the Same in the Treatment and/or Prevention of Cardiovascular Diseases. U.S. Patent 14,767,133, 11 March 2014. [Google Scholar]
Sample Availability: Samples of the compounds 1–3 are available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Shen, C.; Huang, Y.-X.; Li, Y.-N.; Liu, X.-F.; Liu, X.-M.; Liu, J.-H. A New Strategy for Rapidly Screening Natural Inhibitors Targeting the PCSK9/LDLR Interaction In Vitro. Molecules 2018, 23, 2397. https://doi.org/10.3390/molecules23092397
Li L, Shen C, Huang Y-X, Li Y-N, Liu X-F, Liu X-M, Liu J-H. A New Strategy for Rapidly Screening Natural Inhibitors Targeting the PCSK9/LDLR Interaction In Vitro. Molecules. 2018; 23(9):2397. https://doi.org/10.3390/molecules23092397
Chicago/Turabian StyleLi, Li, Chen Shen, Ya-Xuan Huang, Ya-Nan Li, Xiu-Feng Liu, Xu-Ming Liu, and Ji-Hua Liu. 2018. "A New Strategy for Rapidly Screening Natural Inhibitors Targeting the PCSK9/LDLR Interaction In Vitro" Molecules 23, no. 9: 2397. https://doi.org/10.3390/molecules23092397
APA StyleLi, L., Shen, C., Huang, Y.-X., Li, Y.-N., Liu, X.-F., Liu, X.-M., & Liu, J.-H. (2018). A New Strategy for Rapidly Screening Natural Inhibitors Targeting the PCSK9/LDLR Interaction In Vitro. Molecules, 23(9), 2397. https://doi.org/10.3390/molecules23092397




