Hepatic Metabolism of Sakuranetin and Its Modulating Effects on Cytochrome P450s and UDP-Glucuronosyltransferases
Abstract
1. Introduction
2. Results
2.1. Phase I Metabolism of Sakuranetin
2.2. Uridine 5′-Diphosphoglucuronic Acid-Dependent Phase II Metabolism of Sakuranetin
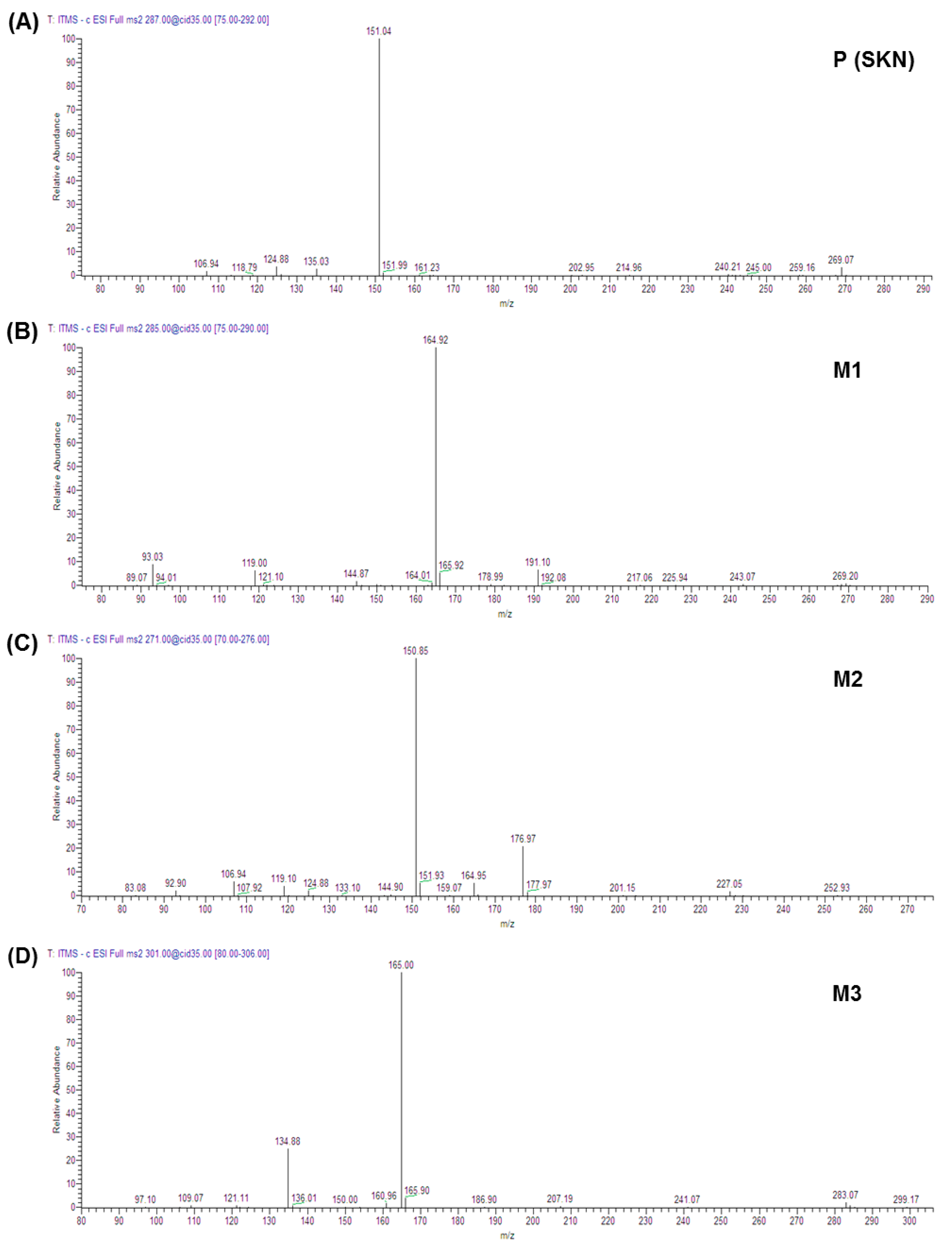
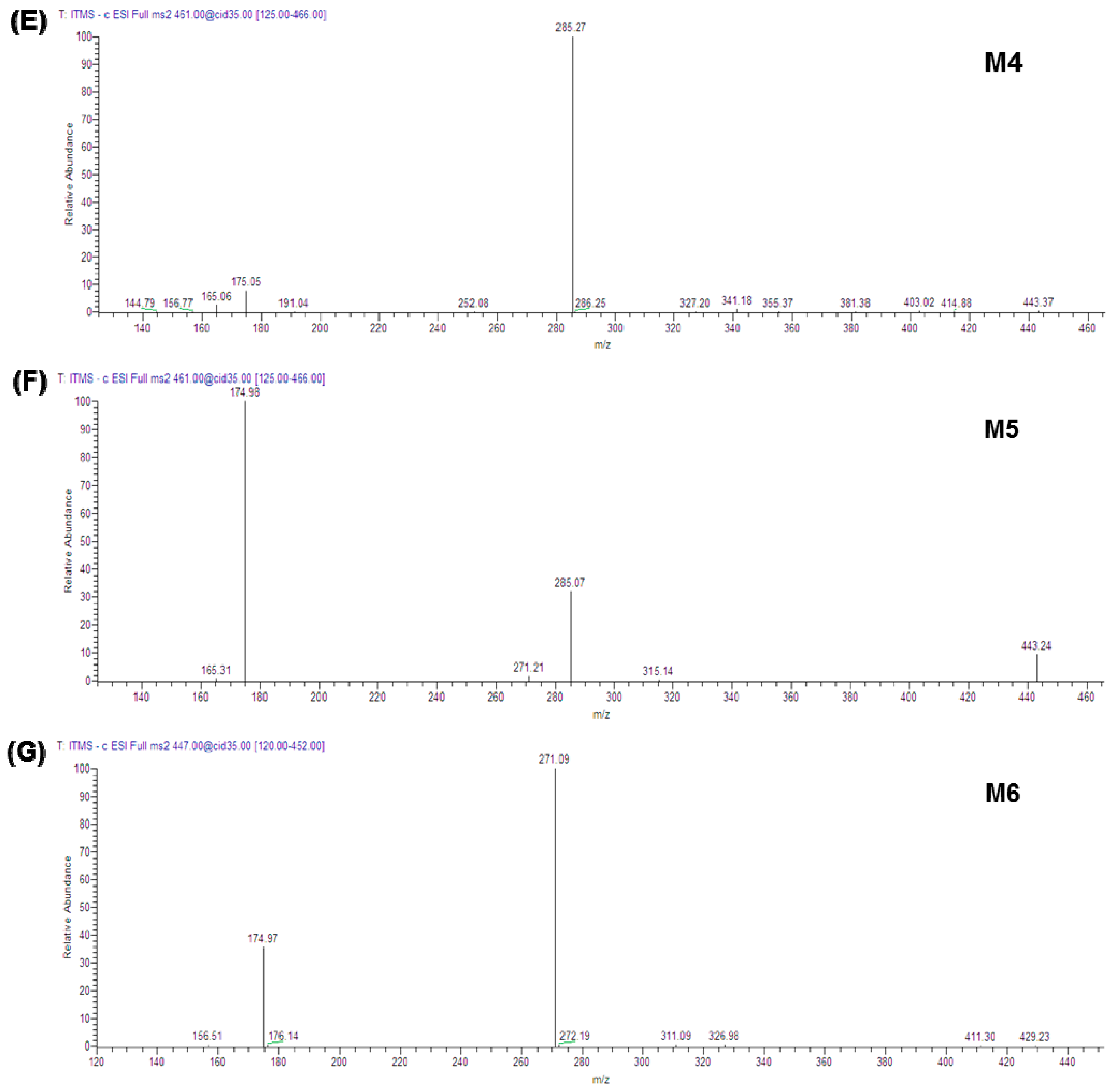
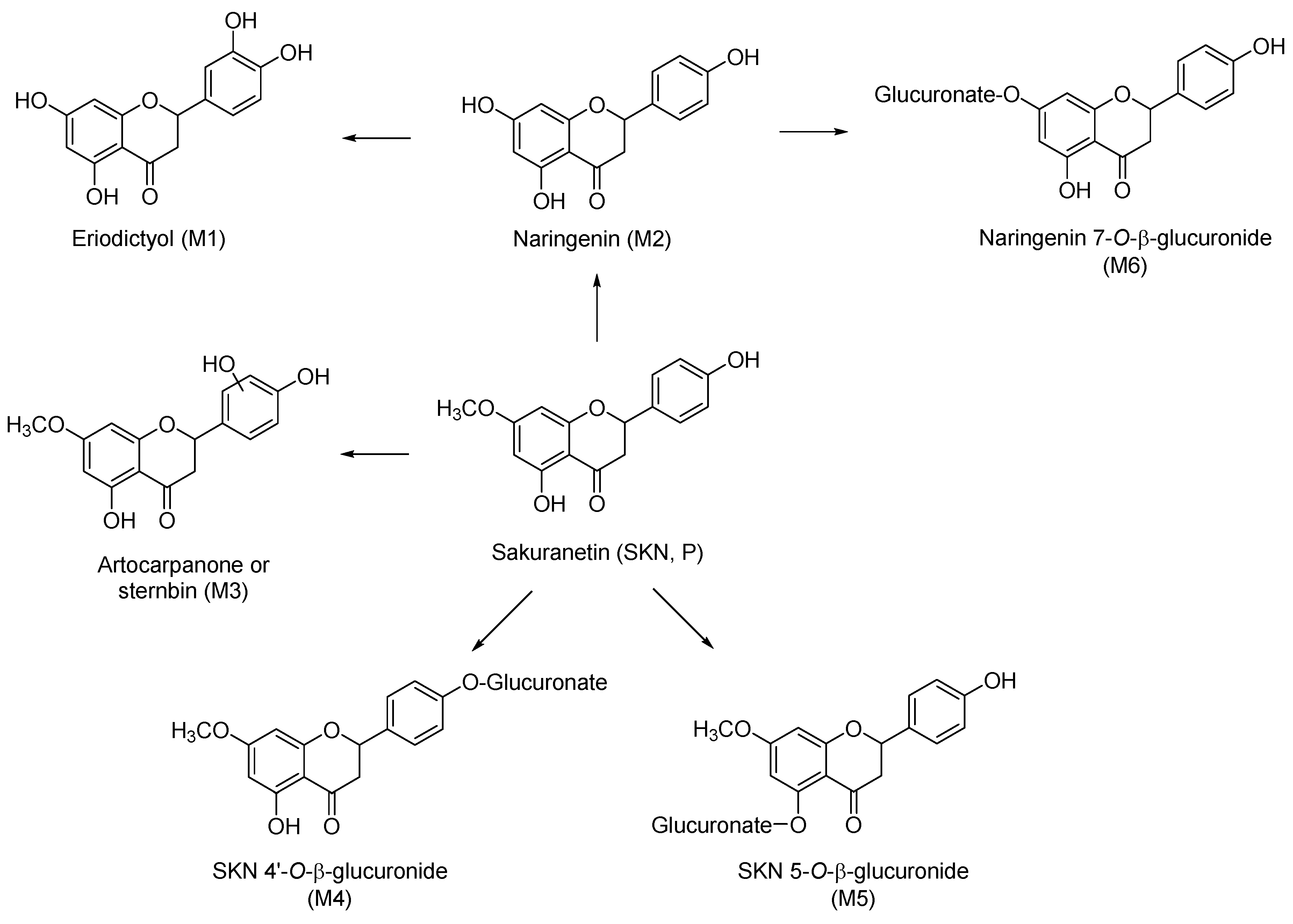
2.3. Metabolites of Sakuranetin
2.4. Inhibition of the Activities of Cytochrome P450 and UDP-Glucuronosyltransferases by Sakuranetin
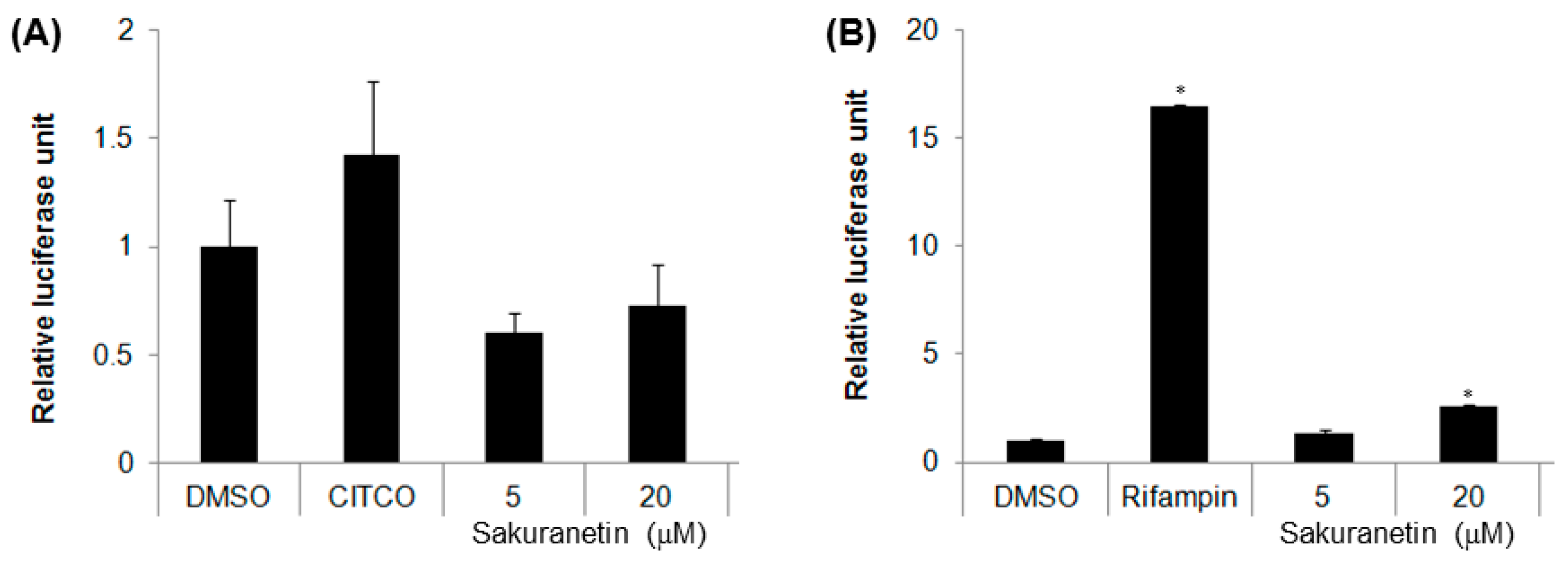
2.5. Induction of CYP2B6 and CYP3A4 Promoter Activities via CAR and the Pregnane X Receptor
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Phase I and Phase II Metabolism
microsomal protein) × (mg microsomal protein concentration/g of liver)
4.3. Cytochrome P450 Inhibition Assays
4.4. UDP-Glucuronosyltransferase Inhibition Assays
4.5. HPLC Instrumentation
4.6. HPLC-Electrospray Mass Spectrometry Analysis
4.7. Cytochrome P450 Promoter Luciferase Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, L.; Kong, Y.; Wu, D.; Zhang, H.; Wu, J.; Chen, J.; Ding, J.; Hu, L.; Jiang, H.; Shen, X. Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: Crystal structure characterization with enzymatic inhibition assay. Protein Sci. 2008, 17, 1971–1978. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, L.; Pinheiro, N.M.; Olivo, C.R.; Choqueta-Toledo, A.; Grecco, S.S.; Lopes, F.D.; Caperuto, L.C.; Martins, M.A.; Tiberio, I.F.; Camara, N.O.; et al. A flavanone from Baccharis retusa (Asteraceae) prevents elastase-induced emphysema in mice by regulating nf-kappab, oxidative stress and metalloproteinases. Respir. Res. 2015, 16, 79. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.Y.; Kang, H. Sakuranetin inhibits inflammatory enzyme, cytokine, and costimulatory molecule expression in macrophages through modulation of JNK, p38, and STAT1. Evid.-Based Complement. Altern. Med. 2016, 2016, 9824203. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, M.; Kinoshita, H.; Okuno, Y. Antimutagenic activity of sakuranetin from Prunus jamasakura. J. Food Sci. 2003, 68, 52–56. [Google Scholar] [CrossRef]
