Comparative Analysis of the Complete Chloroplast Genomes of Four Aconitum Medicinal Species
Abstract
1. Introduction
2. Results and Discussion
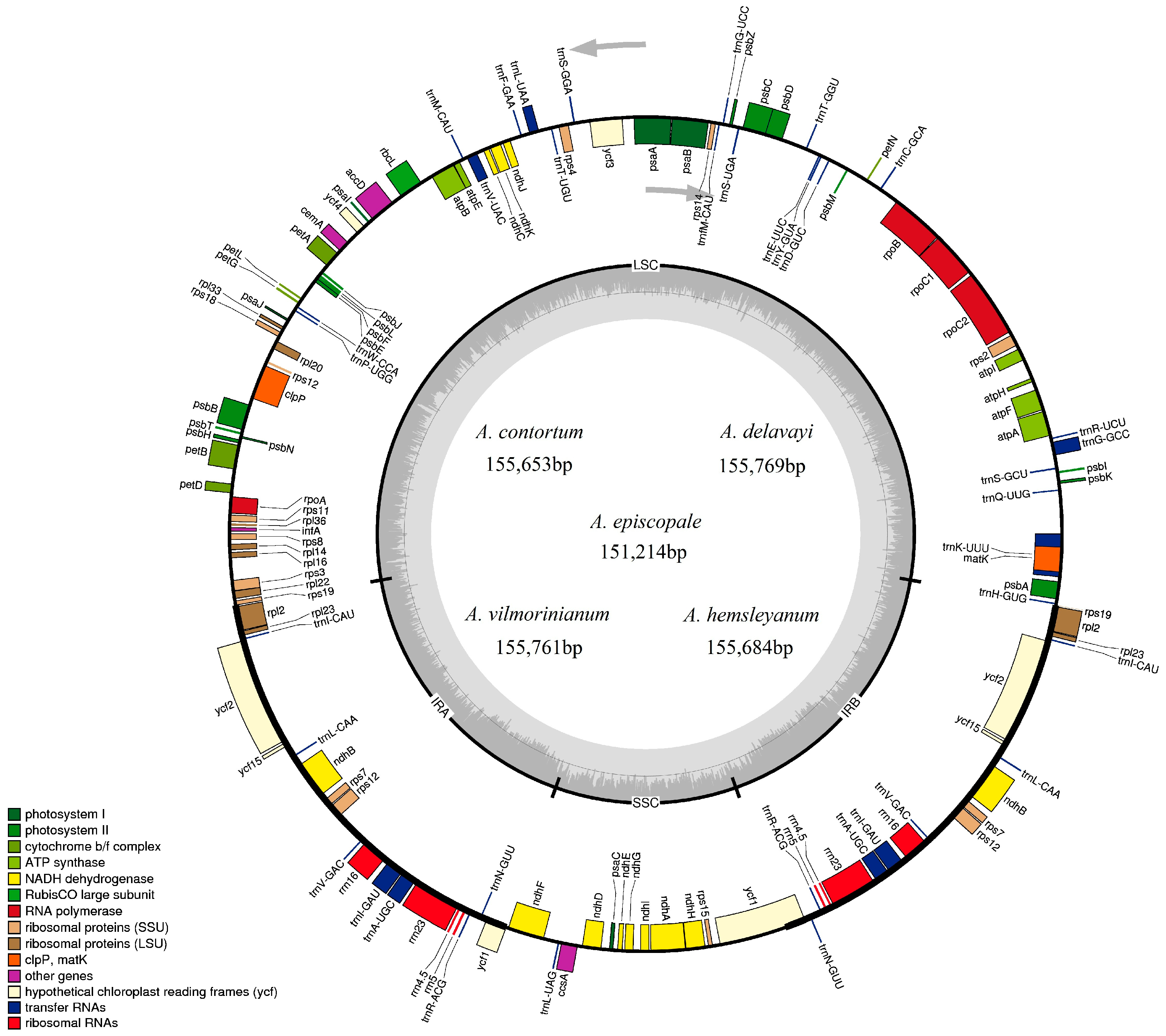
2.1. Chloroplast Genome Features
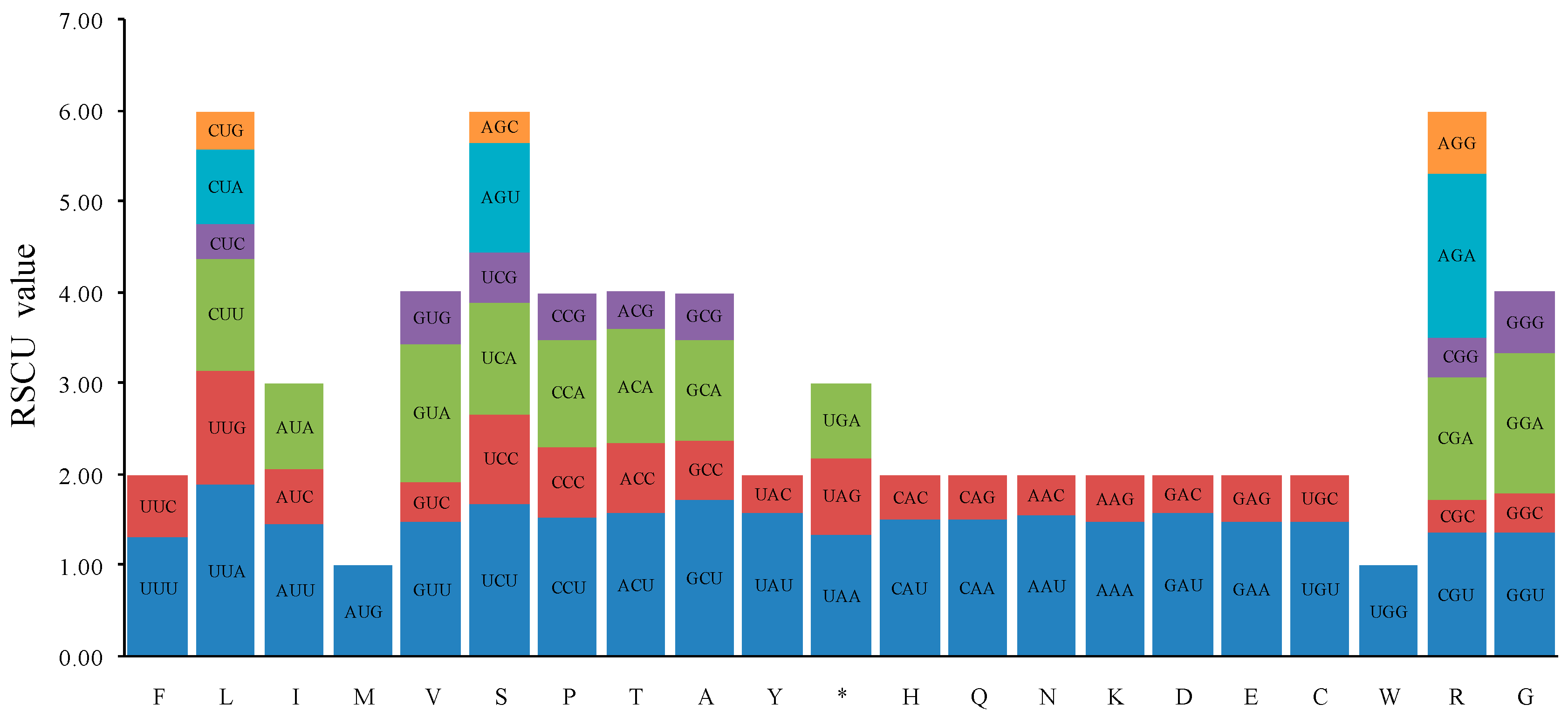
2.2. Codon Usage
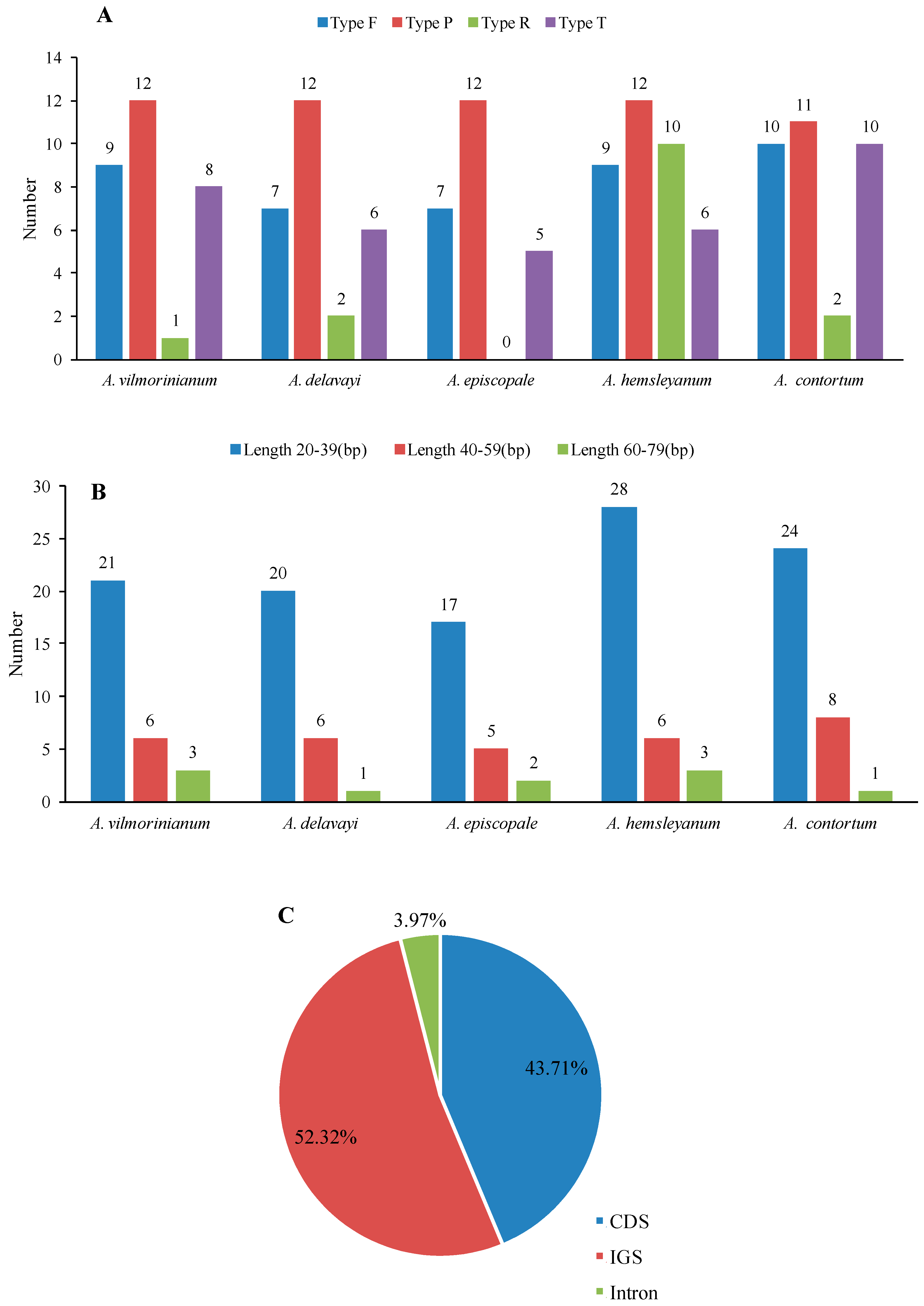
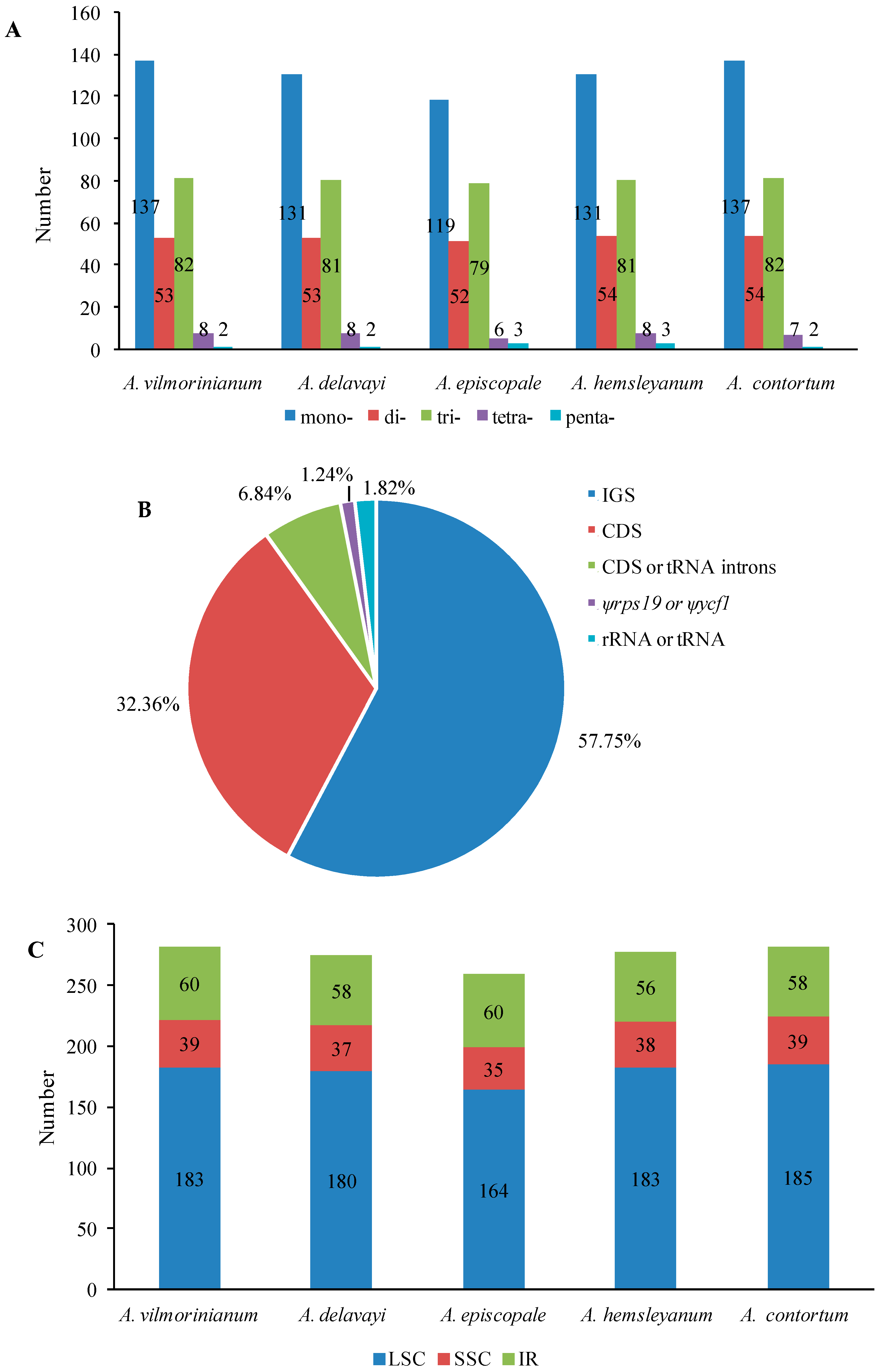
2.3. Repeat and SSR Analyses
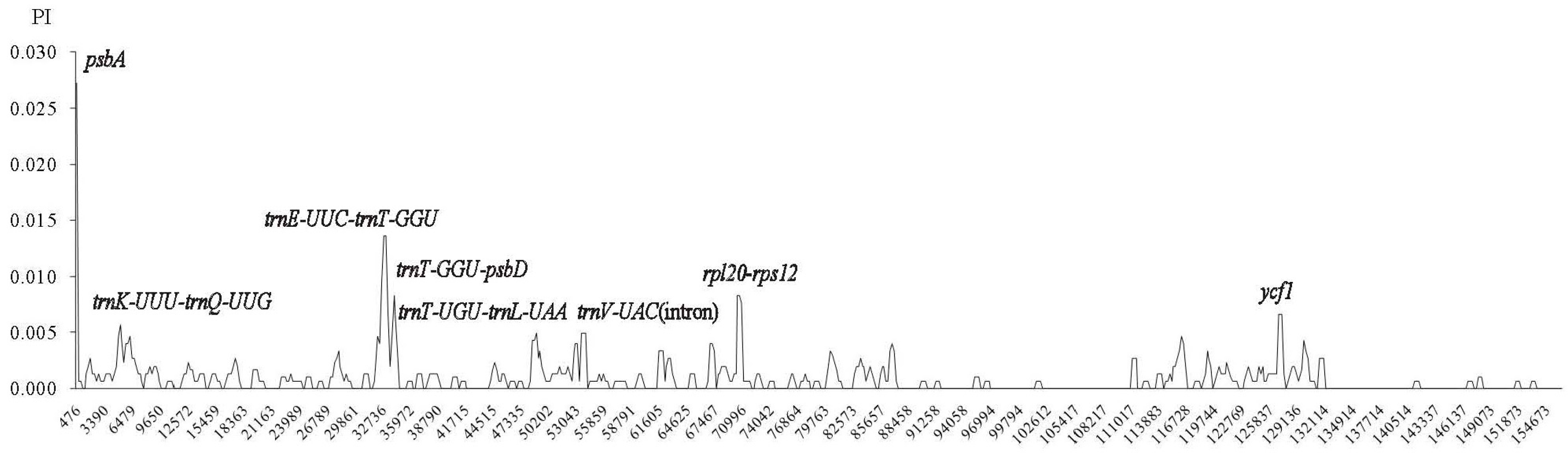
2.4. Comparative Chloroplast Genomic Analysis
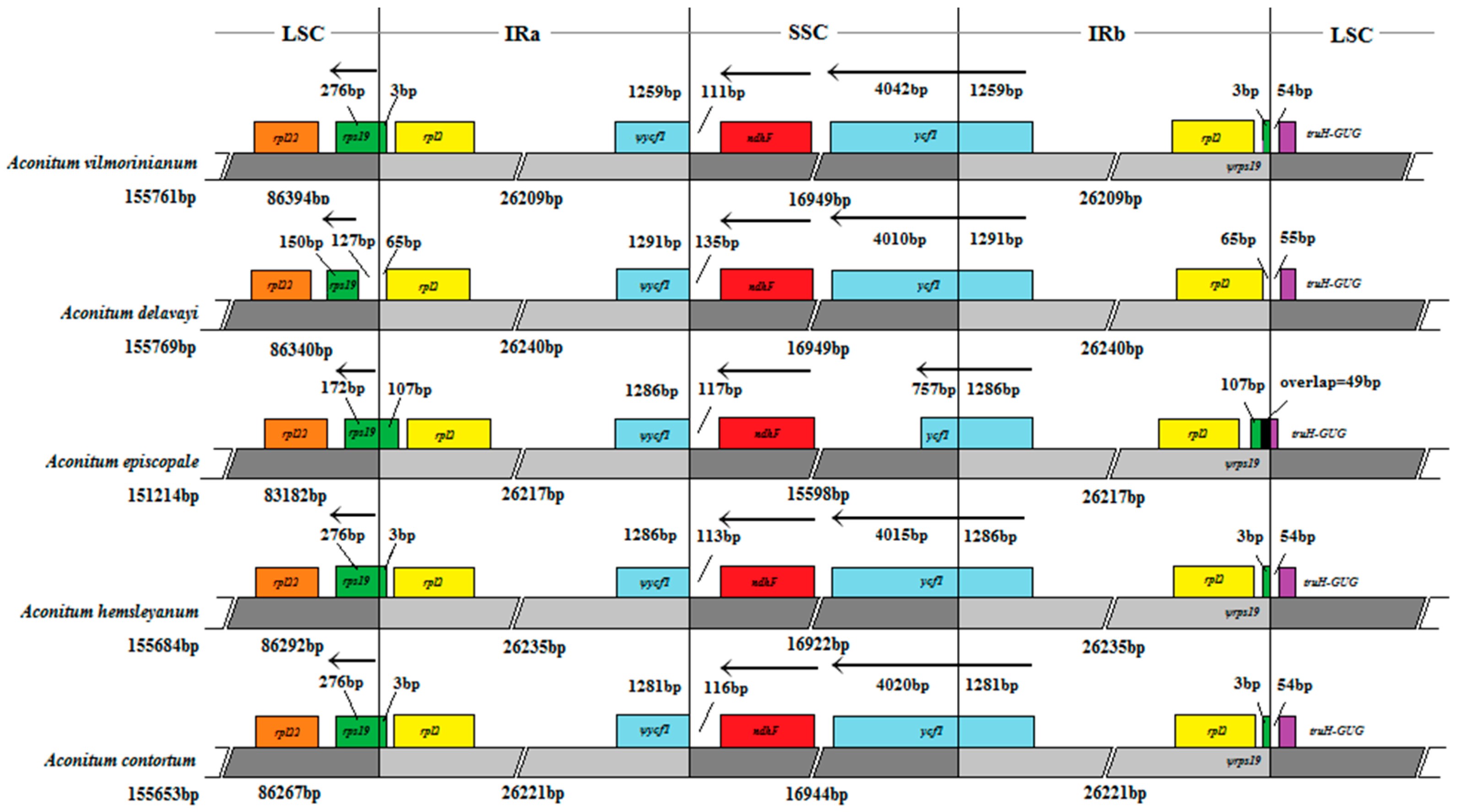
2.5. IR Expansion and Contraction
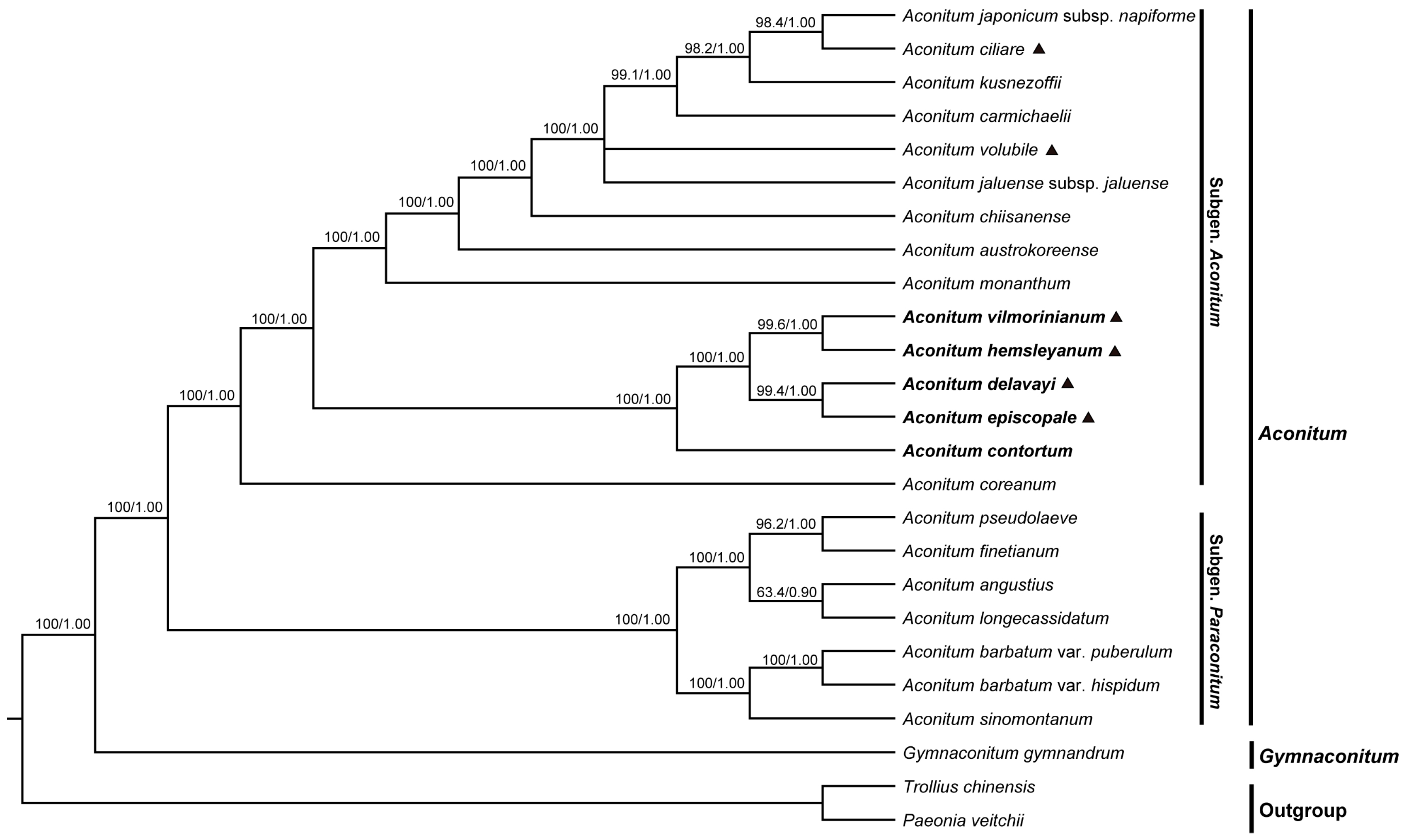
2.6. Phylogenetic Analyses
3. Materials and Methods
3.1. Plant Material, DNA Extraction and Sequencing
3.2. Chloroplast Genome Assembly and Annotation
3.3. Structure of Genome and Genome Comparison
3.4. Phylogenetic Analyses
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M.; dePamphilis, C.W.; Muller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Tonti-Filippini, J.; Nevill, P.G.; Dixon, K.; Small, I. What can we do with 1000 plastid genomes? Plant J. 2017, 90, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Park, I.; Kim, W.J.; Yeo, S.M.; Choi, G.; Kang, Y.M.; Piao, R.; Moon, B.C. The complete chloroplast genome sequences of Fritillaria ussuriensis Maxim. and Fritillaria cirrhosa D. Don and comparative analysis with other Fritillaria species. Molecules 2017, 22, 982. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.B.; Yu, H.; Wang, J.H.; Lei, W.J.; Gao, J.H.; Qiu, X.P.; Wang, J.S. The complete chloroplast genome sequences of the medicinal plant Forsythia suspensa (Oleaceae). Int. J. Mol. Sci. 2017, 18, 2288. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.G.; Chen, X.L.; Cui, Y.X.; Sun, W.; Li, Y.H.; Wang, Y.; Song, J.Y.; Yao, H. Molecular structure and phylogenetic analyses of complete chloroplast genomes of two Aristolochia medicinal species. Int. J. Mol. Sci. 2017, 18, 1839. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.Q.; Drew, B.T.; Yang, J.B.; Gao, L.M.; Li, D.Z. Comparative chloroplast genomes of eleven Schima (Theaceae) species: Insights into DNA barcoding and phylogeny. PLoS ONE 2017, 12, e0178026. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Dong, W.P.; Li, W.Q.; Lu, Y.Z.; Xie, X.M.; Jin, X.B.; Shi, J.P.; He, K.H.; Suo, Z.L. Comparative analysis of six Lagerstroemia complete chloroplast genomes. Front. Plant Sci. 2017, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.H.; Tembrock, L.R.; Zheng, S.Y.; Wu, Z.Q. The complete chloroplast genome of Catha edulis: A comparative analysis of genome features with related species. Int. J. Mol. Sci. 2018, 19, 525. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Q.; Kadota, Y. Aconitum; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MI, USA, 2001; Volume 6, pp. 149–222. [Google Scholar]
- Cheng, Z.Y.; Jiang, Z.H. The medicinal plant resources of Chinese Aconitum and Delphinium. Acta Bot. Yunn. 1988, 10, 63–76. [Google Scholar]
- Wu, Z.Y.; Zhou, T.Y.; Xiao, P.G. Compendium of Xinhua Bentgrass (I); Shanghai Scientific & Technical Publishers: Shanghai, China, 1988. [Google Scholar]
- Kong, H.H.; Liu, W.Z.; Yao, G.; Gong, W. A comparison of chloroplast genome sequences in Aconitum (Ranunculaceae): A traditional herbal medicinal genus. PeerJ 2017, 5, e4018. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Park, I.; Kim, W.J.; Yang, S.; Yeo, S.M.; Li, H.L.; Moon, B.C. The complete chloroplast genome sequence of Aconitum coreanum and Aconitum carmichaelii and comparative analysis with other Aconitum species. PLoS ONE 2017, 12, e0184257. [Google Scholar] [CrossRef] [PubMed]
- Niitsu, K.; Ikeya, Y.; Mitsuhashi, H.; Cheng, S.Y.; Li, H.L. Studies on the Alkaloids from Aconitum contortum (I). Heterocycles 1990, 31, 1517–1524. [Google Scholar] [CrossRef]
- Yang, Q.E. A study on botanical origin of the Chinese herbal drug ‘Dula’. Acta Bot. Yunn. 1990, 12, 247–253. [Google Scholar]
- Xiao, P.G.; Wang, F.P.; Gao, F.; Yan, L.P.; Chen, D.L.; Liu, Y. A pharmacophylogenetic study of Aconitum L. (Ranunculaceae) from China. Acta Phytotaxon. Sin. 2006, 44, 1–46. [Google Scholar] [CrossRef]
- Liang, J.Y.; Yan, Z.Y.; Chen, X.; Wan, D.G.; Yang, X.C. Rearch on the anti-arrhythmic effect of Yi nationality herb ‘Dula’. Li Shi Zhen Med. Mater. Med. Res. 2008, 19, 2108–2109. [Google Scholar]
- Li, X.P.; He, J.; He, S.L.; Meng, J. Research progress of Aconitum vilmorinianum. J. West China Forest. Sci. 2017, 46, 1–7. [Google Scholar]
- Wang, W.T. Aconitum. In Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1979; Volume 27, pp. 113–326. [Google Scholar]
- Zhang, F.M.; Ge, S.; Chen, W.L. Phylogeny of the Aconitum delavayi complex (Ranunculaceae) based on evidence from nuclear ribosomal ITS sequences. Acta Phytotaxon. Sin. 2003, 41, 220–228. [Google Scholar]
- Zhang, F.M.; Chen, W.L.; Yang, Q.E.; Ge, S. Genetic differentiation and relationship of populations in the Aconitum delavayi complex (Ranunculaceae) and their taxonomic implications. Plant Syst. Evol. 2005, 254, 39–48. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, F.M.; Yang, Q.E. Phylogeny of Aconitum subgenus Aconitum (Ranunculaceae) inferred from ITS sequences. Plant Syst. Evol. 2005, 252, 11–25. [Google Scholar] [CrossRef]
- He, J.; Wong, K.L.; Shaw, P.C.; Wang, H.; Li, D.Z. Identification of the medicinal plants in Aconitum L. by DNA barcoding technique. Planta Med. 2010, 76, 1622–1628. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.E. Taxonomic notes on some species of Aconitum L. (Ranunculaceae) from Yunnan, China. Acta Phytotaxon. Sin. 1999, 37, 545–590. [Google Scholar]
- Chen, X.C.; Li, Q.S.; Li, Y.; Qian, J.; Han, J.P. Chloroplast genome of Aconitum barbatum var. puberulum (Ranunculaceae) derived from CCS reads using the PacBio RS platform. Front. Plant Sci. 2015, 6, 42. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.W.; Feng, D.J.; Song, G.S.; Wei, X.L.; Chen, L.; Wu, X.L.; Li, X.G.; Zhu, Z. The first intron of rice EPSP synthase enhances expression of foreign gene. Sci. China Ser. C 2003, 46, 561. [Google Scholar] [CrossRef] [PubMed]
- Morton, B.R. Selection on the codon bias of chloroplast and cyanelle genes in different plant and algal lineages. J. Mol. Evol. 1998, 46, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.J.; Lv, S.Z.; Zhang, Y.X.; Du, X.H.; Wang, L.; Biradar, S.S.; Tan, X.F.; Wan, F.H.; Song, W.N. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora). PLoS ONE 2012, 7, e36869. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Song, J.Y.; Gao, H.H.; Zhu, Y.J.; Xu, J.; Pang, X.H.; Yao, H.; Sun, C.; Li, X.E.; Li, C.Y.; et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE 2013, 8, e57607. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. Chloroplast evolution: Secondary symbiogenesis and multiple losses. Curr. Biol. 2002, 12, 62–64. [Google Scholar] [CrossRef]
- Yi, X.; Gao, L.; Wang, B.; Su, Y.J.; Wang, T. The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): Evolutionary comparison of Cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in Gymnosperms. Genome Biol. Evol. 2013, 5, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.P.; Xu, C.; Cheng, T.; Lin, K.; Zhou, S.L. Sequencing angiosperm plastid genomes made easy: A complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol. Evol. 2013, 5, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.C.; Zhou, T.; Duan, D.; Yang, J.; Feng, L.; Zhao, G.F. Comparative analysis of the complete chloroplast genomes of Five Quercus species. Front. Plant Sci. 2016, 7, 959. [Google Scholar] [CrossRef] [PubMed]
- Perdereau, A.C.; Kelleher, C.T.; Douglas, G.C.; Hodkinson, T.R. High levels of gene flow and genetic diversity in Irish populations of Salix caprea L. inferred from chloroplast and nuclear SSR markers. BMC Plant Biol. 2014, 14, 202. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lin, F.R.; Huang, P.; Guo, W.Y.; Zheng, Y.Q. Complete chloroplast genome sequence of Decaisnea insignis: Genome organization, genomic resources and comparative analysis. Sci. Rep. 2017, 7, 10073. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.F.; Wu, M.L.; Liao, B.S.; Liu, Z.X.; Bai, R.; Xiao, S.M.; Li, X.W.; Zhang, B.L.; Xu, J.; Chen, S.L. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules 2017, 22, 1330. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Leese, F.; Mayer, C.; Held, C. Isolation of microsatellites from unknown genomes using known genomes as enrichment templates. Limnol. Oceanogr. Methods 2008, 6, 412–426. [Google Scholar] [CrossRef]
- Li, Q.; Wan, J.M. SSRHunter: Development of a local searching software for SSR sites. Hereditas 2005, 27, 808–810. [Google Scholar] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Sequence data of A. vilmorinianum, A. delavayi, A. episcopale, A. hemsleyanum and A. contortum have been deposited into GenBank and are available from the authors. |

| Characteristics | A. vil | A. del | A. epi | A. hem | A. con |
|---|---|---|---|---|---|
| Location | Wuding | Heqing | Dali | Qiaojia | Dali |
| Voucher specimens | LCF1 | 1395 | 1379 | QJ6 | 895 |
| GenBank numbers | MG678799 | MG678802 | MG678801 | MG678800 | MG678803 |
| Total clean reads | 8,783,602 | 24,899,740 | 22,334,862 | 14,157,482 | 19,869,478 |
| Number of contigs | 2540 | 1468 | 1190 | 1161 | 1749 |
| Contigs used for constructing the cp genome | 4 | 3 | 3 | 3 | 3 |
| N50 of contigs (bp) | 1612 | 369 | 1939 | 1966 | 1953 |
| Cp genome coverage (×) | 56.4 | 159.9 | 147.7 | 90.9 | 127.7 |
| Total cp DNA size (bp) | 155,761 | 155,769 | 151,214 | 155,684 | 155,653 |
| LSC size (bp) | 86,394 | 86,340 | 83,182 | 86,292 | 86,267 |
| IR size (bp) | 26,209 | 26,240 | 26,217 | 26,235 | 26,221 |
| SSC size (bp) | 16,949 | 16,949 | 15,598 | 16,922 | 16,944 |
| Total number of genes | 132 | 131 | 131 | 132 | 132 |
| Number of different protein-coding genes | 78 | 78 | 78 | 78 | 78 |
| Number of different tRNA genes | 30 | 30 | 29 | 30 | 30 |
| Number of different rRNA genes | 4 | 4 | 4 | 4 | 4 |
| Number of duplicated genes | 20 | 19 | 20 | 20 | 20 |
| Total number of pseudogenes | 2 | 1 | 2 | 2 | 2 |
| GC content (%) | 38.1 | 38.1 | 38.3 | 38.1 | 38.1 |
| GC content of LSC (%) | 36.2 | 36.2 | 36.4 | 36.2 | 36.2 |
| GC content of IR (%) | 43.0 | 43.0 | 42.9 | 43.0 | 43.0 |
| GC content of SSC (%) | 32.5 | 32.6 | 32.9 | 32.6 | 32.6 |
| Category | Grope of Genes | Name of Genes |
|---|---|---|
| Transcription and translation | Ribosomal proteins (LSU) | rpl2*(×2), rpl14, rpl16, rpl20, rpl22, rpl23(×2), rpl33, rpl36 |
| Ribosomal proteins (SSU) | rps2, rps3, rps4, rps7(×2), rps8, rps11, rps12**(×2), rps14, rps15, rps18, ψrps19 | |
| RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | |
| Translational initiation factor | infA | |
| rRNA genes | rrn16(×2), rrn23(×2), rrn4.5(×2), rrn5(×2) | |
| tRNA genes | trnA-UGC*(×2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC*, trnG-UCC, trnH-GUG, trnI-CAU(×2), trnI-GAU*(×2), trnK-UUU*, trnL-CAA(×2), trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU(×2), trnP-UGG, trnQ-UUG, trnR-ACG(×2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(×2), trnV-UAC*, trnW-CCA, trnY-GUA | |
| Photosynthesis | Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| NADH oxidoreductase | ndhA*, ndhB*(×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Cytochrome b6/f complex | petA*, petB*, petD, petG, petL, petN | |
| ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
| Rubisco large subunit | rbcL | |
| ATP-dependent protease subunit gene | clpP** | |
| Other genes | Maturase | matK |
| Envelop membrane protein | cemA | |
| Subunit Acetyl- CoA-Carboxylate | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Unknown | Conserved Open reading frames | ψycf1, ycf2(×2), ycf3**, ycf4, ycf15(×2) |
| No. | Size (bp) | Units | Type | Location Region |
|---|---|---|---|---|
| 1 | 30 | TAAAC(A)GGAA(G)AGAGAGGGATTCGAACCCTCG | F | IGS(psbI,trnS-GCU), IGS(psbC,trnS-UGA) |
| 2 | 52 | AGAAAAAGAATTGCAATAGCTAAATGG(A)TGA(G)TGA(C)GCAATATCGGTCAGCCATA | F | psaB(CDS),psaA(CDS) |
| 3 | 39 | CAGAACCGTACATGAGATTTTCACCTCATACGGCTCCTC | F | ycf3(intron), IGS(rps12,trnV-GAC) |
| 4 | 31 | CC(G)ATATTGATGATAGTGAC(G)GATATT(C)GATGA | F | ycf2(CDS) |
| 5 | 42 | TGGTTGTTCGCCGTTCAAGAATTCTTGAACGGCGAACAACCA | F | ycf15(CDS) |
| 6 | 31 | ATCATCG(A)ATATCC(G)TCACTATCATCAATATC(G)G | F | ycf2(CDS) |
| 7 | 32 | GAGATTTTATTTCG(A)AATTTGAAATAAAATCTC | P | IGS(psbI,trnS-GCU) |
| 8 | 30 | ACGGAAAGAGAGGGATTCGAACCCTCGGTA | P | IGS(psbI,trnS-GCU), IGS(trnS-GGA,rps4) |
| 9 | 30 | AA(C)GGAG(A)AGAGAGGGATTCGAACCCTCGA(G)TA | P | IGS(trnSUGA,psbZ), IGS(trnS-GGA,rps4) |
| 10 | 39 | CAGAACCGTACATGAGATTTTCACCTCATACGGCTCCTC | P | ycf3(intron), IGS(trnV-GAC,rps12) |
| 11 | 72 | GTAAGAATAAGAACTCAATGGACCTTGCCCCTCG(A)AATTT(C)GAGGGGCAAGGTCCATTGAGTTCTTATTCTTAC | P | IGS(petA,psbJ) |
| 12 | 48 | ATGTATCTAGGGACTAGTCGCTTC(G)C(G)AAGCGACTAGTCCCTAGATACAT | P | IGS(petD,rpoA) |
| 13 | 31 | CCATATTGATGATAGTGACGATATTGATGAT | P | ycf2(CDS) |
| 14 | 31 | CGATATTGATGATAGTGAGGATATCGATGAT | P | ycf2(CDS) |
| 15 | 42 | TGGTTGTTCGCCGTTCAAGAATTCTTGAACGGCGAACAACCA | P | ycf15(CDS) |
| 16 | 42 | TGGTTGTTCGCCGTTCAAGAATTCTTGAACGGCGAACAACCA | P | ycf15(CDS) |
| 17 | 38 | TACACATGAAGTAAAGAAA×2 | T | IGS(trnS-GCU,trnG-GCC) |
| 18 | 26 | TTTTATAGTTAAA×2 | T | clpP(intron) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, J.; Li, X.; Li, H.; Yang, J.; Wang, H.; He, J. Comparative Analysis of the Complete Chloroplast Genomes of Four Aconitum Medicinal Species. Molecules 2018, 23, 1015. https://doi.org/10.3390/molecules23051015
Meng J, Li X, Li H, Yang J, Wang H, He J. Comparative Analysis of the Complete Chloroplast Genomes of Four Aconitum Medicinal Species. Molecules. 2018; 23(5):1015. https://doi.org/10.3390/molecules23051015
Chicago/Turabian StyleMeng, Jing, Xuepei Li, Hongtao Li, Junbo Yang, Hong Wang, and Jun He. 2018. "Comparative Analysis of the Complete Chloroplast Genomes of Four Aconitum Medicinal Species" Molecules 23, no. 5: 1015. https://doi.org/10.3390/molecules23051015
APA StyleMeng, J., Li, X., Li, H., Yang, J., Wang, H., & He, J. (2018). Comparative Analysis of the Complete Chloroplast Genomes of Four Aconitum Medicinal Species. Molecules, 23(5), 1015. https://doi.org/10.3390/molecules23051015




