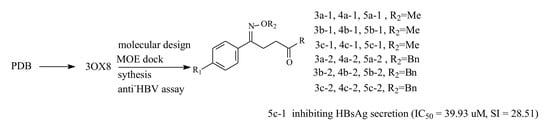
Discovery of Oxime Ethers as Hepatitis B Virus (HBV) Inhibitors by Docking, Screening and In Vitro Investigation
Abstract
1. Introduction
2. Results and Discussion
2.1. Molecular Docking Study
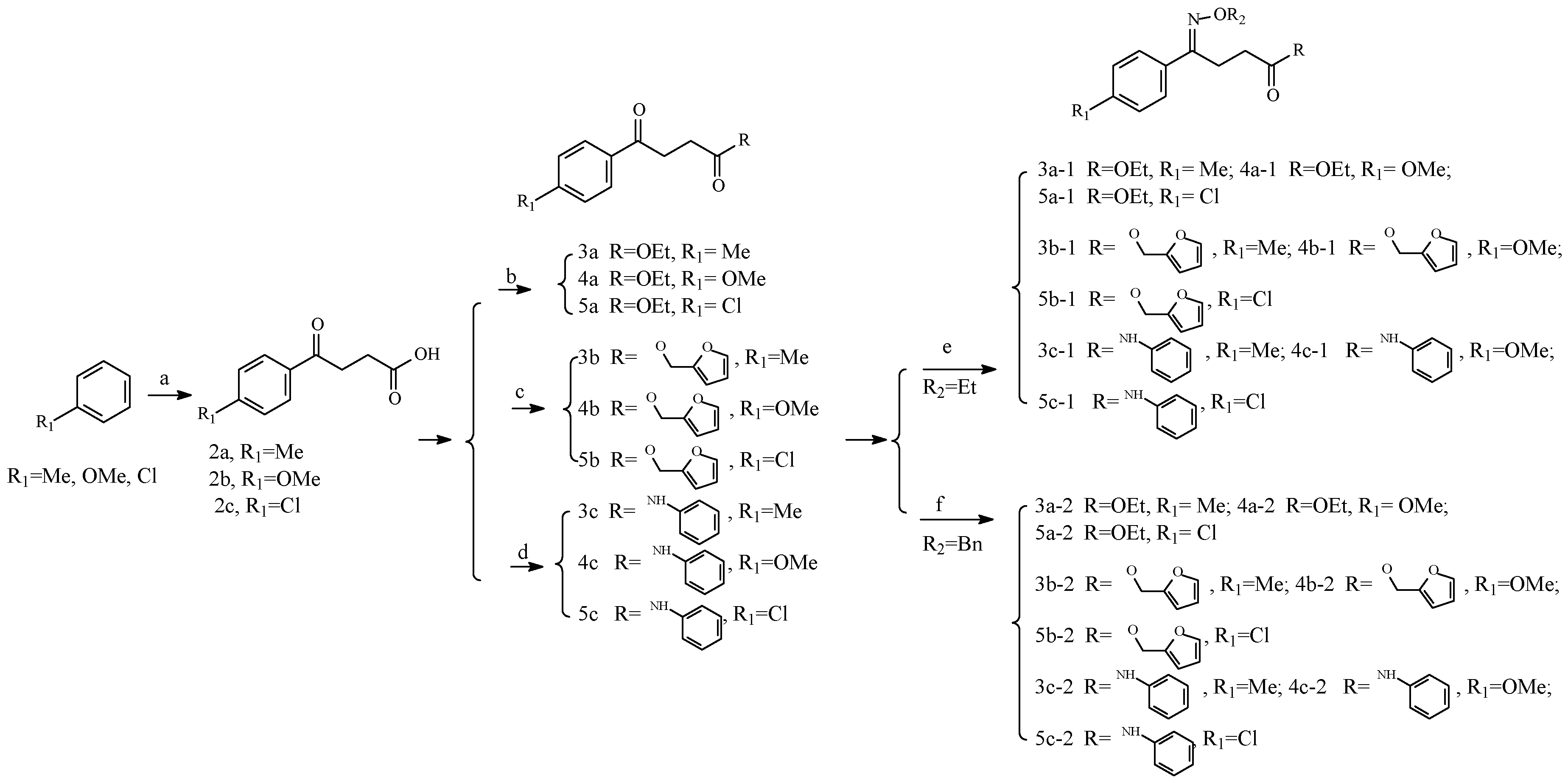
2.2. Chemistry
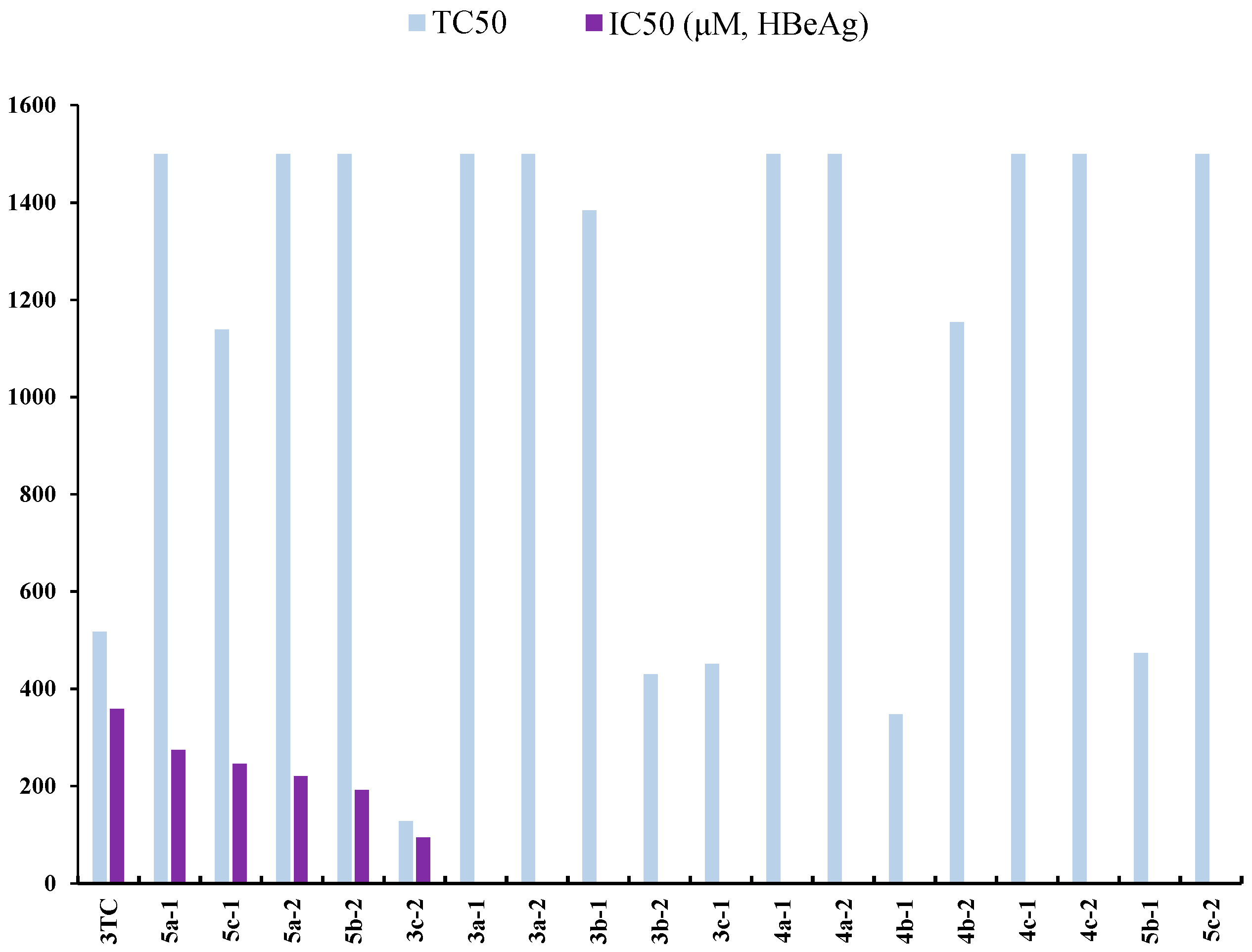
2.3. Anti-Hepatitis B Virus (HBV) Activity
2.4. Structure–Activity Relationship (SAR)
3. Methods
3.1. Molecular Docking
3.2. Synthesis Methods
3.2.1. Materials and Methods
3.2.2. Chemistry
3.3. Bio-Evaluation Methods
3.3.1. Cells and Cell Culture
3.3.2. Drug Treatment
3.3.3. Cell Toxicity
3.3.4. Method for HBsAg and HBeAg Inhibition Assays
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lok, A.S. Chronic hepatitis B. N. Engl. J. Med. 2002, 346, 1682–1683. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Hepatitis B Fact Sheet N°204; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Zhang, E.; Kosinska, A.; Lu, M.; Yan, H.; Roggendorf, M. Current status of immunomodulatory therapy in chronic hepatitis B, fifty years after discovery of the virus: Search for the “magic bullet” to kill cccDNA. Antivir. Res. 2015, 123, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Block, T.M.; Rawat, S.; Brosgart, C.L. Chronic hepatitis B: A wave of new therapies on the horizon. Antivir. Res. 2015, 121, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Scaglione, S.J.; Lok, A.S.F. Effectiveness of hepatitis B treatment in clinical practice. Gastroenterology 2012, 142, 1360–1368. [Google Scholar] [CrossRef] [PubMed]
- Clercq, E.D.; Férir, G.; Kaptein, S.; Neyts, J. Antiviral treatment of chronic hepatitis B virus (HBV) infections. Viruses 2010, 2, 1279–1305. [Google Scholar] [CrossRef] [PubMed]
- Pradere, U.; Garnier-Amblard, E.C.; Coats, S.J.; Amblard, F.; Schinazi, R.F. Synthesis of nucleoside phosphate and phosphonate prodrugs. Chem. Rev. 2014, 114, 9154–9218. [Google Scholar] [CrossRef] [PubMed]