- Shimizu, T.; Lin, F.; Hasegawa, M.; Okada, K.; Nojiri, H.; Yamane, H. Purification and identification of naringenin 7-O-methyltransferase, a key enzyme in biosynthesis of flavonoid phytoalexin sakuranetin in rice. J. Biol. Chem. 2012, 287, 19315–19325. [Google Scholar] [CrossRef] [PubMed]
- Asahina, Y. Ueber das Sakuranin, ein neues Glykosid der Rinde von Prunus Pseudo-Cerasus Lindl. var. Sieboldi Maxim. Arch. Pharm. 1908, 246, 259–272. [Google Scholar] [CrossRef]
- Tuchinda, P.; Reutrakul, V.; Claeson, P.; Pongprayoon, U.; Sematong, T.; Santisuk, T.; Taylor, W.C. Anti-inflammatory cyclohexenyl chalcone derivatives in Boesenbergia pandurata. Phytochemistry 2002, 59, 169–173. [Google Scholar] [CrossRef]
- Rakwal, R.; Hasegawa, M.; Kodama, O. A methyltransferase for synthesis of the flavanone phytoalexin sakuranetin in rice leaves. Biochem. Biophys. Res. Commun. 1996, 222, 732–735. [Google Scholar] [CrossRef] [PubMed]
- Grecco, S.D.S.; Ferreira, M.J.P.; Romoff, P.; Favero, O.A.; Lago, J.H.G. Phenolic derivatives from Baccharis retusa DC. (Asteraceae). Biochem. Syst. Ecol. 2012, 42, 21–24. [Google Scholar] [CrossRef]
- Moon, Y.J.; Wang, X.; Morris, M.E. Dietary flavonoids: Effects on xenobiotic and carcinogen metabolism. Toxicol. In Vitro 2006, 20, 187–210. [Google Scholar] [CrossRef] [PubMed]
- Dahan, A.; Altman, H. Food-drug interaction: Grapefruit juice augments drug bioavailability mechanism, extent and relevance. Eur. J. Clin. Nutr. 2004, 58, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ubeaud, G.; Hagenbach, J.; Vandenschrieck, S.; Jung, L.; Koffel, J.C. In vitro inhibition of simvastatin metabolism in rat and human liver by naringenin. Life Sci. 1999, 65, 1403–1412. [Google Scholar] [CrossRef]
- Nielsen, S.E.; Breinholt, V.; Justesen, U.; Cornett, C.; Dragsted, L.O. In vitro biotransformation of flavonoids by rat liver microsomes. Xenobiotica 1998, 28, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Breinholt, V.M.; Offord, E.A.; Brouwer, C.; Nielsen, S.E.; Brosen, K.; Friedberg, T. In vitro investigation of cytochrome p450-mediated metabolism of dietary flavonoids. Food Chem. Toxicol. 2002, 40, 609–616. [Google Scholar] [CrossRef]
- Otake, Y.; Walle, T. Oxidation of the flavonoids galangin and kaempferide by human liver microsomes and cyp1a1, cyp1a2, and cyp2c9. Drug Metab. Dispos. 2002, 30, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Katsumata, S.; Hamana, K.; Horie, K.; Toshima, H.; Hasegawa, M. Identification of sternbin and naringenin as detoxified metabolites from the rice flavanone phytoalexin sakuranetin by Pyricularia oryzae. Chem. Biodivers. 2017, 14, e1600240. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.-R.S.; Galal, A.M.; Ahmed, M.S.; Mossa, G.S. O-Demethylation and sulfation of 7-methoxylated flavanones by Cunninghamella elegans. Chem. Pharm. Bull. 2003, 51, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Knowlden, J.M.; Gee, J.M.; Robertson, J.F.; Ellis, I.O.; Nicholson, R.I. A possible divergent role for the oestrogen receptor a and b subtypes in clinical breast cancer. Int. J. Cancer 2000, 89, 209–212. [Google Scholar] [CrossRef]
- Davies, B.; Morris, T. Physiological parameters in laboratory animals and humans. Pharm. Res. 1993, 10, 1093–1095. [Google Scholar] [CrossRef] [PubMed]
- Iwatsubo, T.; Hirota, N.; Ooie, T.; Suzuki, H.; Shimada, N.; Chiba, K.; Ishizaki, T.; Green, C.E.; Tyson, C.A.; Sugiyama, Y. Prediction of in vivo drug metabolism in the human liver from in vitro metabolism data. Pharmacol. Ther. 1997, 73, 147–171. [Google Scholar] [CrossRef]
- Nikolic, D.; van Breemen, R.B. New metabolic pathways for flavanones catalyzed by rat liver microsomes. Drug Metab. Dispos. 2004, 32, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Bredsdorff, L.; Nielsen, I.L.F.; Rasmussen, S.E.; Cornett, C.; Barron, D.; Bouisset, F.; Offord, E.; Williamson, G. Absorption, conjugation and excretion of the flavanones, naringenin and hesperetin from α-rhamnosidase-treated orange juice in human subjects. Br. J. Nutr. 2010, 103, 1602–1609. [Google Scholar] [CrossRef] [PubMed]
- Manach, C.; Scalbert, A.; Morand, C.; Remesy, C.; Jimenez, L. Polyphenols: Food sources and bioavailability. Am. J. Clin. Nutr. 2004, 79, 727–747. [Google Scholar] [CrossRef] [PubMed]
- Martens, S.; Mithofer, A. Flavones and flavone synthases. Phytochemistry 2005, 66, 2399–2407. [Google Scholar] [CrossRef] [PubMed]
- Schijlen, E.G.; Ric de Vos, C.H.; van Tunen, A.J.; Bovy, A.G. Modification of flavonoid biosynthesis in crop plants. Phytochemistry 2004, 65, 2631–2648. [Google Scholar] [CrossRef] [PubMed]
- Iwashina, T. The structure and distribution of the flavonoids in plants. J. Plant Res. 2000, 113, 287–299. [Google Scholar] [CrossRef]
- Lewinsohn, E.; Britsch, L.; Mazur, Y.; Gressel, J. Flavanone glycoside biosynthesis in citrus: Chalcone synthase, UDP-glucose: Flavanone-7-O-glucosyl-transferase and -rhamnosyl-transferase activities in cell-free extracts. Plant Physiol. 1989, 91, 1323–1328. [Google Scholar] [CrossRef] [PubMed]
- Manach, C.; Morand, C.; Gil-Izquierdo, A.; Bouteloup-Demange, C.; Remesy, C. Bioavailability in humans of the flavanones hesperidin and narirutin after the ingestion of two doses of orange juice. Eur. J. Clin. Nutr. 2003, 57, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Silberberg, M.; Gil-Izquierdo, A.; Combaret, L.; Remesy, C.; Scalbert, A.; Morand, C. Flavanone metabolism in healthy and tumor-bearing rats. Biomed. Pharmacother. 2006, 60, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Hsiu, S.L.; Huang, T.Y.; Hou, Y.C.; Chin, D.H.; Chao, P.D. Comparison of metabolic pharmacokinetics of naringin and naringenin in rabbits. Life Sci. 2002, 70, 1481–1489. [Google Scholar] [CrossRef]
- Radominska-Pandya, A.; Bratton, S.; Little, J.M. A historical overview of the heterologous expression of mammalian udp-glucuronosyltransferase isoforms over the past twenty years. Curr. Drug Metab. 2005, 6, 141–160. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yin, O.Q.; Zuo, Z.; Chow, M.S. Pharmacokinetics and modeling of quercetin and metabolites. Pharm. Res. 2005, 22, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Brett, G.M.; Hollands, W.; Needs, P.W.; Teucher, B.; Dainty, J.R.; Davis, B.D.; Brodbelt, J.S.; Kroon, P.A. Absorption, metabolism and excretion of flavanones from single portions of orange fruit and juice and effects of anthropometric variables and contraceptive pill use on flavanone excretion. Br. J. Nutr. 2009, 101, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Terao, J.; Yamaguchi, S.; Shirai, M.; Miyoshi, M.; Moon, J.H.; Oshima, S.; Inakuma, T.; Tsushida, T.; Kato, Y. Protection by quercetin and quercetin 3-O-beta-d-glucuronide of peroxynitrite-induced antioxidant consumption in human plasma low-density lipoprotein. Free Radic. Res. 2001, 35, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.P.E.; Abd El Mohsen, M.M.; Rice-Evans, C. Cellular uptake and metabolism of flavonoids and their metabolites: Implications for their bioactivity. Arch. Biochem. Biophys. 2004, 423, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-L.; Xiao, N.; Li, X.-W.; Fan, Y.; Alolga, R.N.; Sun, X.-Y.; Wang, S.-L.; Li, P.; Qi, L.-W. Pharmacokinetic comparison between quercetin and quercetin 3-O-β-glucuronide in rats by UHPLC-MS/MS. Sci. Rep. 2016, 6, 35460. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Park, Y.B.; Bae, K.H.; Bok, S.H.; Kwon, Y.K.; Lee, E.S.; Choi, M.S. Cholesterol-lowering activity of naringenin via inhibition of 3-hydroxy-3-methylglutaryl coenzyme a reductase and acyl coenzyme a: Cholesterol acyltransferase in rats. Ann. Nutr. Metab. 1999, 43, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Kanno, S.; Tomizawa, A.; Hiura, T.; Osanai, Y.; Shouji, A.; Ujibe, M.; Ohtake, T.; Kimura, K.; Ishikawa, M. Inhibitory effects of naringenin on tumor growth in human cancer cell lines and sarcoma s-180-implanted mice. Biol. Pharm. Bull. 2005, 28, 527–530. [Google Scholar] [CrossRef] [PubMed]
- Zierau, O.; Gester, S.; Schwab, P.; Metz, P.; Kolba, S.; Wulf, M.; Vollmer, G. Estrogenic activity of the phytoestrogens naringenin, 6-(1,1-dimethylallyl)naringenin and 8-prenylnaringenin. Planta Med. 2002, 68, 449–451. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, T.I. Naringenin: A partial agonist on estrogen receptor in T47D-kbluc breast cancer cells. Int. J. Clin. Exp. Med. 2013, 6, 890–899. [Google Scholar] [PubMed]
- Chu, L.L.; Pandey, R.P.; Jung, N.; Jung, H.J.; Kim, E.-H.; Sohng, J.K. Hydroxylation of diverse flavonoids by cyp450 bm3 variants: Biosynthesis of eriodictyol from naringenin in whole cells and its biological activities. Microb. Cell Fact. 2016, 15, 135. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Kim, S.; Lee, J.; Park, J.Y.; Zhou, W.; Liu, X.; Kim, S.D.; Song, Y.S.; Jang, C.Y.; Oh, S.R.; et al. Characterization of phase I and phase II hepatic metabolism and reactive intermediates of Larrea nitida Cav. and its lignan compounds. Phytother. Res. 2017, 31, 140–151. [Google Scholar] [CrossRef] [PubMed]
- An, B.H.; Jeong, H.; Zhou, W.; Liu, X.; Kim, S.; Jang, C.Y.; Kim, H.-S.; Sohn, J.; Park, H.-J.; Sung, N.-H.; et al. Evaluation of the biological activity of Opuntia ficus indica as a tissue- and estrogen receptor subtype-selective modulator. Phytother. Res. 2016, 30, 971–980. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors upon request. |
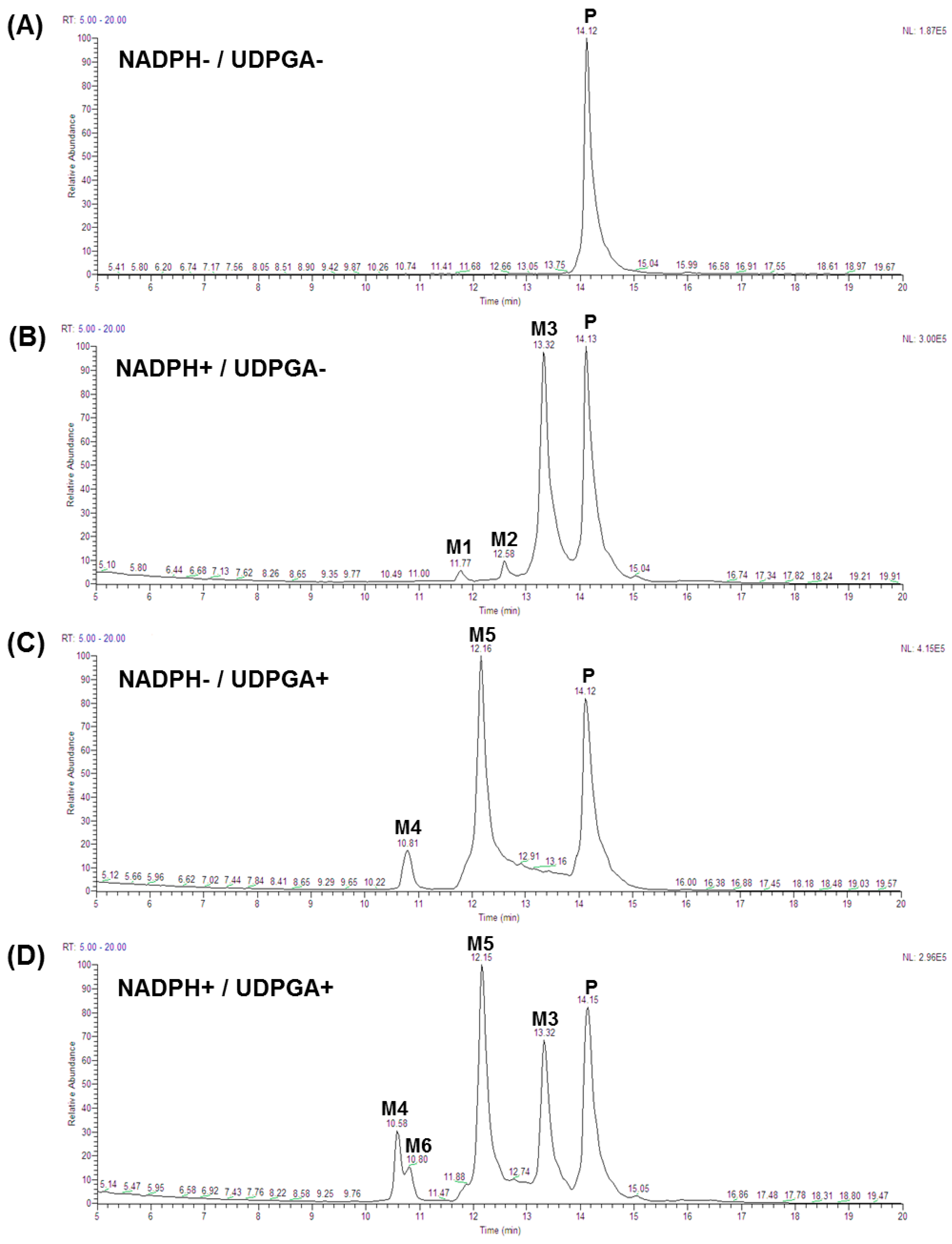
| Species | Amount Remaining at 30 min (%) a | t1/2 (min) b | CLint, hepatic (mL/min/kg) c |
|---|---|---|---|
| Human | 65.5 | >60 | <20.8 |
| Mouse | 4.90 | 5.40 | 565 |
| Rat | 29.0 | 10.1 | 124 |
| Dog | 18.5 | 2.7 | 641 |
| UDPGA | + | + | ||||
|---|---|---|---|---|---|---|
| NADPH | − | + | ||||
| Species | Amount Remaining at 30 min (%) a | t1/2 (min) b | CLint, hepatic (mL/min/kg) c | Amount Remaining at 30 min (%) a | t1/2 (min) | CLint, hepatic (mL/min/kg) |
| Human | 71.3 | >60 | <41.6 | 54.9 | 25.0 | 49.9 |
| Mouse | 1.8 | 3.80 | 802.4 | 0.0 | 0.91 | 3280 |
| Rat | 17.1 | 4.40 | 286.8 | 0.0 | 2.20 | 557 |
| Dog | 42.4 | 24.2 | 71.4 | 0.0 | 5.80 | 300 |
| Label | Precursor Ion, m/z | Major Fragment Ions (Relative Abundance) | Identification |
|---|---|---|---|
| Parent phytochemical (Figure 1A) | |||
| P | [M − H]− 285.22 | 164.92 (100) | SKN a |
| NADPH-dependent phase I metabolites (Figure 1B and Figure 2B) | |||
| M1 | [M − H]− 287.43 | 151.04 (100) | Eriodictyol a |
| M2 | [M − H]− 271.33 | 150.85 (100); 176.97 (21) | Naringenin a |
| M3 | [M − H]− 301.41 | 165.00 (100); 134.88 (25) | Either artocarpanone or sternbin b |
| Phase II glucuronide conjugation metabolites (Figure 1C and Figure 2E,F) | |||
| M4 | [M − H]− 461.52 | 285.27 (100) | Either SKN-5-O-β-glucuronide or SKN-4′-O-β-glucuronide b |
| M5 | [M − H]− 461.61 | 174.98 (100); 285.07 (32); 443.24 (10) | |
| NADPH and UDPGA-dependent metabolites (Figure 1D and Figure 2G) | |||
| M6 | [M − H]− 447.39 | 271.09 (100); 174.97 (36) | Naringenin 7-O-β-glucuronide b |
| CYP Isozyme | Phenotyping Reaction | % Inhibition in Co-Incubation | % Inhibition in Pre-Incubation |
|---|---|---|---|
| 1A2 | Phenacetin O-deethylation (PCOD) | 0.4 ± 0.5 a | 1.5 ± 2.1 |
| 2B6 | Bupropion hydroxylation (BPHY) | −0.6 ± 0.9 | 0.8 ± 0.2 |
| 2C9 | Diclofenac 4′-hydroxylation (DCHY) | 15.3 ± 3.8 | 8.7 ± 1.4 |
| 2C9 | Tolbutamide 6-hydroxylation (TOLHY) | 2.0 ± 0.6 | 10.1 ± 1.2 |
| 2D6 | Dextromethorphan O-demethylation (DEXOD) | −0.6 ± 3.1 | 3.5 ± 2.6 |
