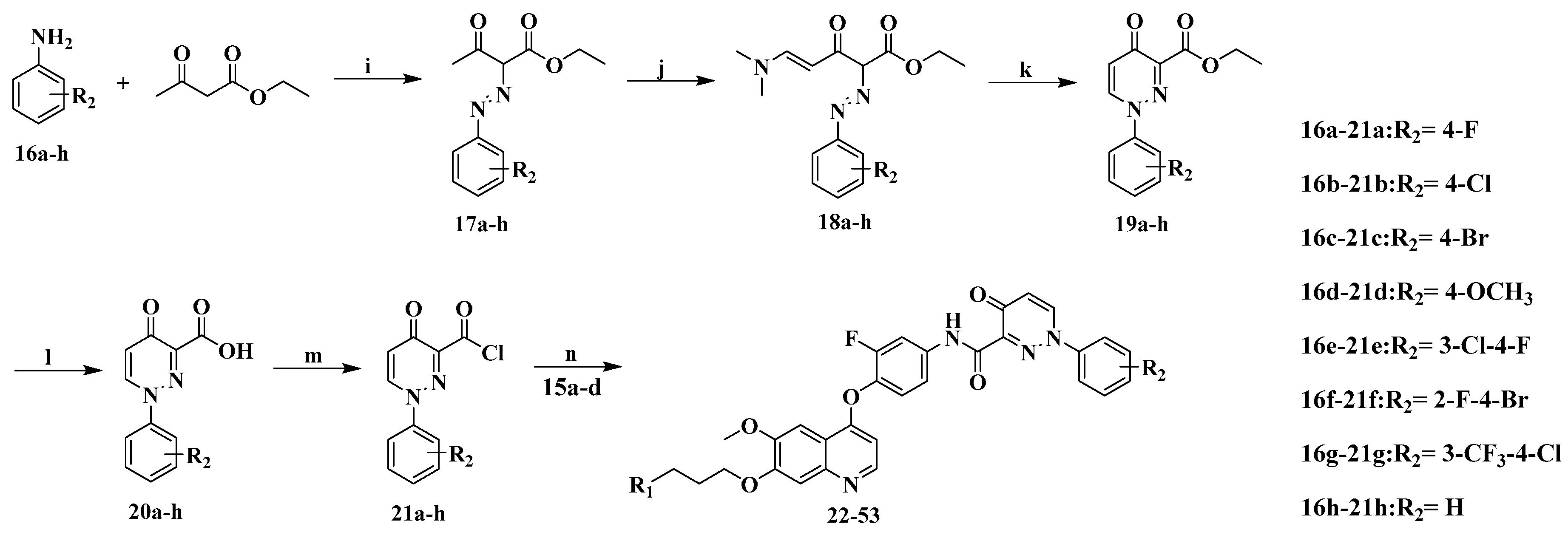
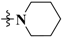3.2.7. Preparation of Compounds 22–53
A solution of phenylpyrdazinone carbonyl chloride 21a–h (0.82 mmol) in dichloromethane (10 mL) was added drop-wise to a solution of aniline 15a–d (0.41 mmol) and diisopropylethylamine (0.49 mmol) in dichloromethane (10 mL) in an ice bath. Upon completion of the addition, the reaction mixture was removed from the ice bath and placed in room temperature for 30 min and monitored by TLC. The mixture was concentrated in vacuum to yield 22–53 which were recrystallized by isopropanol.
N-(3-Fluoro-4-((6-methoxy-7-(3-morpholinopropoxy)quinolin-4-yl)xy)phenyl)-1-4-fluorophenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (22). Light yellow solid. 37.2% yield, m.p: 152.3–153.5 °C. 1H NMR (400 MHz, DMSO) δ 12.02 (s, 1H), 8.99 (d, J = 7.8 Hz, 1H), 8.48 (d, J = 5.2 Hz, 1H), 8.02 (d, J = 12.7 Hz, 1H), 7.86 (dd, J = 8.9, 4.6 Hz, 2H), 7.55 (d, J = 12.2 Hz, 2H), 7.49 (dt, J = 15.2, 7.6 Hz, 3H), 7.40 (s, 1H), 6.93 (d, J = 7.8 Hz, 1H), 6.49 (d, J = 5.2 Hz, 1H), 4.20 (t, J = 6.4 Hz, 2H), 3.95 (s, 3H), 3.59 (t, J = 4.2 Hz, 4H), 2.47 (d, J = 7.0 Hz, 2H), 2.40 (s, 4H), 2.02–1.94 (m, 2H). 13C NMR (101 MHz, DMSO) δ 169.09, 162.83, 159.92, 159.17, 154.74, 152.30, 151.93, 149.58, 148.81, 147.50, 146.37, 142.03, 139.41, 137.05, 136.95, 124.33, 123.87, 123.79, 120.46, 116.62, 116.38, 114.45, 108.53, 102.09, 101.50, 99.02, 66.66, 66.18(2C), 55.76, 54.79, 53.34(2C), 25.66. TOF MS ES+ (m/z): (M + H)+, calcd for C34H31F2N5O6: 644.2321, found, 644.2304.
1-(4-Chlorophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-morpholinopropoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (23). Light yellow solid. 36.2% yield, m.p: 138.2–139.1 °C. 1H NMR (400 MHz, DMSO) δ 11.95 (s, 1H), 9.02 (d, J = 7.9 Hz, 1H), 8.49 (d, J = 5.0 Hz, 1H), 8.02 (d, J = 12.8 Hz, 1H), 7.86 (d, J = 8.5 Hz, 2H), 7.70 (d, J = 8.5 Hz, 2H), 7.55 (d, J = 10.6 Hz, 2H), 7.50 (t, J = 8.7 Hz, 1H), 7.44 (s, 1H), 6.93 (d, J = 7.8 Hz, 1H), 6.51 (d, J = 5.0 Hz, 1H), 4.26 (s, 2H), 3.96 (s, 3H), 3.78 (s, 4H), 3.04 (s, 4H), 2.50–2.48 (m, 2H), 2.20 (s, 2H). TOF MS ES+ (m/z): (M + H)+, calcd for C34H31ClFN5O6: 660.2025, found, 660.2009.
1-(4-Bromophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-morpholinopropoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (24). Light yellow solid. 37.9% yield, m.p: 125.7–127.1 °C. 1H NMR (400 MHz, DMSO) δ 12.03 (s, 1H), 9.12 (d, J = 7.8 Hz, 1H), 8.58 (d, J = 5.2 Hz, 1H), 8.11 (d, J = 12.8 Hz, 1H), 7.91 (q, J = 8.6 Hz, 4H), 7.66 (d, J = 9.9 Hz, 2H), 7.60 (t, J = 8.7 Hz, 2H), 7.52 (s, 1H), 7.03 (d, J = 7.8 Hz, 1H), 6.60 (d, J = 5.1 Hz, 1H), 4.31 (d, J = 5.9 Hz, 2H), 4.05 (s, 3H), 3.76 (s, 4H), 2.76 (s, 4H), 2.60 (s, 2H), 2.15 (s, 2H). TOF MS ES+ (m/z): (M + H)+, calcd for C34H31BrFN5O6: 704.1520, found, 704.1501.
N-(3-Fluoro-4-((6-methoxy-7-(3-morpholinopropoxy)quinolin-4-yl)oxy)phenyl)-1-(4-methoxyphenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (25). White solid. 38.1% yield, m.p: 153.8–155.2 °C. 1H NMR (400 MHz, DMSO) δ 12.18 (s, 1H), 8.95 (d, J = 7.8 Hz, 1H), 8.69 (d, J = 5.8 Hz, 1H), 8.08 (d, J = 12.6 Hz, 1H), 7.77–7.67 (m, 3H), 7.59 (dd, J = 20.1, 9.7 Hz, 3H), 7.15 (d, J = 9.0 Hz, 2H), 6.92 (d, J = 7.8 Hz, 1H), 6.81 (d, J = 4.7 Hz, 1H), 4.33 (s, 2H), 4.02 (s, 3H), 3.84 (s, 3H), 3.49 (s, 4H), 3.12 (s, 4H), 2.50–2.49 (m, 2H), 2.35 (s, 2H). 13C NMR (101 MHz, DMSO) δ 169.48, 160.52, 159.82, 159.74, 155.18, 152.73, 152.11, 149.98, 149.21, 147.60, 146.46, 142.42, 137.63, 136.81, 124.77, 123.44(2C), 121.10, 117.15, 115.14(2C), 109.25, 109.04, 102.70, 101.95, 99.60, 66.58, 63.96, 56.30, 56.10(2C), 54.20, 51.79(2C), 23.59. TOF MS ES+ (m/z): (M + H)+, calcd for C35H34FN5O7: 656.2521, found, 656.2503.
1-(3-Chloro-4-fluorophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-morpholinopropoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (26). Pure white solid. 35.1% yield, m.p: 138.1–140.6 °C. 1H NMR (400 MHz, DMSO) δ 11.84 (s, 1H), 8.93 (d, J = 7.3 Hz, 1H), 8.39 (d, J = 4.6 Hz, 1H), 8.05 (s, 1H), 7.92 (d, J = 12.9 Hz, 1H), 7.77 (s, 1H), 7.61 (t, J = 8.8 Hz, 1H), 7.47 (d, J = 11.6 Hz, 2H), 7.44–7.38 (m, 1H), 7.31 (s, 1H), 6.85 (d, J = 7.5 Hz, 1H), 6.40 (d, J = 4.8 Hz, 1H), 4.11 (s, 2H), 3.87 (s, 3H), 3.50 (s, 4H), 2.37 (d, J = 6.5 Hz, 2H), 2.31 (s, 4H), 1.90 (d, J = 5.5 Hz, 2H). 13C NMR (101 MHz, DMSO) δ 169.67, 160.19, 159.65, 158.56, 156.09, 155.23, 152.78, 152.45, 150.11, 149.26, 147.94, 146.91, 142.28, 140.13, 137.45, 124.72, 124.21, 122.72, 120.84, 118.39, 118.17, 117.17, 115.01, 109.09, 102.62, 99.57, 67.19, 66.70(2C), 56.26, 55.29, 53.85(2C), 26.19. TOF MS ES+ (m/z): (M + H)+, calcd for C34H30ClF2N5O6: 678.1931, found, 678.1908.
1-(4-Bromo-2-fluorophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-morpholinopropoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (27). White solid. 35.7% yield, m.p: 113.7–115.1 °C. 1H NMR (400 MHz, DMSO) δ 11.82 (s, 1H), 8.75 (d, J = 7.7 Hz, 1H), 8.47 (d, J = 5.2 Hz, 1H), 7.97 (d, J = 10.3 Hz, 2H), 7.76 (t, J = 8.3 Hz, 1H), 7.69 (d, J = 8.7 Hz, 1H), 7.53 (s, 2H), 7.51–7.45 (m, 1H), 7.40 (s, 1H), 6.90 (d, J = 7.8 Hz, 1H), 6.48 (d, J = 5.0 Hz, 1H), 4.19 (t, J = 6.1 Hz, 2H), 3.95 (s, 3H), 3.59 (d, J = 3.7 Hz, 4H), 2.46 (t, J = 6.9 Hz, 2H), 2.39 (s, 4H), 2.04–1.93 (m, 2H). TOF MS ES+ (m/z): (M + H)+, calcd for C34H30BrF2N5O6: 722.1426, found, 722.1403.
1-(4-Chloro-3-(trifluoromethyl)phenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-morpholinopropoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (28). Light yellow solid. 47.3% yield, m.p: 203.5–204.7 °C. 1H NMR (400 MHz, DMSO) δ 11.75 (s, 1H), 9.02 (d, J = 7.8 Hz, 1H), 8.37 (d, J = 5.3 Hz, 1H), 8.19 (s, 1H), 8.06 (d, J = 7.1 Hz, 1H), 7.91 (d, J = 9.6 Hz, 2H), 7.49–7.42 (m, 2H), 7.39 (d, J = 9.3 Hz, 1H), 7.30 (s, 1H), 6.85 (d, J = 7.9 Hz, 1H), 6.39 (d, J = 5.2 Hz, 1H), 4.10 (s, 2H), 3.85 (s, 3H), 3.49 (s, 4H), 2.38–2.37 (m, 2H), 2.30 (s, 4H), 1.88 (s, 2H). 13C NMR (101 MHz, DMSO) δ 169.17, 159.80, 159.18, 154.74, 152.28, 151.90, 149.60, 148.83, 148.05, 146.35, 141.80, 141.62, 136.87, 136.41, 133.06, 130.37, 126.69, 124.31, 120.91, 120.86, 120.19, 116.70, 114.52, 108.83, 108.60, 102.17, 99.07, 66.61, 65.84(2C), 55.80, 54.67, 53.09(2C), 25.32. TOF MS ES+ (m/z): (M + H)+, calcd for C35H30ClF4N5O6: 728.1899, found, 728.1875.
N-(3-Fluoro-4-((6-methoxy-7-(3-morpholinopropoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1-phenyl-1,4-dihydropyridazine-3-carboxamide (29). Pure white solid. 39.7% yield, m.p: 118.7–119.8 °C. 1H NMR (400 MHz, DMSO) δ 12.00 (s, 1H), 9.03 (d, J = 7.8 Hz, 1H), 8.47 (d, J = 5.2 Hz, 1H), 8.02 (d, J = 12.7 Hz, 1H), 7.82 (d, J = 7.7 Hz, 2H), 7.63 (t, J = 7.2 Hz, 2H), 7.59–7.47 (m, 4H), 7.40 (s, 1H), 6.93 (d, J = 7.9 Hz, 1H), 6.49 (d, J = 5.3 Hz, 1H), 4.20 (t, J = 6.1 Hz, 2H), 3.95 (s, 3H), 3.59 (s, 4H), 2.50–2.48 (m, 2H), 2.40 (s, 4H), 1.99 (d, J = 6.6 Hz, 2H). TOF MS ES+ (m/z): (M + H)+, calcd for C34H32FN5O6: 626.2415, found, 626.2396.
N-(3-Fluoro-4-((6-methoxy-7-(3-(piperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-1-(4-fluorophenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (30). Light yellow solid. 43.4% yield, m.p: 149.4–151.1 °C. 1H NMR (400 MHz, DMSO) δ 12.03 (s, 1H), 9.00 (d, J = 7.8 Hz, 1H), 8.48 (d, J = 5.1 Hz, 1H), 8.03 (d, J = 12.5 Hz, 1H), 7.87 (dd, J = 8.6, 4.3 Hz, 2H), 7.56 (d, J = 10.6 Hz, 2H), 7.50 (dd, J = 16.0, 7.8 Hz, 3H), 7.41 (s, 1H), 6.94 (d, J = 7.8 Hz, 1H), 6.50 (d, J = 5.0 Hz, 1H), 4.20 (d, J = 6.1 Hz, 2H), 3.96 (s, 3H), 2.51 (s, 2H), 2.42 (s, 4H), 2.00 (d, J = 6.7 Hz, 2H), 1.53 (s, 4H), 1.41 (s, 2H). 13C NMR (101 MHz, DMSO) δ 169.09, 162.82, 160.37, 159.92, 159.17, 154.74, 152.29, 151.93, 149.59, 148.80, 147.49, 146.38, 142.02, 139.40, 124.31, 123.87, 123.78, 120.46, 116.62(2C), 116.38, 114.46, 108.77, 108.53, 102.09, 99.02, 66.77, 55.76, 54.98, 53.97(2C), 25.91, 25.39(2C), 23.94. TOF MS ES+ (m/z): (M + H)+, calcd for C35H33F2N5O5: 642.2528, found, 642.2511.
1-(4-Chlorophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(piperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (31). White solid. 42.7% yield, m.p: 158.2–159.6 °C. 1H NMR (400 MHz, DMSO) δ 12.05 (s, 1H), 9.11 (d, J = 7.8 Hz, 1H), 8.57 (d, J = 5.1 Hz, 1H), 8.11 (d, J = 12.4 Hz, 1H), 7.95 (d, J = 8.6 Hz, 2H), 7.80 (d, J = 8.6 Hz, 2H), 7.70–7.55 (m, 3H), 7.49 (s, 1H), 7.03 (d, J = 7.8 Hz, 1H), 6.58 (d, J = 4.9 Hz, 1H), 4.27 (d, J = 6.0 Hz, 2H), 4.05 (s, 3H), 2.52 (t, J = 6.8 Hz, 2H), 2.44 (s, 4H), 2.06 (d, J = 6.0 Hz, 2H), 1.60 (s, 4H), 1.48 (s, 2H). 13C NMR (101 MHz, DMSO) δ 169.65, 160.56, 159.75, 155.31, 152.57, 150.20, 149.38, 148.49, 146.95, 142.19, 142.17, 142.16, 137.63, 133.58, 130.20(2C), 124.87, 123.61(2C), 120.95, 117.25, 115.04, 109.35, 109.12, 102.69, 99.63, 67.44, 56.36, 55.65, 54.67(2C), 26.68, 26.17(2C), 24.70. TOF MS ES+ (m/z): (M + H)+, calcd for C35H33ClFN5O5: 658.2233, found, 658.2209.
1-(4-Bromophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(piperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (32). Light yellow solid. 48.3% yield, m.p: 141.3–143.4 °C. 1H NMR (400 MHz, DMSO) δ 11.94 (s, 1H), 9.02 (d, J = 7.8 Hz, 1H), 8.49 (d, J = 5.2 Hz, 1H), 8.02 (d, J = 12.7 Hz, 1H), 7.84 (d, J = 9.0 Hz, 2H), 7.79 (d, J = 8.5 Hz, 2H), 7.56 (s, 2H), 7.50 (t, J = 8.8 Hz, 1H), 7.43 (s, 1H), 6.93 (d, J = 7.9 Hz, 1H), 6.51 (d, J = 5.0 Hz, 1H), 4.23 (s, 2H), 3.96 (s, 3H), 2.80 (s, 4H), 2.12 (s, 2H), 1.64 (s, 4H), 1.47 (s, 2H), 1.23 (s, 2H). TOF MS ES+ (m/z): (M + H)+, calcd for C35H33BrFN5O5: 702.1727, found, 702.1703.
N-(3-Fluoro-4-((6-methoxy-7-(3-(piperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-1-(4-methoxyphenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (33). White solid. 35.2% yield, m.p: 105.4–106.5 °C. 1H NMR (400 MHz, DMSO) δ 12.14 (s, 1H), 8.94 (d, J = 7.7 Hz, 1H), 8.47 (d, J = 4.9 Hz, 1H), 8.02 (d, J = 12.7 Hz, 1H), 7.72 (d, J = 7.7 Hz, 2H), 7.55 (d, J = 8.8 Hz, 2H), 7.49 (t, J = 8.8 Hz, 1H), 7.40 (s, 1H), 7.15 (d, J = 7.7 Hz, 2H), 6.92 (d, J = 7.7 Hz, 1H), 6.49 (d, J = 4.9 Hz, 1H), 4.19 (s, 2H), 3.95 (s, 3H), 3.83 (s, 3H), 2.50 (s, 4H), 2.49–2.40 (m, 2H), 2.01 (d, J = 6.1 Hz, 2H), 1.54 (s, 4H), 1.41 (s, 2H). 13C NMR (101 MHz, DMSO) δ 169.05, 159.97, 159.28, 159.18, 154.73, 152.29, 151.88, 149.57, 148.81, 147.00, 146.35, 141.92, 136.98, 136.36, 124.30, 122.95(2C), 120.66, 116.65, 114.68(2C), 114.49, 108.76, 108.55, 102.10, 99.04, 66.69, 55.77, 55.62, 54.83, 53.76(2C), 25.61, 25.07(2C), 23.65. TOF MS ES+ (m/z): (M + H)+, calcd for C36H36FN5O6: 654.2728, found, 654.2705.
1-(3-Chloro-4-fluorophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(piperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (34). Light yellow solid. 37.9% yield, m.p: 121.7–123.4 °C. 1H NMR (400 MHz, DMSO) δ 11.93 (s, 1H), 9.02 (d, J = 7.8 Hz, 1H), 8.48 (d, J = 5.2 Hz, 1H), 8.17–8.13 (m, 1H), 8.03 (d, J = 13.0 Hz, 1H), 7.90–7.84 (m, 1H), 7.71 (t, J = 9.0 Hz, 1H), 7.60–7.54 (m, 2H), 7.51 (t, J = 8.7 Hz, 1H), 7.40 (s, 1H), 6.94 (d, J = 7.9 Hz, 1H), 6.50 (d, J = 5.2 Hz, 1H), 4.19 (t, J = 6.3 Hz, 2H), 3.96 (s, 3H), 2.43 (t, J = 7.0 Hz, 2H), 2.36 (s, 4H), 2.00–1.93 (m, 2H), 1.55–1.47 (m, 4H), 1.39 (s, 2H). TOF MS ES+ (m/z): (M + H)+, calcd for C35H32ClF2N5O5: 676.2138, found, 676.2109.
1-(4-Bromo-2-fluorophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(piperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (35). White solid. 42.1% yield, m.p: 105.9–107.4 °C. 1H NMR (400 MHz, DMSO) δ 11.79 (s, 1H), 8.74 (d, J = 7.9 Hz, 1H), 8.45 (d, J = 5.1 Hz, 1H), 7.96 (d, J = 11.9 Hz, 2H), 7.74 (t, J = 8.3 Hz, 1H), 7.69 (d, J = 8.9 Hz, 1H), 7.57–7.50 (m, 2H), 7.48 (t, J = 8.8 Hz, 1H), 7.38 (s, 1H), 6.90 (d, J = 7.9 Hz, 1H), 6.47 (d, J = 5.2 Hz, 1H), 4.17 (t, J = 6.3 Hz, 2H), 3.93 (s, 3H), 2.48 (s, 4H), 2.41 (t, J = 7.0 Hz, 2H), 1.99–1.89 (m, 2H), 1.55–1.42 (m, 4H), 1.36 (s, 2H). 13C NMR (101 MHz, DMSO) δ 168.74, 159.81, 159.16, 154.73, 152.27, 151.96, 149.60, 149.03, 148.80, 146.37, 144.52, 136.87, 130.47, 128.70, 128.53, 124.33, 122.89, 120.60, 120.38, 119.69, 116.64, 114.45, 108.75, 108.51, 102.07, 99.02, 66.81, 55.77, 55.05, 54.06(2C), 26.04, 25.54(2C), 24.08. TOF MS ES+ (m/z): (M + H)+, calcd for C35H32BrF2N5O5: 720.1633, found, 720.1612.
1-(4-Chloro-3-(trifluoromethyl)phenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(piperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (36). Light yellow solid. 48.7% yield, m.p: 185.7–186.6 °C. 1H NMR (400 MHz, DMSO) δ 11.77 (s, 1H), 9.01 (d, J = 8.0 Hz, 1H), 8.37 (d, J = 5.1 Hz, 1H), 8.19 (s, 1H), 8.06 (d, J = 8.8 Hz, 1H), 7.90 (d, J = 8.6 Hz, 2H), 7.46 (d, J = 13.6 Hz, 2H), 7.39 (t, J = 8.8 Hz, 1H), 7.30 (s, 1H), 6.84 (d, J = 7.8 Hz, 1H), 6.39 (d, J = 5.0 Hz, 1H), 4.10 (s, 2H), 3.85 (s, 3H), 2.56 (s, 4H), 1.95 (s, 2H), 1.48 (s, 4H), 1.34 (s, 2H), 1.12 (s, 2H). TOF MS ES+ (m/z): (M + H)+, calcd for C36H32ClF4N5O5: 726.2106, found, 726.2097.
N-(3-Fluoro-4-((6-methoxy-7-(3-(piperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1-phenyl-1,4-dihydropyridazine-3-carboxamide (37). Pure white solid. 38.9% yield, m.p: 152.3–153.1 °C. 1H NMR (400 MHz, DMSO) δ 12.02 (s, 1H), 9.03 (d, J = 7.7 Hz, 1H), 8.47 (d, J = 4.6 Hz, 1H), 8.02 (d, J = 12.7 Hz, 1H), 7.81 (d, J = 7.6 Hz, 2H), 7.63 (t, J = 7.3 Hz, 2H), 7.58–7.46 (m, 4H), 7.39 (s, 1H), 6.94 (d, J = 7.7 Hz, 1H), 6.49 (d, J = 4.5 Hz, 1H), 4.18 (s, 2H), 3.95 (s, 3H), 2.43 (t, J = 6.7 Hz, 2H), 2.35 (s, 4H), 1.96 (d, J = 6.0 Hz, 2H), 1.50 (s, 4H), 1.39 (s, 2H). 13C NMR (101 MHz, DMSO) δ 168.65, 159.48, 158.66, 154.24, 151.79, 151.46, 149.10, 148.28, 147.12, 145.89, 142.34, 141.26, 136.56, 129.20(2C), 128.11, 123.79, 120.82(2C), 120.02, 116.14, 113.97, 108.27, 108.04, 101.60, 98.54, 66.32, 55.27, 54.53, 53.54(2C), 25.54, 25.02(2C), 23.56. TOF MS ES+ (m/z): (M + H)+, calcd for C35H34FN5O5: 624.2622, found, 624.2605.