- Chotiyaputta, W.; Lok, A.S. Hepatitis B virus variants. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Cento, V.; Hemert, F.V.; Neumann-Fraune, M.; Mirabelli, C.; Di Maio, V.C.; Salpini, R.; Bertoli, A.; Micheli, V.; Gubertini, G.; Romano, S.; et al. Anti-HBV treatment induces novel reverse transcriptase mutations with reflective effect on HBV S antigen. J. Infect. 2013, 67, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.K.; Cheung, A.M.; O'Rourke, K.; Naylor, C.D.; Detsky, A.S.; Heathcote, J. Effect of alpha-interferon treatment in patients with hepatitis B e antigen-positive chronic hepatitis B: A meta-analysis. Ann. Intern. Med. 1993, 119, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Perrillo, R.P.; Schiff, E.R.; Davis, G.L.; Bodenheimer, H.C., Jr.; Lindsay, K.; Payne, J.; Dienstag, J.L.; O’Brien, C.; Tamburro, C.; van Lie, R.A.W.; et al. A randomized, controlled trial of interferon alfa-2b alone and after prednisone withdrawal for the treatment of chronic hepatitis B. N. Engl. J. Med. 1990, 323, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Dusheiko, G.; Bertoletti, A. Resistance to lamivudine therapy: Is there more than meets the eye. Gut 2011, 54, 4–6. [Google Scholar]
- Naveen, C.S.; Shakya, N.; Mak, M.; Agrawal, B.; Kumar, R. Antiviral Activity of Various 1-(2’-Deoxy-β-d-lyxofuranosyl), 1-(2’-Fluoro-β-d-xylofuranosyl)1-(3’-Fluoro-β-d-arabino- furanosyl), and 2’-Fluoro-2’,3’-didehydro-2’,3’-dideoxyribose Pyrimidine Nucleoside Analogues against Duck Hepatitis B Virus (DHBV) and Human Hepatitis B Virus (HBV) Replication. J. Med. Chem. 2010, 53, 7156–7166. [Google Scholar]
- Lv, Z.L.; Sheng, C.Q.; Wang, T.T.; Zhang, Y.K.; Liu, J.; Feng, J.L.; Sun, H.L.; Zhong, H.Y.; Niu, C.J.; Li, K. Design, Synthesis, and Antihepatitis B Virus Activities of Novel 2-Pyridone Derivatives. J. Med. Chem. 2010, 53, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Matthias, D.; Karen, M.; Rob, D.V.; Tim, H.M.J.; Géry, D.; Norbert de, K. Design, Synthesis, and Antiviral Evaluation of Purine-β-lactam and Purine-aminopropanol Hybrids. J. Med. Chem. 2012, 55, 5637–5641. [Google Scholar]