| 3A4 | Testosterone 6β-hydroxylation (TSTHY) | 15.6 ± 0.6 | 12.6 ± 2.8 |
| UGT Isozyme | Phenotyping Reaction | % Inhibition in Co-Incubation | % Inhibition in Pre-Incubation |
|---|---|---|---|
| 1A1 | 17 β-Estradiol 3-O-glucuronidation (ESG) | 0.4 ± 0.5 | 1.5 ± 2.1 |
| 1A3 | Chenodeoxycholic acid 24-glucuronidation (CDCAG) | −0.6 ± 0.9 | 0.8 ± 0.2 |
| 1A4 | Trifluoperazine N-glucuronidation (TFPG) | 15.3 ± 3.8 | 8.7 ± 1.4 |
| 1A6 | 1-Naphthol β-d-glucuronidation (NPG) | 2.0 ± 0.6 | 10.1 ± 1.2 |
| 1A9 | Mycophenolic acid O-glucuronidation (MPAG) | −0.6 ± 3.1 | 3.5 ± 2.6 |
| 2B7 | Zidovudine 5′-glucuronidation (AZTG) | 15.6 ± 0.6 | 12.6 ± 2.8 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, H.; Lee, J.; Kim, S.; Yeo, Y.Y.; So, H.; Wu, H.; Song, Y.S.; Jang, C.-Y.; Kim, H.-D.; Kim, M.J.; et al. Hepatic Metabolism of Sakuranetin and Its Modulating Effects on Cytochrome P450s and UDP-Glucuronosyltransferases. Molecules 2018, 23, 1542. https://doi.org/10.3390/molecules23071542
Jeong H, Lee J, Kim S, Yeo YY, So H, Wu H, Song YS, Jang C-Y, Kim H-D, Kim MJ, et al. Hepatic Metabolism of Sakuranetin and Its Modulating Effects on Cytochrome P450s and UDP-Glucuronosyltransferases. Molecules. 2018; 23(7):1542. https://doi.org/10.3390/molecules23071542
Chicago/Turabian StyleJeong, Hyesoo, Jimin Lee, Soolin Kim, Yoo Yeon Yeo, Hyunyoung So, Honghua Wu, Yun Seon Song, Chang-Young Jang, Hee-Doo Kim, Min Jung Kim, and et al. 2018. "Hepatic Metabolism of Sakuranetin and Its Modulating Effects on Cytochrome P450s and UDP-Glucuronosyltransferases" Molecules 23, no. 7: 1542. https://doi.org/10.3390/molecules23071542
APA StyleJeong, H., Lee, J., Kim, S., Yeo, Y. Y., So, H., Wu, H., Song, Y. S., Jang, C.-Y., Kim, H.-D., Kim, M. J., & Chang, M. (2018). Hepatic Metabolism of Sakuranetin and Its Modulating Effects on Cytochrome P450s and UDP-Glucuronosyltransferases. Molecules, 23(7), 1542. https://doi.org/10.3390/molecules23071542