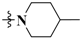
N-(3-Fluoro-4-((6-methoxy-7-(3-(4-methylpiperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-1-(4-fluorophenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (38). White solid. 35.8% yield, m.p: 210.1–211.4 °C. 1H NMR (400 MHz, DMSO) δ 12.02 (s, 1H), 9.00 (d, J = 7.8 Hz, 1H), 8.49 (s, 1H), 8.03 (d, J = 12.9 Hz, 1H), 7.88 (d, J = 4.8 Hz, 2H), 7.64–7.45 (m, 5H), 7.40 (s, 1H), 6.94 (d, J = 7.4 Hz, 1H), 6.50 (s, 1H), 4.19 (s, 2H), 3.96 (s, 3H), 2.87 (d, J = 10.5 Hz, 2H), 2.46 (s, 2H), 1.97 (s, 2H), 1.91 (d, J = 10.2 Hz, 2H), 1.59 (d, J = 11.5 Hz, 2H), 1.33 (s, 1H), 1.15 (d, J = 12.2 Hz, 2H), 0.89 (d, J = 6.0 Hz, 3H). TOF MS ES+ (m/z): (M + H)+, calcd for C36H35F2N5O5: 656.2685, found, 656.2673.
1-(4-Chlorophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(4-methylpiperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (39). Light yellow solid. 51.4% yield, m.p: 220.6–222.1 °C. 1H NMR (400 MHz, DMSO) δ 11.96 (s, 1H), 9.02 (d, J = 7.8 Hz, 1H), 8.48 (d, J = 4.4 Hz, 1H), 8.02 (d, J = 12.9 Hz, 1H), 7.86 (d, J = 7.5 Hz, 2H), 7.70 (d, J = 7.5 Hz, 2H), 7.59–7.46 (m, 3H), 7.39 (s, 1H), 6.94 (d, J = 7.9 Hz, 1H), 6.49 (d, J = 4.6 Hz, 1H), 4.18 (s, 2H), 3.96 (s, 3H), 2.85 (d, J = 10.3 Hz, 2H), 2.44 (t, J = 6.4 Hz, 2H), 1.97 (d, J = 6.2 Hz, 2H), 1.88 (t, J = 11.2 Hz, 2H), 1.57 (d, J = 11.7 Hz, 2H), 1.30 (s, 1H), 1.16 (t, J = 11.5 Hz, 2H), 0.89 (d, J = 6.0 Hz, 3H). 13C NMR (101 MHz, DMSO) δ 169.67, 160.51, 159.74, 155.31, 152.87, 152.57, 150.22, 149.38, 148.42, 146.98, 142.18, 137.59, 136.96, 133.59, 130.21(2C), 124.87, 123.60(2C), 120.97, 117.24, 115.06, 109.36, 109.14, 102.71, 99.66, 67.43, 56.37, 55.25, 54.02(2C), 34.57(2C), 30.93, 26.79, 22.34. TOF MS ES+ (m/z): (M + H)+, calcd for C36H35ClFN5O5: 672.2389, found, 672.2367.
1-(4-Bromophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(4-methylpiperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (40). Light yellow solid. 49.8% yield, m.p: 220.7–221.4 °C. 1H NMR (400 MHz, DMSO) δ 11.94 (s, 1H), 9.03 (d, J = 7.4 Hz, 1H), 8.49 (s, 1H), 8.02 (d, J = 12.3 Hz, 1H), 7.84 (d, J = 8.7 Hz, 2H), 7.80 (d, J = 8.0 Hz, 2H), 7.55 (s, 2H), 7.50 (s, 1H), 7.40 (s, 1H), 6.94 (d, J = 7.8 Hz, 1H), 6.51 (s, 1H), 4.20 (s, 2H), 3.96 (s, 3H), 2.92 (s, 2H), 2.53 (s, 2H), 2.00 (s, 4H), 1.61 (d, J = 11.7 Hz, 2H), 1.36 (s, 1H), 1.17 (d, J = 12.0 Hz, 2H), 0.90 (d, J = 5.8 Hz, 3H). TOF MS ES+ (m/z): (M + H)+, calcd for C36H35BrFN5O5: 716.1884, found, 716.1863.
N-(3-Fluoro-4-((6-methoxy-7-(3-(4-methylpiperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-1-(4-methoxyphenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (41). Light yellow solid. 40.3% yield, m.p: 145.1–146.3 °C. 1H NMR (400 MHz, DMSO) δ 12.12 (s, 1H), 8.94 (d, J = 7.7 Hz, 1H), 8.47 (d, J = 5.3 Hz, 1H), 8.02 (d, J = 12.9 Hz, 1H), 7.73 (d, J = 8.7 Hz, 2H), 7.54 (s, 2H), 7.49 (t, J = 8.3 Hz, 1H), 7.39 (s, 1H), 7.16 (d, J = 7.4 Hz, 2H), 6.92 (d, J = 8.0 Hz, 1H), 6.50 (s, 1H), 4.18 (s, 2H), 3.95 (s, 3H), 3.84 (s, 3H), 2.85 (d, J = 9.0 Hz, 2H), 2.44 (d, J = 6.5 Hz, 2H), 1.97 (d, J = 6.9 Hz, 2H), 1.87 (t, J = 11.4 Hz, 2H), 1.57 (d, J = 11.9 Hz, 2H), 1.30 (s, 1H), 1.19–1.11 (m, 2H), 0.88 (d, J = 6.3 Hz, 3H). TOF MS ES+ (m/z): (M + H)+, calcd for C37H38FN5O6: 668.2884, found, 668.2836.
1-(3-Chloro-4-fluorophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(4-methylpiperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (42). Light yellow solid. 41.5% yield, m.p: 112.5–113.7 °C. 1H NMR (400 MHz, DMSO) δ 11.94 (s, 1H), 9.03 (d, J = 7.8 Hz, 1H), 8.49 (d, J = 5.1 Hz, 1H), 8.15 (d, J = 4.0 Hz, 1H), 8.03 (d, J = 12.7 Hz, 1H), 7.88 (d, J = 9.0 Hz, 1H), 7.71 (t, J = 8.8 Hz, 1H), 7.57 (d, J = 13.1 Hz, 2H), 7.51 (t, J = 8.8 Hz, 1H), 7.42 (s, 1H), 6.94 (d, J = 7.8 Hz, 1H), 6.50 (d, J = 5.0 Hz, 1H), 4.22 (s, 2H), 3.97 (s, 3H), 3.10 (s, 2H), 2.75 (s, 2H), 2.09 (s, 2H), 1.67 (d, J = 12.3 Hz, 2H), 1.30 (s, 1H), 1.25 (d, J = 12.3 Hz, 4H), 0.91 (d, J = 6.3 Hz, 3H). TOF MS ES+ (m/z): (M + H)+, calcd for C36H34ClF2N5O5: 690.2295, found, 690.2273.
1-(4-Bromo-2-fluorophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(4-methylpiperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (43). White solid. 35.3% yield, m.p: 115.8–117.3 °C. 1H NMR (400 MHz, DMSO) δ 11.78 (s, 1H), 8.74 (d, J = 7.9 Hz, 1H), 8.46 (s, 1H), 7.96 (d, J = 12.2 Hz, 2H), 7.79–7.66 (m, 2H), 7.52 (s, 2H), 7.51–7.44 (m, 1H), 7.37 (s, 1H), 6.90 (d, J = 7.8 Hz, 1H), 6.47 (d, J = 5.1 Hz, 1H), 4.17 (t, J = 5.9 Hz, 2H), 3.93 (s, 3H), 2.85 (d, J = 10.6 Hz, 2H), 2.45 (s, 2H), 1.99–1.93 (m, 2H), 1.90 (d, J = 11.9 Hz, 2H), 1.56 (d, J = 12.3 Hz, 2H), 1.22 (d, J = 10.2 Hz, 1H), 1.13 (d, J = 12.3 Hz, 2H), 0.87 (d, J = 6.3 Hz, 3H). 13C NMR (101 MHz, DMSO) δ 168.76, 159.78, 159.16, 154.73, 153.28, 151.96, 149.60, 148.97, 148.78, 146.38, 136.87, 130.37, 128.71, 128.70, 128.52, 124.32, 122.97, 122.89, 120.61, 120.38, 119.71, 116.64, 108.76, 108.52, 102.08, 99.03, 66.80, 55.76, 54.67, 53.43(2C), 33.93(2C), 30.33, 26.16, 21.77. TOF MS ES+ (m/z): (M + H)+, calcd for C36H34BrF2N5O5: 734.1790, found, 734.1773.
1-(4-Chloro-3-(trifluoromethyl)phenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(4-methylpiperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (44). Light yellow solid. 38.7% yield, m.p: 137.7–138.6 °C. 1H NMR (400 MHz, DMSO) δ 11.77 (s, 1H), 9.02 (d, J = 7.9 Hz, 1H), 8.37 (d, J = 5.3 Hz, 1H), 8.19 (s, 1H), 8.06 (d, J = 8.4 Hz, 1H), 7.90 (d, J = 8.5 Hz, 2H), 7.50–7.42 (m, 2H), 7.39 (t, J = 8.9 Hz, 1H), 7.30 (s, 1H), 6.84 (d, J = 8.2 Hz, 1H), 6.39 (d, J = 5.3 Hz, 1H), 4.09 (s, 2H), 3.85 (s, 3H), 2.91 (s, 2H), 1.99 (d, J = 48.7 Hz, 4H), 1.53 (d, J = 10.0 Hz, 2H), 1.30 (s, 1H), 1.12 (s, 4H), 0.79 (d, J = 6.3 Hz, 3H). 13C NMR (101 MHz, DMSO) δ 169.66, 160.26, 159.66, 155.22, 152.77, 152.34, 150.03, 149.30, 148.44, 146.84, 142.26, 142.08, 137.47, 136.98, 133.53, 130.85, 127.13, 124.79, 121.39, 121.33, 120.70, 117.17, 115.00, 109.06, 102.61, 101.99, 99.53, 67.12, 56.25, 54.85, 53.52(2C), 33.67(2C), 30.28, 26.04, 22.04. TOF MS ES+ (m/z): (M + H)+, calcd for C37H34ClF4N5O5: 740.2263, found, 740.2237.
N-(3-Fluoro-4-((6-methoxy-7-(3-(4-methylpiperidin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1-phenyl-1,4-dihydropyridazine-3-carboxamide (45). Pure white solid. 44.1% yield, m.p: 148.5–150.1 °C. 1H NMR (400 MHz, DMSO) δ 12.02 (s, 1H), 9.03 (d, J = 7.7 Hz, 1H), 8.47 (d, J = 5.1 Hz, 1H), 8.02 (d, J = 13.1 Hz, 1H), 7.82 (d, J = 7.8 Hz, 2H), 7.63 (t, J = 7.7 Hz, 2H), 7.51 (dd, J = 17.2, 8.4 Hz, 4H), 7.39 (s, 1H), 6.94 (d, J = 7.8 Hz, 1H), 6.49 (d, J = 4.8 Hz, 1H), 4.18 (s, 2H), 3.95 (s, 3H), 2.86 (d, J = 10.9 Hz, 2H), 2.45 (t, J = 6.7 Hz, 2H), 2.01–1.94 (m, 2H), 1.89 (t, J = 11.7 Hz, 2H), 1.57 (d, J = 11.2 Hz, 2H), 1.32 (s, 1H), 1.20–1.11 (m, 2H), 0.88 (d, J = 6.2 Hz, 3H). 13C NMR (101 MHz, DMSO) δ 169.63, 160.54, 159.68, 155.25, 152.44, 150.13, 149.33, 148.26, 146.90, 143.37, 142.29, 137.48, 136.85, 130.23(2C), 129.13, 124.81, 121.86(2C), 121.00, 117.17, 115.02, 109.29, 109.12, 102.67, 99.60, 67.27, 56.31, 55.03, 53.76(2C), 34.12(2C), 30.58, 26.42, 22.16. TOF MS ES+ (m/z): (M + H)+, calcd for C36H36FN5O5: 638.2779, found, 638.2754.
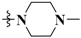
N-(3-Fluoro-4-((6-methoxy-7-(3-(4-methylpiperazin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-1-(4-fluorophenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (46). White solid. 36.1% yield, m.p: 184.6–185.7 °C. 1H NMR (400 MHz, DMSO) δ 12.03 (s, 1H), 9.00 (d, J = 7.7 Hz, 1H), 8.48 (d, J = 4.7 Hz, 1H), 8.02 (d, J = 12.5 Hz, 1H), 7.88 (s, 2H), 7.55 (s, 2H), 7.54–7.44 (m, 3H), 7.40 (s, 1H), 6.94 (d, J = 7.7 Hz, 1H), 6.50 (s, 1H), 4.19 (s, 2H), 3.96 (s, 3H), 2.47 (s, 2H), 2.42 (d, J = 42.1 Hz, 8H), 2.17 (s, 3H), 1.97 (s, 2H). 13C NMR (101 MHz, DMSO) δ 168.57, 162.32, 159.87, 159.44, 158.67, 154.24, 151.44, 149.09, 148.31, 147.05, 145.88, 141.53, 138.91, 136.44, 123.82, 123.38(2C), 123.30, 119.95, 116.12(2C), 115.88, 113.96, 108.03, 101.61, 98.53, 66.20, 55.27, 54.02(2C), 53.73, 51.91(2C), 44.90, 25.46. TOF MS ES+ (m/z): (M + H)+, calcd for C35H34F2N6O5: 657.2637, found, 657.2612.
1-(4-Chlorophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(4-methylpiperazin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (47). White solid. 59.1% yield, m.p: 163.5–165.3 °C. 1H NMR (400 MHz, DMSO) δ 11.95 (s, 1H), 9.03 (d, J = 7.1 Hz, 1H), 8.48 (d, J = 4.5 Hz, 1H), 8.02 (d, J = 13.1 Hz, 1H), 7.86 (d, J = 7.2 Hz, 2H), 7.71 (d, J = 8.3 Hz, 2H), 7.59–7.47 (m, 3H), 7.40 (s, 1H), 6.94 (d, J = 7.8 Hz, 1H), 6.50 (s, 1H), 4.19 (s, 2H), 3.96 (s, 3H), 2.46 (d, J = 6.6 Hz, 2H), 2.35–2.22 (s, 8H), 2.15 (s, 3H), 1.97 (s, 2H). TOF MS ES+ (m/z): (M + H)+, calcd for C35H34ClFN6O5: 673.2341, found, 673.2315.
1-(4-Bromophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(4-methylpiperazin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (48). White solid. 38.2% yield, m.p: 159.7–161.2 °C. 1H NMR (400 MHz, DMSO) δ 11.95 (s, 1H), 9.03 (d, J = 7.9 Hz, 1H), 8.49 (d, J = 5.2 Hz, 1H), 8.03 (d, J = 12.6 Hz, 1H), 7.82 (m, J = 8.9 Hz, 4H), 7.57 (d, J = 10.4 Hz, 2H), 7.54–7.48 (m, 1H), 7.42 (s, 1H), 6.94 (d, J = 7.8 Hz, 1H), 6.51 (d, J = 5.1 Hz, 1H), 4.21 (s, 2H), 3.97 (s, 3H), 2.78 (s, 4H), 2.60 (s, 4H), 2.47 (s, 2H), 2.03 (d, J = 6.1 Hz, 2H), 1.23 (s, 3H). TOF MS ES+ (m/z): (M + H)+, calcd for C35H34BrFN6O5: 717.1836, found, 717.1804.
1-(4-((l1-Oxidanyl)-l5-methyl)phenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(4-methylpiperazin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (49). Light yellow solid. 47.5% yield, m.p: 167.5–169.1 °C. 1H NMR (400 MHz, DMSO) δ 12.02 (s, 1H), 8.83 (d, J = 7.7 Hz, 1H), 8.36 (d, J = 4.0 Hz, 1H), 7.91 (d, J = 12.9 Hz, 1H), 7.62 (d, J = 8.8 Hz, 2H), 7.44 (d, J = 10.3 Hz, 2H), 7.39 (t, J = 8.7 Hz, 1H), 7.28 (s, 1H), 7.05 (d, J = 8.7 Hz, 2H), 6.82 (d, J = 7.9 Hz, 1H), 6.39 (s, 1H), 4.08 (s, 2H), 3.84 (s, 3H), 3.73 (s, 3H), 2.35 (d, J = 6.8 Hz, 2H), 2.37–2.14 (s, 8H), 2.06 (s, 3H), 1.86 (s, 2H). 13C NMR (101 MHz, DMSO) δ 168.79, 159.80, 159.17, 155.77, 154.70, 153.23, 152.26, 151.91, 149.56, 148.93, 148.78, 146.33, 144.46, 137.06, 128.67, 128.49, 124.27, 122.96, 120.57, 120.34, 119.70, 116.66, 114.45, 108.46, 102.05, 99.00, 66.65, 66.18(3C), 55.74, 54.79, 53.34(3C), 25.65. TOF MS ES+ (m/z): (M + H)+, calcd for C36H37FN6O6: 669.2837, found, 669.2801.
1-(3-Chloro-4-fluorophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(4-methylpiperazin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (50). Light yellow solid. 35.4% yield, m.p: 144.7–146.2 °C. 1H NMR (400 MHz, DMSO) δ 11.92 (s, 1H), 9.02 (d, J = 7.9 Hz, 1H), 8.48 (d, J = 5.1 Hz, 1H), 8.14 (dd, J = 6.3, 2.7 Hz, 1H), 8.02 (d, J = 12.6 Hz, 1H), 7.94–7.83 (m, 1H), 7.71 (t, J = 9.0 Hz, 1H), 7.61–7.54 (m, 2H), 7.51 (t, J = 8.8 Hz, 1H), 7.40 (s, 1H), 6.94 (d, J = 7.9 Hz, 1H), 6.50 (d, J = 5.2 Hz, 1H), 4.19 (t, J = 6.2 Hz, 2H), 3.96 (s, 3H), 2.51 (s, 2H), 2.49–2.45 (m, 2H), 2.35 (s, 4H), 2.16 (s, 3H), 2.02–1.93 (m, 2H), 1.37–1.17 (m, 2H). 13C NMR (101 MHz, DMSO) δ 169.66, 160.22, 159.66, 158.57, 156.09, 155.23, 152.47, 150.12, 149.27, 148.00, 146.88, 142.31, 140.17, 137.44, 137.03, 124.75, 124.24, 122.75, 120.84, 118.40, 118.18, 117.19, 115.00, 109.04, 102.62, 99.57, 67.26, 56.26, 55.24(2C), 54.82, 53.19(2C), 46.18, 26.54. TOF MS ES+ (m/z): (M + H)+, calcd for C35H33ClF2N6O5: 691.2247, found, 691.2255.
1-(4-Bromo-2-fluorophenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(4-methylpiperazin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (51). White solid. 39.8% yield, m.p: 103.1–104.7 °C. 1H NMR (400 MHz, DMSO) δ 11.82 (s, 1H), 8.76 (dd, J = 7.8, 1.8 Hz, 1H), 8.48 (d, J = 5.2 Hz, 1H), 8.00 (s, 1H), 7.97 (d, J = 1.9 Hz, 1H), 7.77 (t, J = 8.4 Hz, 1H), 7.71 (d, J = 8.5 Hz, 1H), 7.55 (d, J = 8.8 Hz, 2H), 7.49 (t, J = 8.8 Hz, 1H), 7.40 (s, 1H), 6.91 (d, J = 7.9 Hz, 1H), 6.49 (d, J = 5.1 Hz, 1H), 4.20 (t, J = 6.4 Hz, 2H), 3.96 (s, 3H), 2.67 (s, 4H), 2.57 (s, 4H), 2.39 (s, 3H), 2.05–1.95 (m, 2H), 1.23 (s, 2H). 13C NMR (101 MHz, DMSO) δ 169.21, 160.34, 159.67, 156.32, 155.21, 153.78, 152.76, 152.43, 150.10, 149.60, 149.32, 146.85, 144.99, 129.19, 129.05, 124.79, 123.46, 121.10, 120.87, 120.16, 117.16, 114.99, 109.04, 102.62, 99.56, 67.16, 56.29, 54.55(3C), 52.32(2C), 45.29, 26.33. TOF MS ES+ (m/z): (M + H)+, calcd for C35H33BrF2N6O5: 735.1742, found, 735.1713.
1-(4-Chloro-3-(trifluoromethyl)phenyl)-N-(3-fluoro-4-((6-methoxy-7-(3-(4-methylpiperazin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1,4-dihydropyridazine-3-carboxamide (52). Light yellow solid. 43.7% yield, m.p: 211.7–212.6 °C. 1H NMR (400 MHz, DMSO) δ 11.77 (s, 1H), 9.03 (t, J = 8.1 Hz, 1H), 8.37 (d, J = 5.2 Hz, 1H), 8.19 (s, 1H), 8.06 (d, J = 8.7 Hz, 1H), 7.93–7.88 (m, 2H), 7.48–7.42 (m, 2H), 7.42–7.36 (m, 1H), 7.30 (d, J = 7.9 Hz, 1H), 6.85 (t, J = 8.0 Hz, 1H), 6.38 (d, J = 5.2 Hz, 1H), 4.09 (d, J = 6.5 Hz, 2H), 3.85 (d, J = 7.9 Hz, 3H), 2.42 (d, J = 6.3 Hz, 4H), 2.40 (s, 4H), 2.20 (d, J = 7.6 Hz, 3H), 1.89 (d, J = 6.9 Hz, 2H), 1.12 (s, 2H). 13C NMR (101 MHz, DMSO) δ 169.16, 159.80, 159.17, 154.73, 152.28, 151.94, 149.61, 148.80, 148.03, 146.38, 141.60, 139.90, 136.99, 136.53, 133.04, 132.81, 130.34, 126.64, 125.45, 124.27, 120.86, 120.19, 116.70, 114.51, 108.58, 102.14, 99.08, 66.67, 55.78, 54.02(3C), 51.78(2C), 44.76, 25.85. TOF MS ES+ (m/z): (M + H)+, calcd for C36H33ClF4N6O5: 741.2215, found, 741.2216.
N-(3-Fluoro-4-((6-methoxy-7-(3-(4-methylpiperazin-1-yl)propoxy)quinolin-4-yl)oxy)phenyl)-4-oxo-1-phenyl-1,4-dihydropyridazine-3-carboxamide (53). Pure white solid. 45.2% yield, m.p: 151.3–152.1°C. 1H NMR (400 MHz, DMSO) δ 12.02 (s, 1H), 9.03 (d, J = 7.8 Hz, 1H), 8.47 (d, J = 4.8 Hz, 1H), 8.02 (d, J = 12.0 Hz, 1H), 7.82 (d, J = 7.4 Hz, 2H), 7.63 (t, J = 7.4 Hz, 2H), 7.51 (dd, J = 16.5, 8.1 Hz, 4H), 7.39 (s, 1H), 6.94 (d, J = 7.7 Hz, 1H), 6.49 (d, J = 4.7 Hz, 1H), 4.18 (s, 2H), 3.95 (s, 3H), 2.45 (d, J = 6.7 Hz, 2H), 2.42–2.22 (s, 8H), 2.15 (s, 3H), 1.97 (d, J = 5.9 Hz, 2H). 13C NMR (101 MHz, DMSO) δ 169.14, 160.02, 159.18, 154.75, 152.30, 151.97, 149.63, 148.81, 147.69, 146.40, 142.87, 141.78, 137.07, 129.72(2C), 128.62, 124.30, 121.35(2C), 120.52, 116.67, 114.50, 108.79, 108.57, 102.14, 99.08, 66.74, 55.80, 54.62(2C), 54.27, 52.53(2C), 45.52, 26.02. TOF MS ES+ (m/z): (M + H)+, calcd for C35H35FN6O5: 639.2731, found, 639.2689.