- Lv, Z.G.; He, W.; Tian, X.H.; Kang, J.F.; Liu, Y.X.; Yu, W.Q.; Peng, Y.M.; Zheng, L.Y.; Wang, Q.D.; Yu, W.Q.; et al. Design, synthesis, and biological evaluation of new N4-Substituted 20-deoxy-20-fluoro-40-azido cytidine derivatives as potent anti-HBV Agents. Eur. J. Med. Chem. 2015, 101, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Imoto, S.; Kohgo, S.; Tokuda, R.; Kumamoto, H.; Aoki, M.; Amano, M.; Kuwata-Higashi, N.; Mitsuya, H.; Haraguchi, K. Design, Synthesis, and Evaluation of Anti-HBV Activity of Hybrid Molecules of Entecavir and Adefovir: Exomethylene Acycloguanine Nucleosides and Their Monophosphate Derivatives. Nuclioside Nucleotides Nucleic Acid 2015, 34, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Xu, X.Q.; Guan, H.; Wang, L.L.; Wu, Q.; Zhao, G.M.; Li, S. A new series of HAPs as anti-HBV agents targeting at capsid assembly. Bioorg. Med. Chem. Lett. 2014, 24, 4247–4249. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.M.; Han, Y.X.; Wang, Y.P.; Li, Y.H.; Shan, Y.Q.; Li, X.; Peng, Z.G.; Bi, C.W.; Zhang, T.; Du, N.N.; et al. Design and Synthesis of Oxymatrine Analogues Overcoming Drug Resistance in Hepatitis B Virus through Targeting Host Heat Stress Cognate 70. J. Med. Chem. 2011, 54, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.J.; Chuang, H.; Liang, Y.C.; Lin, H.H.; Horng, J.C.; Kuo, Y.C.; Chen, C.W.; Tsai, F.Y.; Yen, S.C.; Chou, S.C.; et al. Design, Synthesis, and Bioevaluation of Paeonol Derivatives as Potential Anti-HBV Agents. Eur. J. Med. Chem. 2015, 90, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.J.; Chen, H.; Ma, Y.B.; Huang, X.Y.; Chen, J.J.; Geng, C.A.; Zhang, X.M.; Chen, J.J. Design, Synthesis and Biological Evaluation of Caudatin Analogs as Potent Hepatitis B Virus Inhibitors. Med. Chem. 2015, 11, 165–179. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lv, J.J.; Yu, S.; Wang, Y.F.; Wang, D.; Xu, M.; Zhang, Y.J.; van Lie, R.A.W. Anti-Hepatitis B Virus Norbisabolane Sesquiterpenoids from Phyllanthus acidus and the Establishment of Their Absolute Configurations Using Theoretical Calculations. J. Org. Chem. 2014, 79, 5432–5447. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.H.; Cheng, P.; Jiang, Z.Y.; Ma, Y.B.; Zhang, X.M.; Zhang, F.X.; Yang, L.M.; Zheng, Y.T.; Chen, J.J. Periglaucines A−D, Anti−HBV and −HIV−1 Alkaloids from Pericampylus glaucus. J. Nat. Prod. 2008, 71, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Kortagere, S.; Xu, J.P.; Mankowski, M.K.; Ptak, R.G.; Cocklin, S. Structure−Activity Relationships of a Novel Capsid Targeted Inhibitor of HIV-1 Replication. J. Chem. Inf. Model. 2014, 54, 3080–3090. [Google Scholar] [CrossRef] [PubMed]
- Niet, D.A.; Stelma, F.; Jansen, L.; Sinnige, M.J.; Remmerswaal, E.B.M.; Takkenberg, R.B.; Kootstra, N.A.; Reedink, H.W. Restoration of T cell function in chronic hepatitis B patients upon treatment with interferon based combination therapy. J. Hepatology 2015, 64, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, Y.; Zhao, B.; Deng, M.M.; Liu, J.; Li, X.; Hou, J.W.; Gui, M.M.; Zhang, S.J.; Li, X.D.; et al. A new unconventional HLA-A2-restricted epitope from HBV core protein elicits antiviral cytotoxic T lymphocytes. Protein Cell 2014, 5, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Chu, Y.W.; Zhang, R.H.; Xu, H.B.; Wang, Y.; Xiong, S.D. Endoplasmic reticulum targeting sequence enhances HBV-specific cytotoxic T lymphocytes induced by a CTL epitope-based DNA vaccine. Virology 2005, 334, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.B.; Liu, H.; Shi, S.; Zou, L.B.; Zou, Z.H. A novel dendritic-cell-targeting DNA vaccine for hepatitis B induces T cell and humoral immune responses and potentiates the antivirus activity in HBV transgenic mice. Immunol. Lett. 2015, 168, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Q.; Ge, J.; Ren, S.L.; Zhou, T.; Sun, Y.; Sun, H.L. Hepatitis B surface antigen (HBsAg) and core antigen (HBcAg) combine CpG oligodeoxynucletides as a novel therapeutic vaccine for chronic hepatitis B infection. Vaccine 2015, 33, 4247–4254. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wei, W.X.; Shi, K.C.; Cao, X.; Zhou, M.; Liu, Z.P. In vitro and in vivo anti-hepatitis B virus activities of the lignan niranthin isolated from Phyllanthus niruri L. J. Ethnopharmacol. 2014, 155, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wei, W.X.; Li, