Comparative Transcriptome Profiling Reveals Defense-Related Genes against Meloidogyne incognita Invasion in Tobacco
Abstract
1. Introduction
2. Results
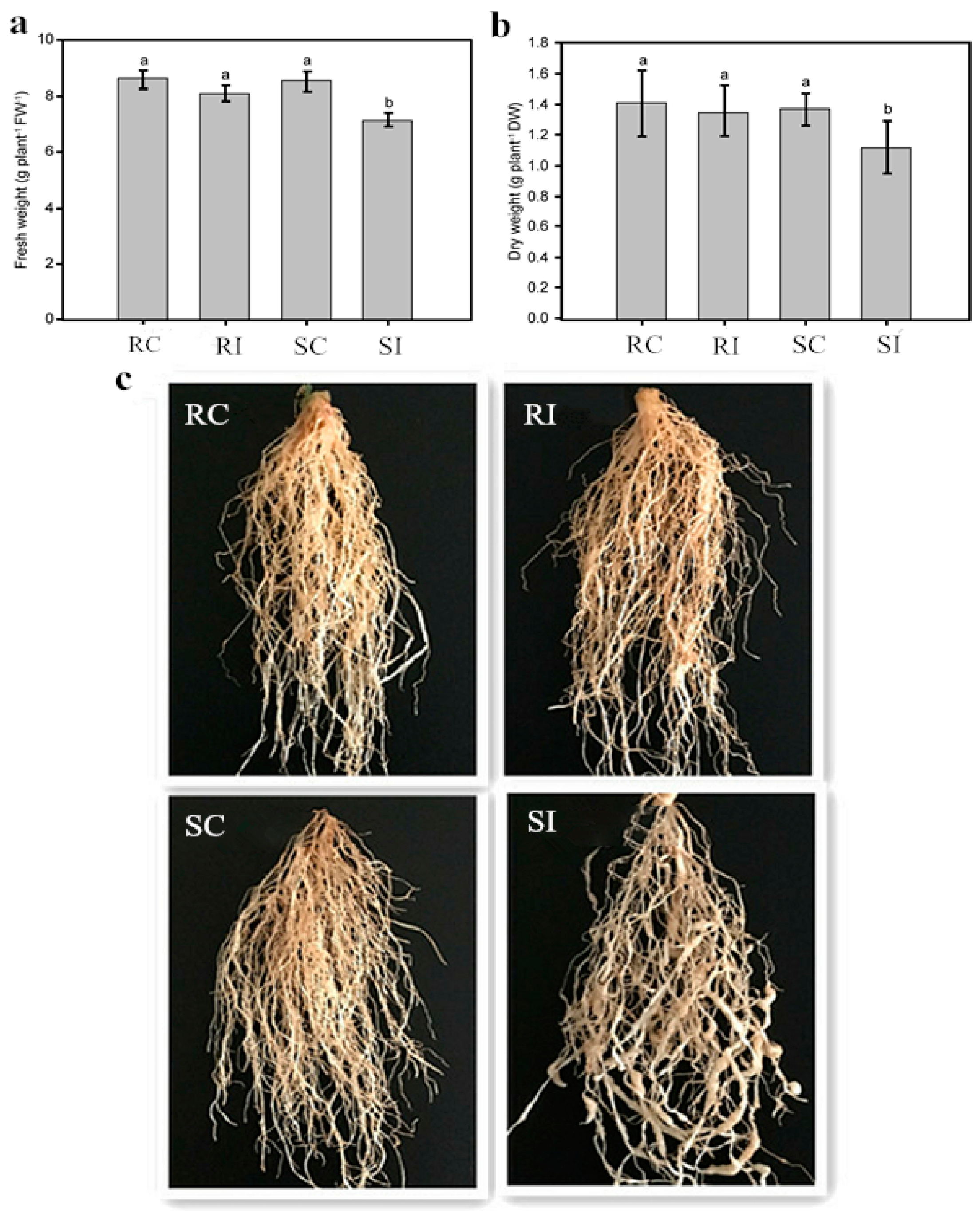
2.1. Effects of Nematode Infection on the Phenotypic Traits of Different Tobacco Genotypes
2.2. Resistance and Susceptibility of the Two Genotypes
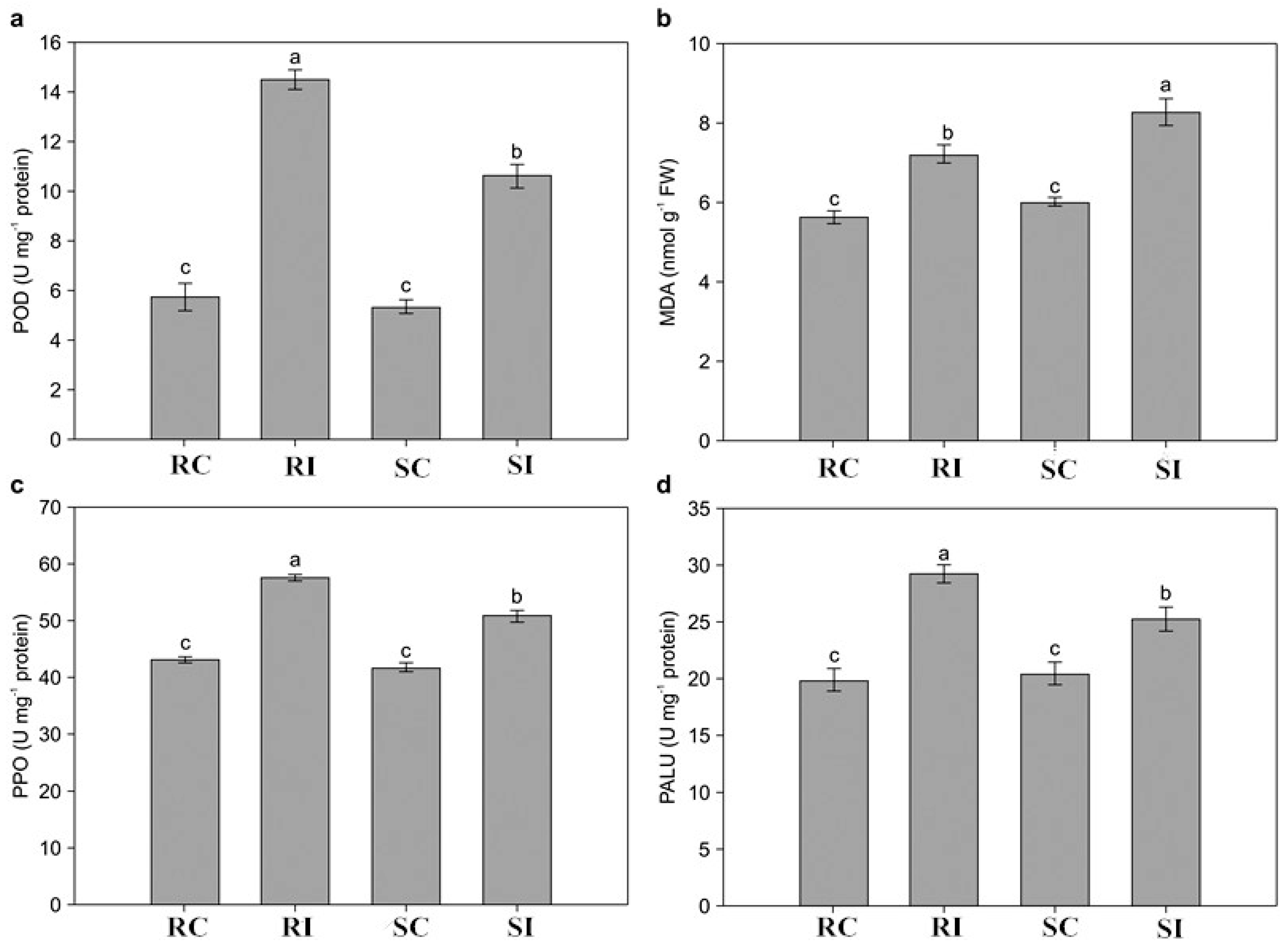
2.3. Determination of Physiological Changes in Response to M. incognita
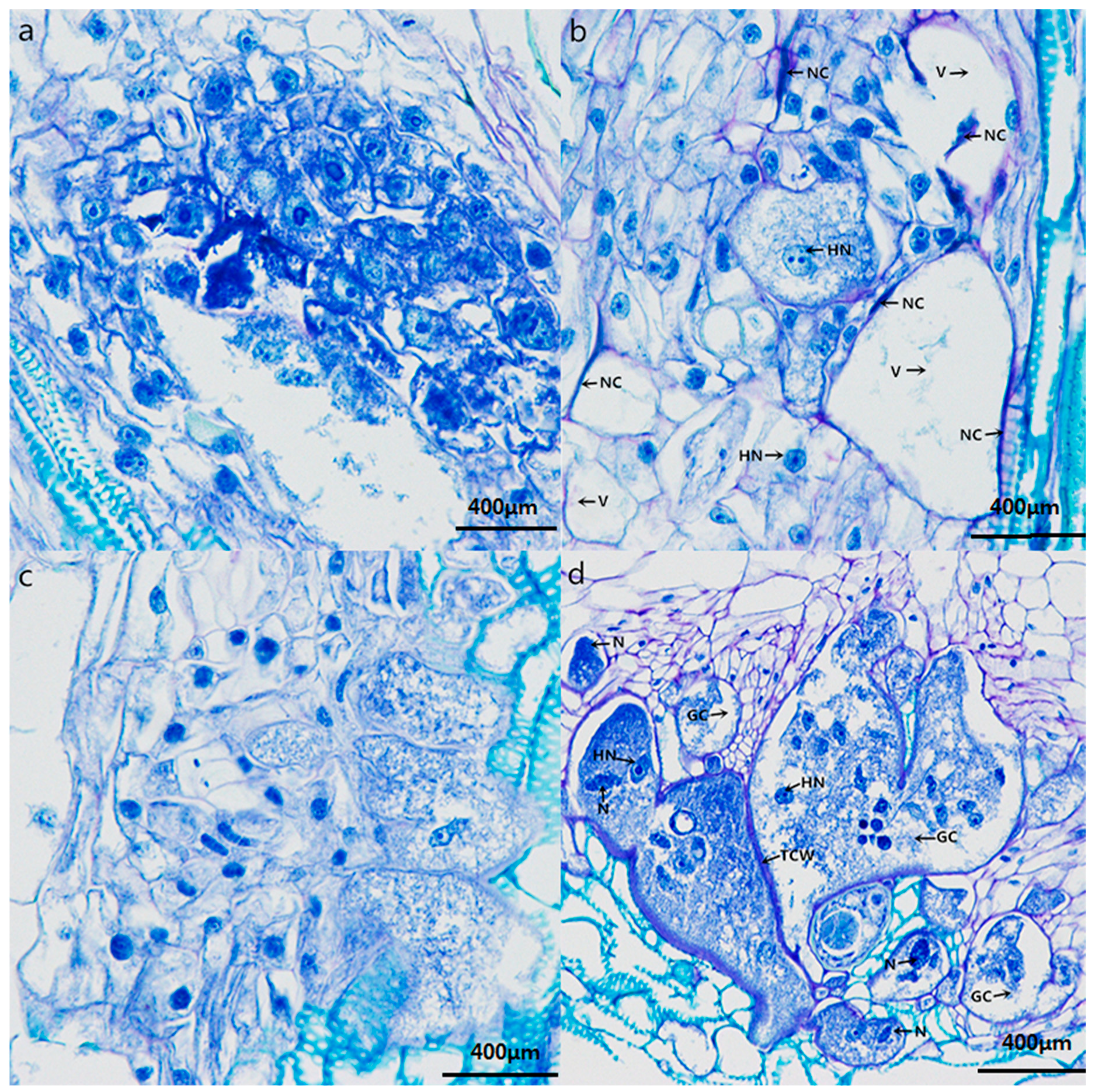
2.4. Histopathological Response to Nematode Infection
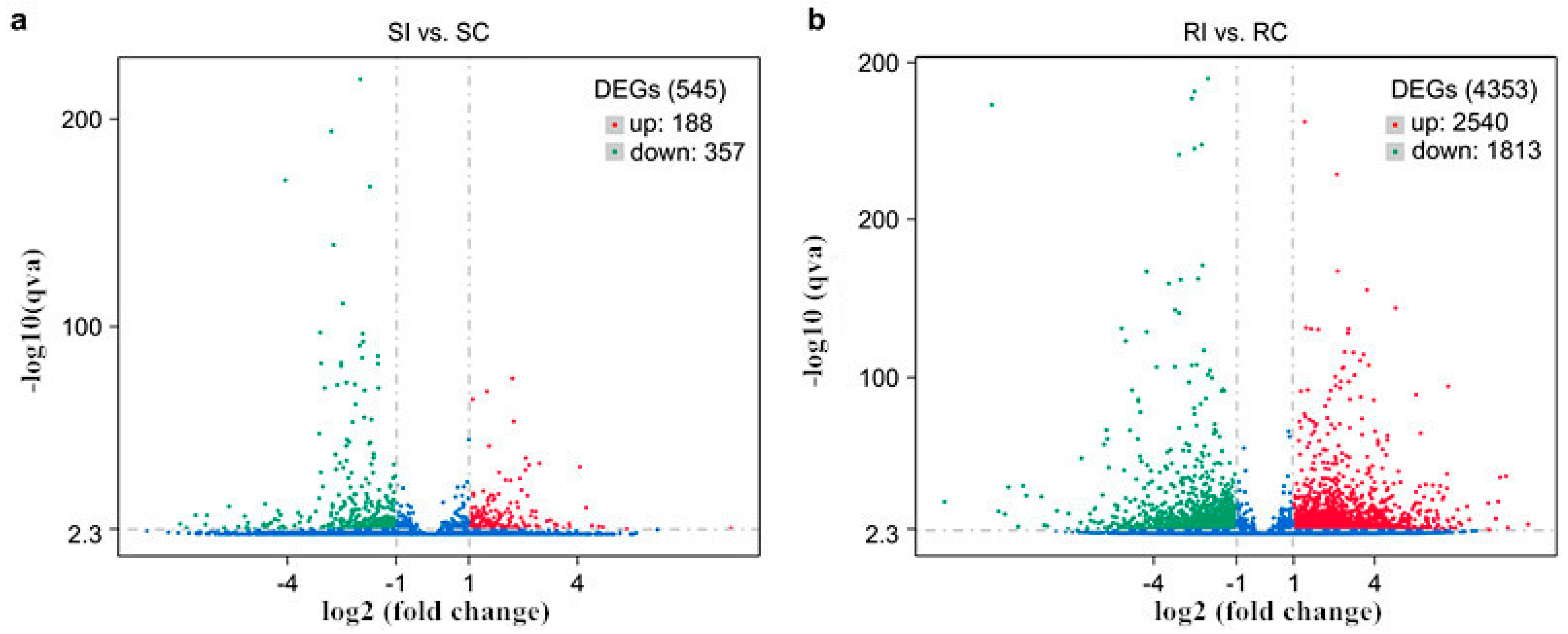
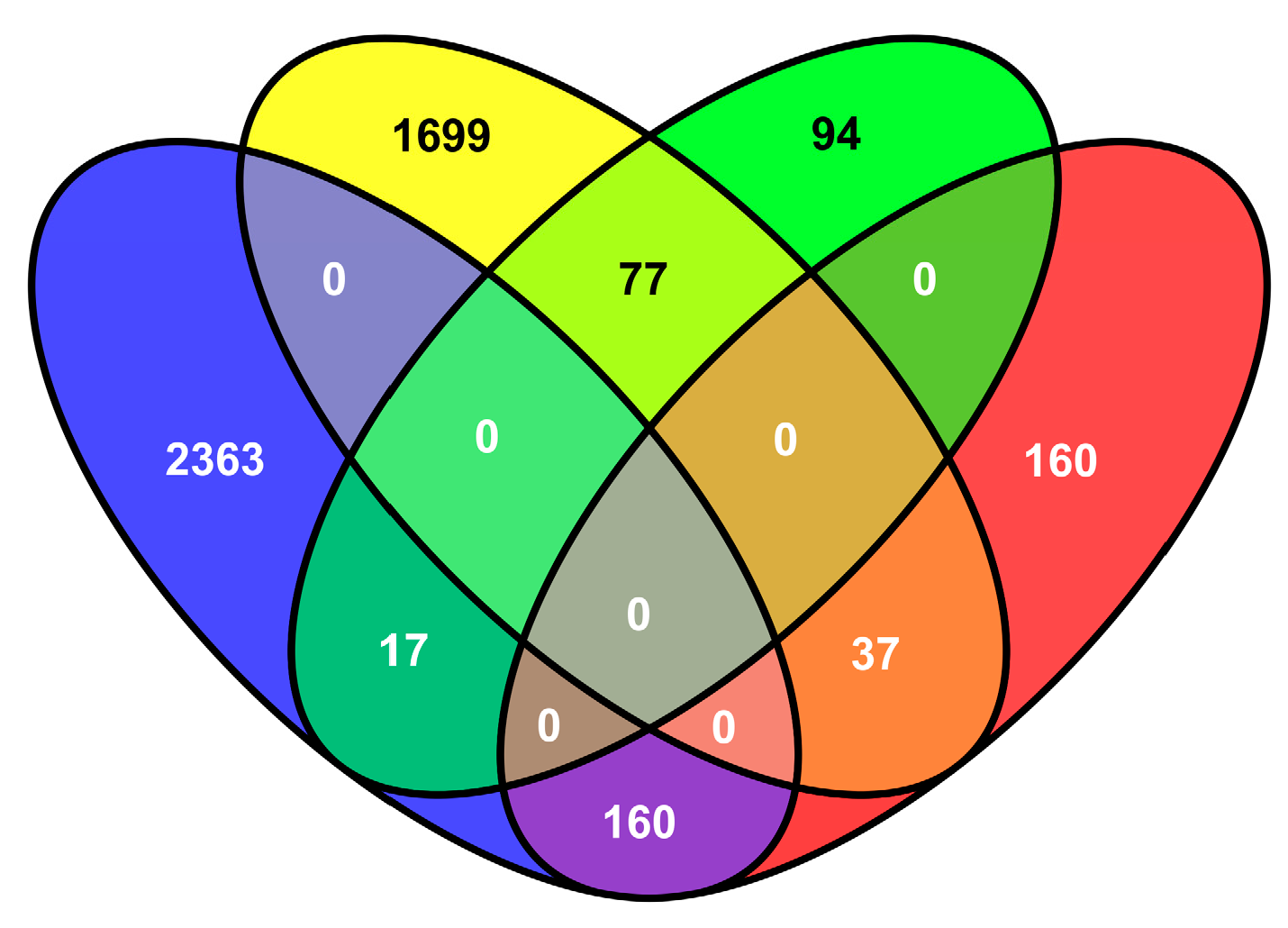
2.5. Transcriptional Changes in Tobacco in Response to M. incognita
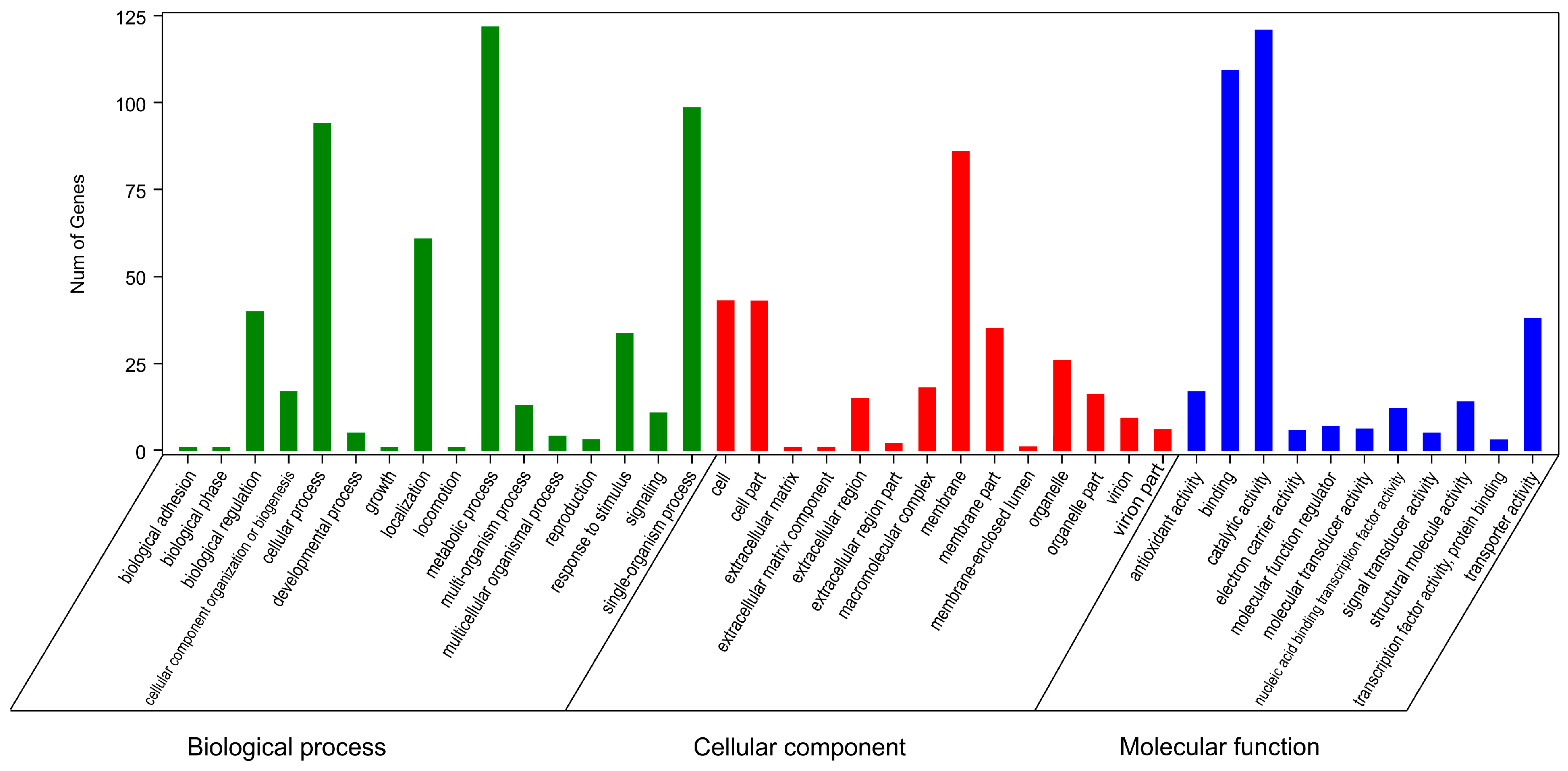
2.6. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analyses of Resistance-Related DEGs
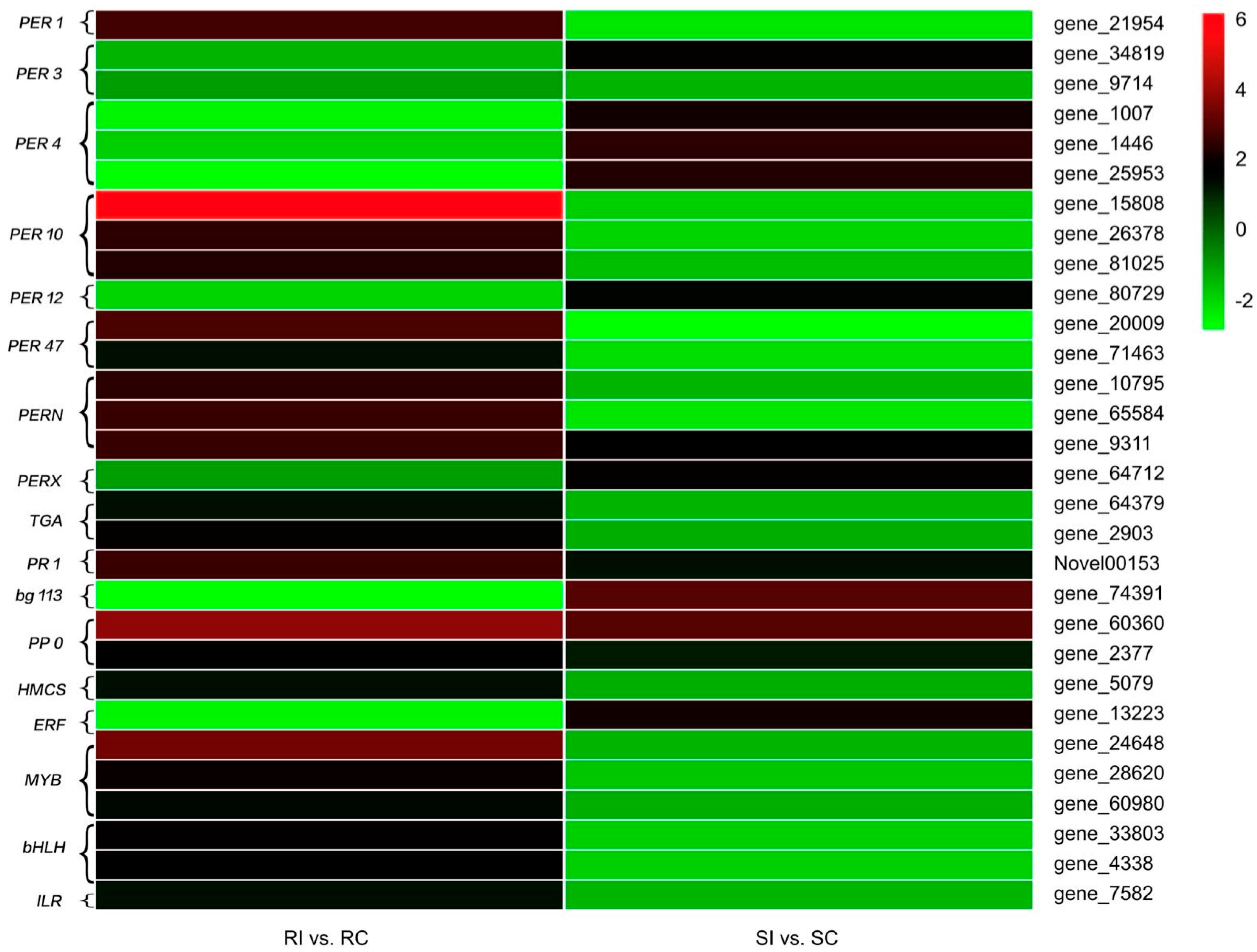
2.7. Antioxidant Enzymes
2.8. Signal Transduction
2.9. Secondary Metabolism
2.10. Transcription Factors
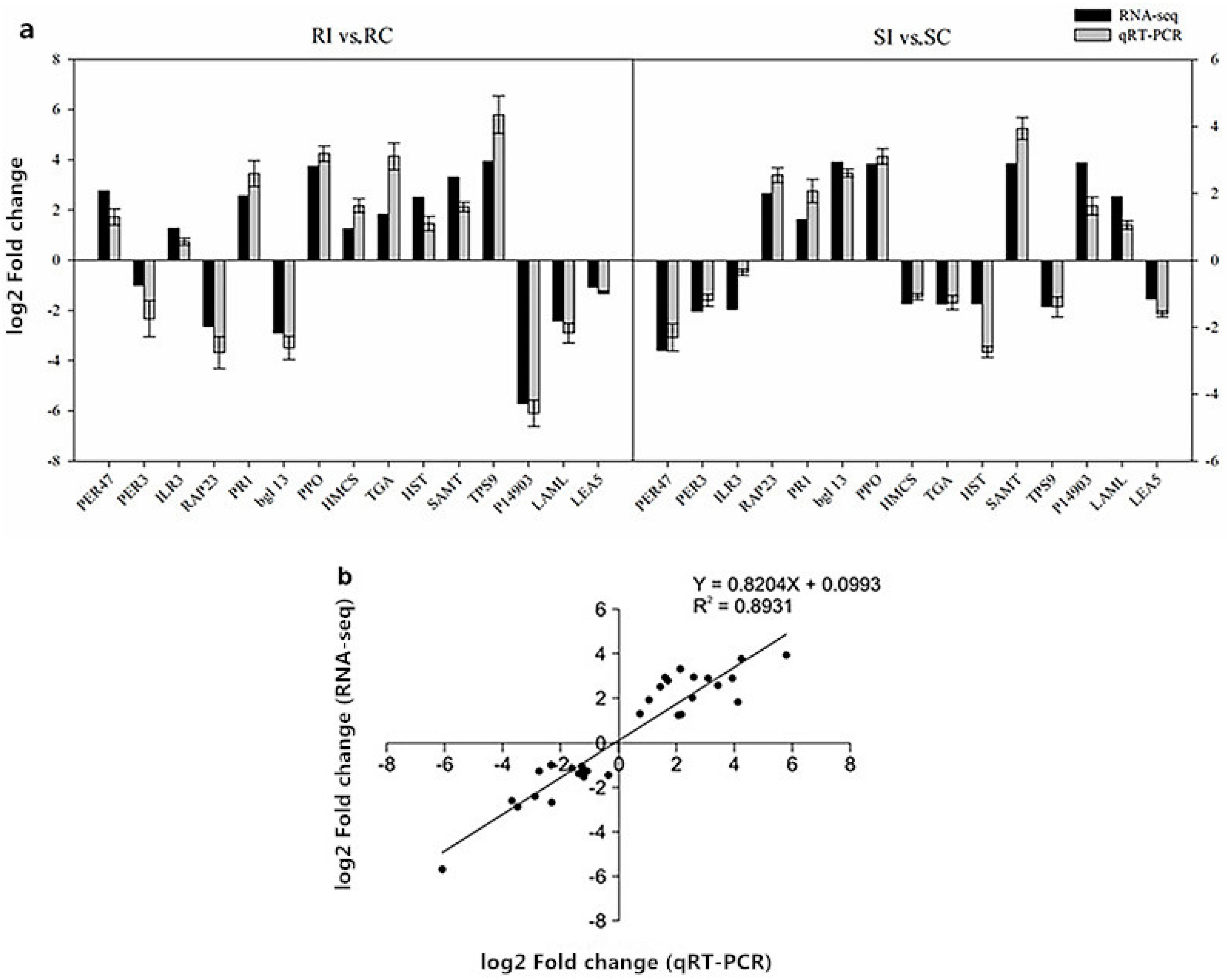
2.11. Verification of RNA-Seq Data by Quantitative Real-Time Polymerase Chain Reaction
3. Discussion
4. Materials and Methods
4.1. Preparation of Tobacco Material and Nematode Inoculation
4.2. Determination of Physiological Parameters and Resistance to M. incognita
4.3. Histological Experiments
4.4. Illumina Sequencing Andreads Mapping to the Reference Genome
4.5. Gene Annotation and Data Analysis
4.6. Validation of RNA-Seq Data by qRT-PCR
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jacquet, M.; Bongiovanni, M.; Martinez, M.; Verschave, P.; Wajnberg, E.; Castagnone-Sereno, P. Variation in resistance to the root-knot nematode Meloidogyne incognita in tomato genotypes bearing the Mi gene. Plant Pathol. 2005, 54, 93–99. [Google Scholar] [CrossRef]
- Garcia, G.M.; Stalker, H.T.; Shroeder, E.; Kochert, G. Identification of RAPD, SCAR, and RFLP markers tightly linked to nematode resistance genes introgressed from Arachiscardenasii into Arachishypogaea. Genome 1996, 39, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Ammiraju, J.; Veremis, J.; Huang, X.; Roberts, P.A.; Kaloshian, I. The heat-stable root-knot nematode resistance gene Mi-9 from Lycopersiconperuvianum is localized on the short arm of chromosome 6. Theor. Appl. Genet. 2003, 106, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Djian-Caporalino, C.; Pijarowski, L.; Fazari, A.; Samson, M.; Gaveau, L.; O’Byrne, C.; Lefebvre, V.; Caranta, C.; Palloix, A.; Abad, P. High-resolution genetic mapping of the pepper (Capsicum annuum L.) resistance loci Me3 and Me4 conferring heat-stable resistance to root-knot nematodes (Meloidogyne spp.). Theor. Appl. Genet. 2001, 103, 592–600. [Google Scholar] [CrossRef]
- Abad, P.; Favery, B.; Rosso, M.N.; Castagnone-Sereno, P. Root-knot nematode parasitism and host response: Molecular basis of a sophisticated interaction. Mol. Plant Pathol. 2003, 4, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Y.; Du, Y.; Chen, S.; Tang, H. Dynamic metabonomic responses of tobacco (Nicotianatabacum) plants to salt stress. J. Proteome Res. 2011, 10, 1904–1914. [Google Scholar] [CrossRef] [PubMed]
- Hamamouch, N.; Li, C.; Seo, P.J.; Park, C.M.; Davis, E.L. Expression of Arabidopsis pathogenesis-related genes during nematode infection. Mol. Plant Pathol. 2011, 12, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.L.; Haegeman, A.; Kikuchi, T. Degradation of the Plant Cell Wall by Nematodes; Springer: Dordrecht, The Netherlands, 2011; pp. 255–272. [Google Scholar]
- Liu, P.P.; Yang, Y.; Pichersky, E.; Klessiq, D.F. Altering expression of benzoic acid/salicylic acid carboxyl methyltransferase 1 compromises systemic acquired resistance and PAMP-triggered immunity in Arabidopsis. Mol. Plant–Microbe Interact. 2010, 23, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, B.C.; Oelmuller, R. Reactive oxygen species generation and signaling in plants. Plant Signal. Behav. 2012, 7, 1621–1633. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, K.K.; Atamian, H.S.; Kaloshian, I.; Eulgem, T. WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J. 2010, 63, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Cooper, W.R.; Jia, L.; Goggin, L. Effects of jasmonate-induced defenses on root-knot nematode infection of resistant and susceptible tomato cultivars. J. Chem. Ecol. 2005, 31, 1953–1967. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Fei, Z.; Giovannoni, J.; Rose, J.K. Catalyzing plant science research with RNA-seq. Front. Plant Sci. 2013, 4, 66. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Zenoni, S.; Delledonne, M. Characterization of transcriptional complexity during berry development in Vitisvinifera using RNA-Seq. Plant Physiol. 2010, 152, 1787–1795. [Google Scholar] [CrossRef] [PubMed]
- Santini, L.; MunhozCde, F.; Bonfim, M.F., Jr.; Brandao, M.M.; Inomoto, M.M.; Vieira, M.L. Host transcriptional profiling at early and later stages of the compatible interaction between Phaseolus vulgaris and Meloidogyne incognita. Phytopathology 2015, 106, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.Y.; Qi, Y.H.; Cao, S.F.; Wei, B.Q.; Zhang, H.S. Histopathology combined with transcriptome analyses reveals the mechanism of resistance to Meloidogyne incognita in Cucumismetuliferus. J. Plant Physiol. 2017, 212, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Shukla, N.; Yadav, R.; Kaur, P.; Rasmussen, S.; Goel, S.; Agarwal, M.; Jagannath, A.; Gupta, R.; Kumar, A. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanumlycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol. Plant Pathol. 2017, 19, 615–633. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jinkai, S.; Xianchao, Z. Evaluation of resistance of tobacco germplasm to root-knot nematodes. J. Shenyang Agric. Univ. 2001, 32, 183–185. [Google Scholar]
- Xu, M.L.; Lu, X.P.; Wang, S.H. The study on resistance of tobacco germplasm to root-knot nematodes. Tobacco Sci. Technol. 1998, 3, 42–43. [Google Scholar]
- Postnikova, O.A.; Hult, M.; Shao, J.; Skantar, A.; Nemchinov, L.G. Transcriptome analysis of resistant and susceptible alfalfa cultivars infected with root-knot nematode Meloidogyne incognita. PLoS ONE 2015, 10, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Bindschedler, L.V.; Dewdney, J.; Blee, K.A.; Stone, J.M.; Asai, T.; Plotnikov, J.; Denoux, C.; Hayes, T.; Gerrish, C.; Davies, D.R.; et al. Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 2006, 47, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Ahammed, G.J.; Wu, C.; Fan, S.Y.; Zhou, Y.H. Crosstalk among jasmonate, salicylate and ethylene signaling pathways in plant disease and immune responses. Curr. Protein Pept. Sci. 2015, 16, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, J.; Yin, L.; Zhang, Y.; Qu, J.; Lu, J. Comparative transcriptome analysis reveals defense-related genes and pathways against downy mildew in Vitisamurensis, grapevine. Plant Physiol. Biochem. 2015, 95, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lindemose, S.; O’Shea, C.; Jensen, M.K.; Skriver, K. Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 2013, 14, 5842–5878. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.B.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Szakasits, D.; Heinen, P.; Wieczorek, K.; Hofmann, J.; Wagner, F.; Kreil, D.P.; Sykacek, P.; Grundler, F.M.; Bohlmann, H. The transcriptome of syncytia induced by the cyst nematode Heteroderaschachtii in Arabidopsis roots. Plant J. Cell Mol. Biol. 2009, 57, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, P.M.; Guimaraes, L.A.; Morgante, C.V.; Silva, O.B., Jr.; Araujo, A.C.; Martins, A.C.; Saraiva, M.A.; Oliveira, T.N.; Togawa, R.C.; Leal-Bertioli, S.C.; et al. Root transcriptome analysis of wild peanut reveals candidate genes for nematode resistance. PLoS ONE 2015, 10, e0140937. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Chittem, K.; Brueggeman, R.; Osorno, J.M.; Richards, J.; Nelson, B.D., Jr. Comparative transcriptomeanalysis of resistant and susceptible common bean genotypes in response to soybean cyst nematode infection. PLoS ONE 2016, 11, e0159338. [Google Scholar] [CrossRef] [PubMed]
- Chong, K.P.; Atong, M.; Rossall, S. The role of syringic acid in the interaction between oil palm and Ganodermaboninense, the causal agent of basal stem rot. Plant Pathol. 2012, 61, 953–963. [Google Scholar] [CrossRef]
- Simonetti, E.; Veronico, P.; Melillo, M.T.; Delibes, A.; Andrés, M.F.; López-Braña, I. Analysis of class III peroxidase genes expressed in roots of resistant and susceptible wheat lines infected by Heteroderaavenae. Mol. Plant–Microbe Interact. 2009, 22, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, H.M.; Hosseini, P.; Alkharouf, N.W.; Hussein, E.H.; Gamal El-Din Ael, K.; Aly, M.A.; Matthews, B.F. Analysis of gene expression in soybean (Glycine max) roots in response to the root knot nematode Meloidogyne incognita using microarrays and KEGG pathways. BMC Genom. 2011, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hewezi, T.; Howe, P.; Maier, T.R.; Hussey, R.S.; Mitchum, M.G.; Davis, E.L.; Baum, T.J. Cellulose binding protein from the parasitic nematode Heteroderaschachtii interacts with Arabidopsis pectin methylesterase: Cooperative cell wall. Plant Cell 2008, 20, 3080–3093. [Google Scholar] [CrossRef] [PubMed]
- Bolwell, G.P.; Bindschedler, L.V.; Blee, K.A.; Butt, V.S.; Davies, D.R.; Gardner, S.L.; Gerrish, C.; Minibayeva, F. Theapoplastic oxidative burst in response to biotic stress in plants: A three-component system. J. Exp. Bot. 2002, 53, 1367–1376. [Google Scholar] [PubMed]
- Zhu, J.J.; Zhang, J.L.; Liu, H.C.; Cao, K.F. Photosynthesis, non-photochemical pathways and activities of antioxidant enzymes in a resilient evergreen oak under different climatic conditions from a valley-savanna in Southwest China. Physiol. Plant. 2009, 135, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, K.C.; Ganguly, A.K.; Dasgupta, D.R. Development of peroxidase (E.C.1.11.1.7) activities in susceptible and resistant cultivars of cowpea inoculated with the root-knot nematode, Meloidogyne incognita. Indian J. Nematol. 1986, 16, 252–256. [Google Scholar]
- Dhandaydham, M.; Charles, L.; Zhu, H.; Starr, J.L.; Huguet, T.; Cook, D.R.; Prosperi, J.M.; Opperman, C. Characterization of root-knot nematode resistance in Medicago truncatula. J. Nematol. 2008, 40, 46–54. [Google Scholar] [PubMed]
- Albuquerque, E.V.S.; Carneiro, R.M.D.G.; Costa, P.M.; Gomes, A.C.M.M.; Santos, M.; Pereira, A.A.; Nicole, M.; Fernandez, D.; Grossi-de-Sa, M.F. Resistance to Meloidogyne incognita, expresses a hypersensitive-like response in Coffea arabica. Eur. J. Plant Pathol. 2010, 127, 365–373. [Google Scholar] [CrossRef]
- Williamson, V.M.; Kumar, A. Nematode resistance in plants: The battle underground. Trends Genet. 2006, 22, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; De Mason, D.A.; Ehlers, J.D.; Close, T.J.; Roberts, P.A. Histological characterization of root-knot nematode resistance in cowpea and its relation to reactive oxygen species modulation. J. Exp. Bot. 2008, 59, 1305–1313. [Google Scholar] [CrossRef] [PubMed]
- Raskin, I. Role of salicylic acid in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 439–463. [Google Scholar] [CrossRef]
- Guo, J.; Shi, G.; Guo, X.; Zhang, L.; Xu, W.; Wang, Y.; Su, Z.; Hua, J. Transcriptome analysis reveals that distinct metabolic pathways operate in salt-tolerant and salt-sensitive upland cotton varieties subjected to salinity stress. Plant Sci. 2015, 238, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Molinari, S.; Fanelli, E.; Leonetti, P. Expression of tomato salicylic acid (SA)-responsive pathogenesis-related genes in Mi-1-mediated and SA-induced resistance to root-knot nematodes. Mol. Plant Pathol. 2014, 15, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Journot-Catalino, N.; Somssich, I.E.; Roby, D.; Kroj, T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 2006, 18, 3289–3302. [Google Scholar] [CrossRef] [PubMed]
- Rao, L.; Perez, D.; White, E. Lamin proteolysis facilitates nuclear events during apoptosis. J. Cell Biol. 1996, 135, 1441–1455. [Google Scholar] [CrossRef] [PubMed]
- Yingsanga, P.; Srilaong, V.; Kanlayanarat, S.; Noichinda, S.; McGlasson, W.B. Relationship between browning and related enzymes (PAL, PPO and POD) in rambutan fruit (Nepheliumlappaceum Linn.) cvs. Rongrien and See-Chompoo. Postharvest Biol. Technol. 2008, 49, 164–168. [Google Scholar] [CrossRef]
- Zhou, W.; Leul, M. Uniconazole-induced alleviation of freezing injury in relation to changes in hormonal balance, enzyme activities and lipid peroxidation in winter rape. Plant Growth Regul. 1998, 26, 41–47. [Google Scholar] [CrossRef]
- Li, X.; Zhao, W.; Zhou, X.; Feng, J.; Gao, Y.; Yao, X.; Liu, Y.; Liu, J.; Yang, R.; Zhao, F.; et al. The use of toluidine blue staining combined with paraffin sectioning and the optimization of freeze-thaw counting methods for analysing root-knot nematodes in tomato. Hortic. Env. Biotechnol. 2017, 58, 620–626. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Educ. Behav. Stat. 2000, 25, 60–83. [Google Scholar] [CrossRef]
- Lu, J.; Du, Z.X.; Kong, J.; Chen, L.N.; Qiu, Y.H.; Li, G.F.; Meng, X.H.; Zhu, S.F. Transcriptome analysis of Nicotiana tabacum infected by cucumber mosaic virus during systemic symptom development. PLoS ONE 2012, 7, e43447. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, G.W.; Delaney, S.K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotianatabacum) during development and abiotic stress. Mol. Genet. Genom. 2010, 283, 233–241. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds RC, RI, SC and SI are available from the authors. |
| Treatment | Raw Reads | Clean Reads | Total Mapped Reads (%) | Reads Mapped to Multiple Locations (%) | Uniquely Mapped Reads (%) |
|---|---|---|---|---|---|
| RC | 56,013,982 | 53,593,540 | 45,054,720 (84.07%) | 732,943 (1.37%) | 44,321,777 (82.7%) |
| RI | 46,922,130 | 45,634,502 | 37,457,092 (82.08%) | 676,444 (1.48%) | 36,780,648 (80.6%) |
| SC | 44,485,484 | 43,237,234 | 35,906,665 (83.05%) | 654,003 (1.51%) | 35,252,662 (81.53%) |
| SI | 47,795,944 | 46,339,310 | 38,154,998 (82.34%) | 701,137 (1.51%) | 37,453,861 (80.83%) |
| Average | 48,804,385 | 47,201,146 | 39,143,369 (82.89%) | 691,132 (1.47%) | 38,627,522 (81.42%) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Xing, X.; Tian, P.; Zhang, M.; Huo, Z.; Zhao, K.; Liu, C.; Duan, D.; He, W.; Yang, T. Comparative Transcriptome Profiling Reveals Defense-Related Genes against Meloidogyne incognita Invasion in Tobacco. Molecules 2018, 23, 2081. https://doi.org/10.3390/molecules23082081
Li X, Xing X, Tian P, Zhang M, Huo Z, Zhao K, Liu C, Duan D, He W, Yang T. Comparative Transcriptome Profiling Reveals Defense-Related Genes against Meloidogyne incognita Invasion in Tobacco. Molecules. 2018; 23(8):2081. https://doi.org/10.3390/molecules23082081
Chicago/Turabian StyleLi, Xiaohui, Xuexia Xing, Pei Tian, Mingzhen Zhang, Zhaoguang Huo, Ke Zhao, Chao Liu, Duwei Duan, Wenjun He, and Tiezhao Yang. 2018. "Comparative Transcriptome Profiling Reveals Defense-Related Genes against Meloidogyne incognita Invasion in Tobacco" Molecules 23, no. 8: 2081. https://doi.org/10.3390/molecules23082081
APA StyleLi, X., Xing, X., Tian, P., Zhang, M., Huo, Z., Zhao, K., Liu, C., Duan, D., He, W., & Yang, T. (2018). Comparative Transcriptome Profiling Reveals Defense-Related Genes against Meloidogyne incognita Invasion in Tobacco. Molecules, 23(8), 2081. https://doi.org/10.3390/molecules23082081




