Design, Synthesis, and In Vitro Evaluation of Novel Histone H3 Peptide-Based LSD1 Inactivators Incorporating α,α-Disubstituted Amino Acids with γ-Turn-Inducing Structures
Abstract
1. Introduction
2. Results
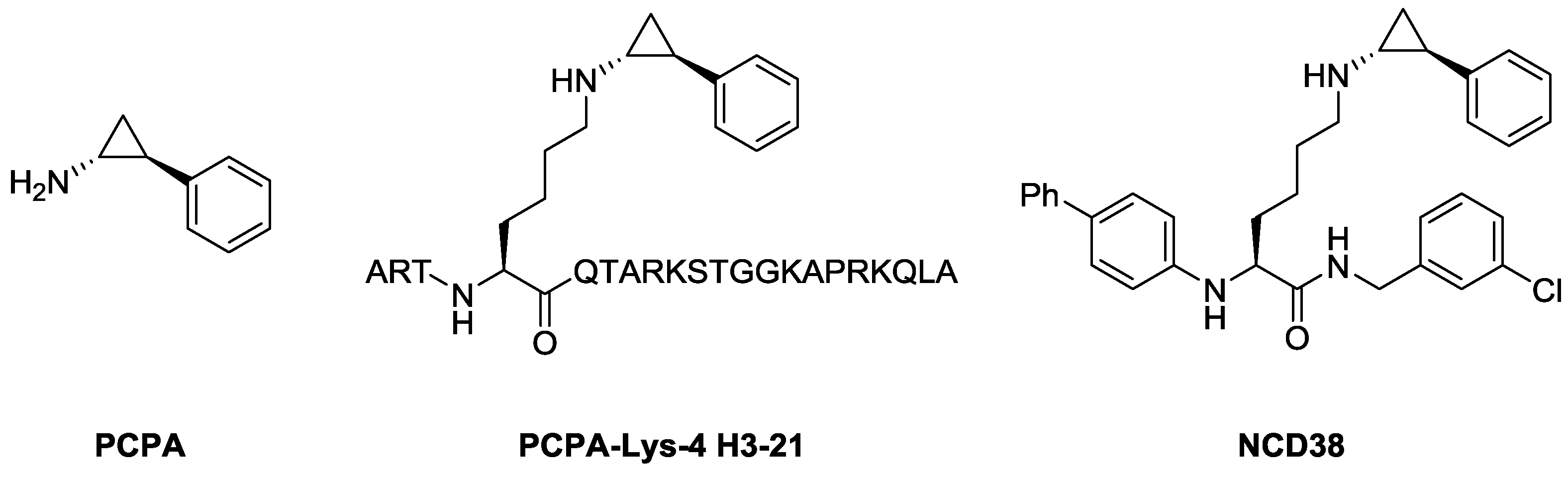
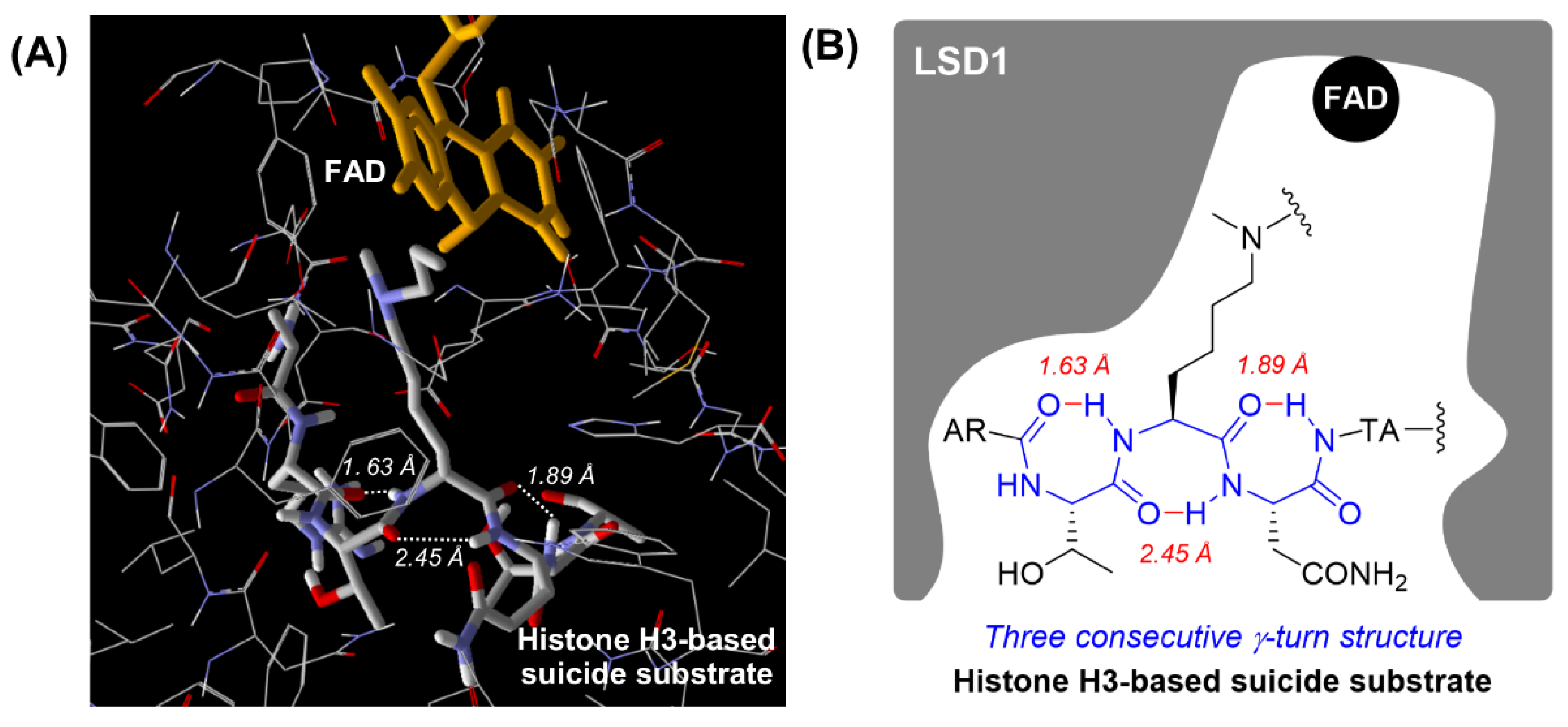
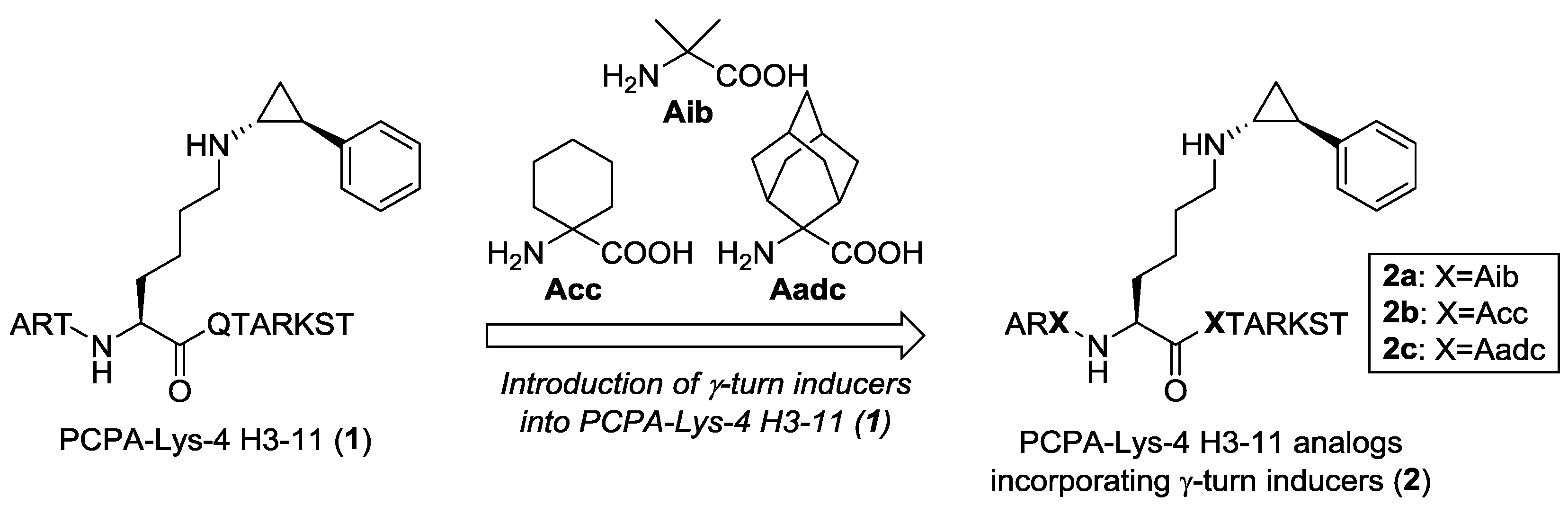
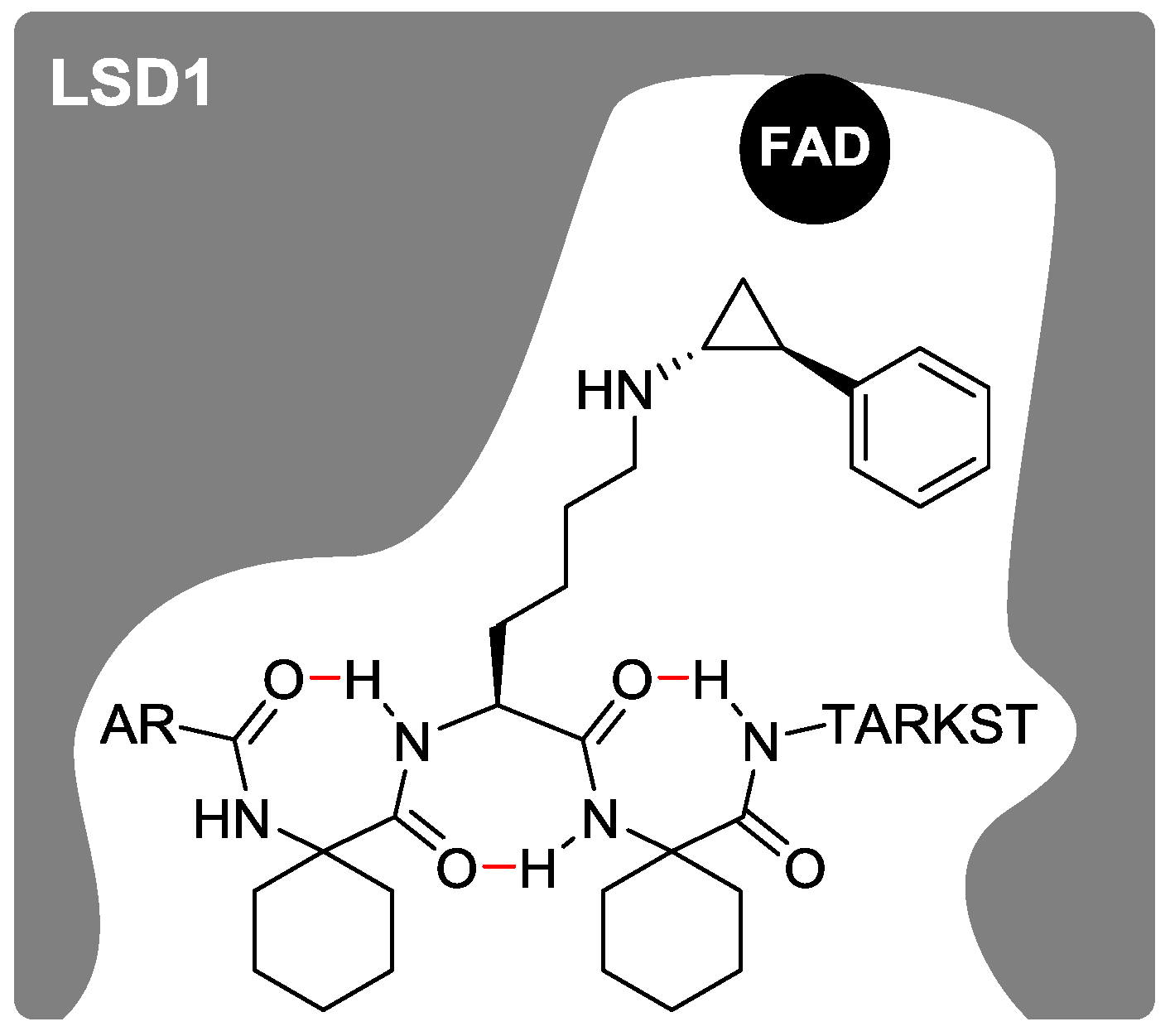
2.1. Design
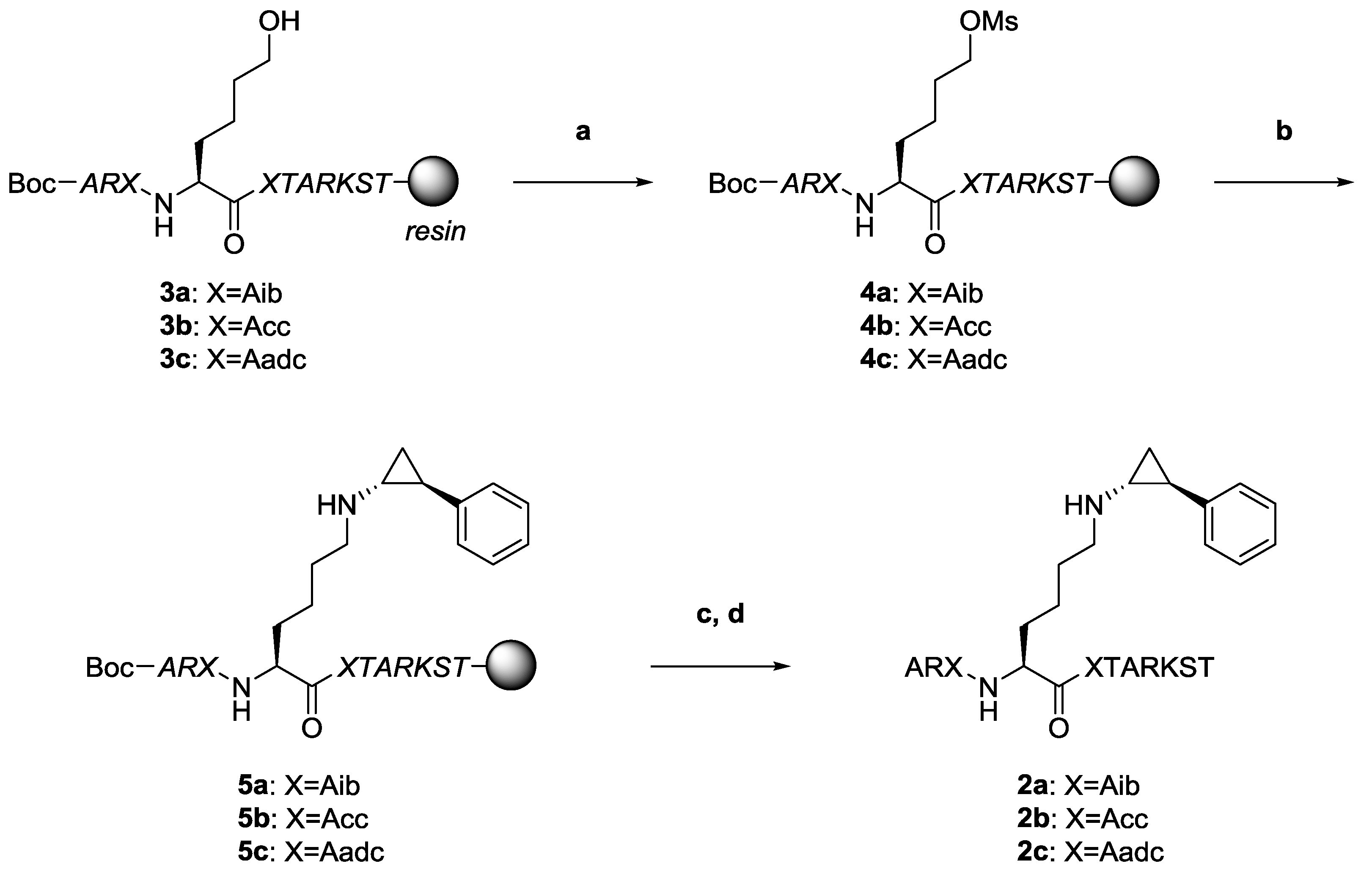
2.2. Synthesis
2.3. In Vitro Evaluation of LSD1 Inhibitory Activity
3. Discussion
4. Materials and Methods
4.1. General Methods
4.2. Synthesis of Peptides with γ-Turn Inducers
4.3. Assay for LSD1 Inhibitory Activity
4.4. Assay for MAO Inhibitory Activity
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Greer:, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Metzger, E.; Wissmann, M.; Yin, N.; Müller, J.M.; Schneider, R.; Peters, A.H.F.M.; Günther, T.; Buettner, R.; Schüle, R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005, 437, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Sengupta, R.; Espejo, A.B.; Lee, M.G.; Dorsey, J.A.; Richter, M.; Opravil, S.; Shiekhattar, R.; Bedford, M.T.; Jenuwein, T.; et al. p53 is regulated by the lysine demethylase LSD1. Nature 2007, 449, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hevi, S.; Kurash, J.K.; Lei, H.; Gay, F.; Bajko, J.; Su, H.; Sun, W.; Chang, H.; Xu, G.; et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009, 41, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Kontaki, H.; Talianidis, I. Lysine Methylation Regulates E2F1-Induced Cell Death. Mol. Cell 2010, 39, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.G.; Wynder, C.; Cooch, N.; Shiekhattar, R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 2005, 437, 432–435. [Google Scholar] [CrossRef]
- Saleque, S.; Kim, J.; Rooke, H.M.; Orkin, S.H. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol. Cell 2007, 27, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A.; Takezawa, S.; Schüle, R.; Kitagawa, H.; Kato, S. Transrepressive function of TLX requires the histone demethylase LSD1. Mol. Cell. Biol. 2008, 28, 3995–4003. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Wissmann, M.; Yin, N.; Müller, J.M.; Greschik, H.; Fodor, B.D.; Jenuwein, T.; Vogler, C.; Schneider, R.; Günther, T.; Buettner, R.; et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 2007, 9, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Højfeldt, J.W.; Agger, K.; Helin, K. Histone lysine demethylases as targets for anticancer therapy. Nat. Rev. Drug Discov. 2013, 12, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Cui, S.; Engel, J.D.; Tanabe, O. Lysine-specific demethylase 1 is a therapeutic target for fetal hemoglobin induction. Nat. Med. 2013, 19, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Vogel, J.L.; Narayanan, A.; Peng, H.; Kristie, T.M. Inhibition of the histone demethylase LSD1 blocks α-herpesvirus lytic replication and reactivation from latency. Nat. Med. 2009, 15, 1312–1317. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Miyata, N. Lysine Demethylases Inhibitors. J. Med. Chem. 2011, 54, 8236–8250. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, B.; Suzuki, T.; Liu, X.; Zhan, P. Medicinal chemistry insights in the discovery of novel LSD1 inhibitors. Epigenomics 2015, 7, 1379–1396. [Google Scholar] [CrossRef] [PubMed]
- McAllister, T.E.; England, K.S.; Hopkinson, R.J.; Brennan, P.E.; Kawamura, A.; Schofield, C.J. Recent Progress in Histone Demethylase Inhibitors. J. Med. Chem. 2016, 59, 1308–1329. [Google Scholar] [CrossRef] [PubMed]
- Maes, T.; Mascaró, C.; Ortega, A.; Lunardi, S.; Ciceri, F.; Somervaille, T.C.P.; Buesa, C. KDM1 histone lysine demethylases as targets for treatments of oncological and neurodegenerative disease. Epigenomics 2015, 7, 609–626. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, H.P.; Smitheman, K.N.; Kamat, C.D.; Soong, D.; Federowicz, K.E.; Van Aller, G.S.; Schneck, J.L.; Carson, J.D.; Liu, Y.; Butticello, M.; et al. A DNA Hypomethylation Signature Predicts Antitumor Activity of LSD1 Inhibitors in SCLC. Cancer Cell 2015, 28, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Ueda, R.; Suzuki, T.; Mino, K.; Tsumoto, H.; Nakagawa, H.; Hasegawa, M.; Sasaki, R.; Mizukami, T.; Miyata, N. Identification of Cell-Active Lysine Specific Demethylase 1-Selective Inhibitors. J. Am. Chem. Soc. 2009, 131, 17536–17537. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, D.; Suzuki, T.; Mino, K.; Ueda, R.; Khan, M.N.A.; Matsubara, T.; Koseki, K.; Hasegawa, M.; Sasaki, R.; Nakagawa, H.; et al. Synthesis and biological activity of optically active NCL-1, a lysine-specific demethylase 1 selective inhibitor. Bioorg. Med. Chem. 2011, 19, 3702–3708. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, D.; Itoh, Y.; Tsumoto, H.; Kakizawa, T.; Mino, K.; Fukuhara, K.; Nakagawa, H.; Hasegawa, M.; Sasaki, R.; Mizukami, T.; et al. Lysine-Specific Demethylase 1-Selective Inactivators: Protein-Targeted Drug Delivery Mechanism. Angew. Chem. Int. Ed. 2013, 52, 8620–8624. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Ogasawara, D.; Ota, Y.; Mizukami, T.; Suzuki, T. Synthesis, LSD1 Inhibitory Activity, and LSD1 Binding Model of Optically Pure Lysine-PCPA Conjugates. Comput. Struct. Biotechnol. J. 2014, 9, e201402002. [Google Scholar] [CrossRef] [PubMed]
- Ahmed Khan, M.N.; Tsumoto, H.; Itoh, Y.; Ota, Y.; Suzuki, M.; Ogasawara, D.; Nakagawa, H.; Mizukami, T.; Miyata, N.; Suzuki, T. Design, synthesis, and biological activity of N-alkylated analogue of NCL1, a selective inhibitor of lysine-specific demethylase 1. Medchemcomm 2015, 6, 407–412. [Google Scholar] [CrossRef]
- Kakizawa, T.; Ota, Y.; Itoh, Y.; Tsumoto, H.; Suzuki, T. Histone H3 peptide based LSD1-selective inhibitors. Bioorg. Med. Chem. Lett. 2015, 25, 1925–1928. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Aihara, K.; Mellini, P.; Tojo, T.; Ota, Y.; Tsumoto, H.; Solomon, V.R.; Zhan, P.; Suzuki, M.; Ogasawara, D.; et al. Identification of SNAIL1 Peptide-Based Irreversible Lysine-Specific Demethylase 1-Selective Inactivators. J. Med. Chem. 2016, 59, 1531–1544. [Google Scholar] [CrossRef] [PubMed]
- Miyamura, S.; Araki, M.; Ota, Y.; Itoh, Y.; Yasuda, S.; Masuda, M.; Taniguchi, T.; Sowa, Y.; Sakai, T.; Suzuki, T.; et al. C–H activation enables a rapid structure–activity relationship study of arylcyclopropyl amines for potent and selective LSD1 inhibitors. Org. Biomol. Chem. 2016, 14, 8576–8585. [Google Scholar] [CrossRef] [PubMed]
- Kakizawa, T.; Ota, Y.; Itoh, Y.; Suzuki, T. Histone H3 peptides incorporating modified lysine residues as lysine-specific demethylase 1 inhibitors. Bioorg. Med. Chem. Lett. 2018, 28, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Ota, Y.; Miyamura, S.; Araki, M.; Itoh, Y.; Yasuda, S.; Masuda, M.; Taniguchi, T.; Sowa, Y.; Sakai, T.; Itami, K.; et al. Design, synthesis and evaluation of γ-turn mimetics as LSD1-selective inhibitors. Bioorg. Med. Chem. 2018, 26, 775–785. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.M.Z.; McCafferty, D.G. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry 2007, 46, 4408–4416. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Culhane, J.C.; Szewczuk, L.M.; Jalili, P.; Ball, H.L.; Machius, M.; Cole, P.A.; Yu, H. Structural Basis for the Inhibition of the LSD1 Histone Demethylase by the Antidepressant trans-2-Phenylcyclopropylamine. Biochemistry 2007, 46, 8058–8065. [Google Scholar] [CrossRef] [PubMed]
- Sareddy, G.R.; Viswanadhapalli, S.; Surapaneni, P.; Suzuki, T.; Brenner, A.; Vadlamudi, R.K. Novel KDM1A inhibitors induce differentiation and apoptosis of glioma stem cells via unfolded protein response pathway. Oncogene 2017, 36, 2423–2434. [Google Scholar] [CrossRef] [PubMed]
- Sugino, N.; Kawahara, M.; Tatsumi, G.; Kanai, A.; Matsui, H.; Yamamoto, R.; Nagai, Y.; Fujii, S.; Shimazu, Y.; Hishizawa, M.; et al. A novel LSD1 inhibitor NCD38 ameliorates MDS-related leukemia with complex karyotype by attenuating leukemia programs via activating super-enhancers. Leukemia 2017, 31, 2302–2314. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Culhane, J.C.; Szewczuk, L.M.; Gocke, C.B.; Brautigam, C.A; Tomchick, D.R.; Machius, M.; Cole, P.A.; Yu, H. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nat. Struct. Mol. Biol. 2007, 14, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, Y.; Ueda, H.; Nozawa, H.; Ogoshi, H. Adamantyl amino acid as γ-turn inducer for peptide. Tetrahedron Lett. 1997, 38, 7901–7904. [Google Scholar] [CrossRef]
- Youdim, M.B.H.; Edmondson, D.; Tipton, K.F. The therapeutic potential of monoamine oxidase inhibitors. Nat. Rev. Neurosci. 2006, 7, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Whetstine, J.R.; Nottke, A.; Lan, F.; Huarte, M.; Smolikov, S.; Chen, Z.; Spooner, E.; Li, E.; Zhang, G.; Colaiacovo, M.; et al. Reversal of Histone Lysine Trimethylation by the JMJD2 Family of Histone Demethylases. Cell 2006, 125, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Christophersen, P.C.; Fano, M.; Saaby, L.; Yang, M.; Nielsen, H.M.; Mu, H. Characterization of particulate drug delivery systems for oral delivery of Peptide and protein drugs. Curr. Pharm. Des. 2015, 21, 2611–2628. [Google Scholar] [CrossRef] [PubMed]
- Kumarasinghe, I.R.; Woster, P.M. Synthesis and Evaluation of Novel Cyclic Peptide Inhibitors of Lysine-Specific Demethylase 1. ACS Med. Chem. Lett. 2014, 5, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Kumarasinghe, I.R.; Woster, P.M. Cyclic by peptide inhibitors of lysine-specific demethylase 1 with improved potency identified by alanine scanning mutagenesis. Eur. J. Med. Chem. 2018, 148, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Li, J.; Zheng, M.; Zuo, L.; Sun, C.; Zhu, Y.; Fang, L.; Liu, L.; Zhou, X. Stable H3 peptide was delivered by gold nanorods to inhibit LSD1 activation and induce human mesenchymal stem cells differentiation. Oncotarget 2017, 8, 23110–23119. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Karstad, R.; Isaksen, G.; Brandsdal, B.O.; Svendsen, J.S.; Svenson, J. Unnatural amino acid side chains as S1, S1’, and S2’ probes yield cationic antimicrobial peptides with stability toward chymotryptic degradation. J. Med. Chem. 2010, 53, 5558–5566. [Google Scholar] [CrossRef] [PubMed]
- Brimble, M.A.; Kowalczyk, R.; Harris, P.W.R.; Dunbar, P.R.; Muir, V.J. Synthesis of fluorescein-labelled O-mannosylated peptides as components for synthetic vaccines: Comparison of two synthetic strategies. Org. Biomol. Chem. 2008, 6, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Culhane, J.C.; Wang, D.; Yen, P.M.; Cole, P.A. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors. J. Am. Chem. Soc. 2010, 132, 3164–3176. [Google Scholar] [CrossRef] [PubMed]
- Szewczuk, L.M.; Culhane, J.C.; Yang, M.; Majumdar, A.; Yu, H.; Cole, P.A. Mechanistic analysis of a suicide inactivator of histone demethylase LSD1. Biochemistry 2007, 46, 6892–6902. [Google Scholar] [CrossRef] [PubMed]
- Culhane, J.C.; Szewczuk, L.M.; Liu, X.; Da, G.; Marmorstein, R.; Cole, P.A. A mechanism-based inactivator for histone demethylase LSD1. J. Am. Chem. Soc. 2006, 128, 4536–4537. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |
| Compound | IC50 (µM) 1 | ||
|---|---|---|---|
| LSD1 | MAO-A | MAO-B | |
| PCPA | 16.5 ± 1.9 | 6.00 ± 1.38 | 6.54 ± 0.51 |
| 1 | 0.126 ± 0.002 | >10 | >10 |
| 2a | 0.0891 ± 0.0053 | >10 | >10 |
| 2b | 0.0584 ± 0.0025 | >10 | >10 |
| 2c | 0.0724 ± 0.0040 | >10 | >10 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ota, Y.; Kakizawa, T.; Itoh, Y.; Suzuki, T. Design, Synthesis, and In Vitro Evaluation of Novel Histone H3 Peptide-Based LSD1 Inactivators Incorporating α,α-Disubstituted Amino Acids with γ-Turn-Inducing Structures. Molecules 2018, 23, 1099. https://doi.org/10.3390/molecules23051099
Ota Y, Kakizawa T, Itoh Y, Suzuki T. Design, Synthesis, and In Vitro Evaluation of Novel Histone H3 Peptide-Based LSD1 Inactivators Incorporating α,α-Disubstituted Amino Acids with γ-Turn-Inducing Structures. Molecules. 2018; 23(5):1099. https://doi.org/10.3390/molecules23051099
Chicago/Turabian StyleOta, Yosuke, Taeko Kakizawa, Yukihiro Itoh, and Takayoshi Suzuki. 2018. "Design, Synthesis, and In Vitro Evaluation of Novel Histone H3 Peptide-Based LSD1 Inactivators Incorporating α,α-Disubstituted Amino Acids with γ-Turn-Inducing Structures" Molecules 23, no. 5: 1099. https://doi.org/10.3390/molecules23051099
APA StyleOta, Y., Kakizawa, T., Itoh, Y., & Suzuki, T. (2018). Design, Synthesis, and In Vitro Evaluation of Novel Histone H3 Peptide-Based LSD1 Inactivators Incorporating α,α-Disubstituted Amino Acids with γ-Turn-Inducing Structures. Molecules, 23(5), 1099. https://doi.org/10.3390/molecules23051099






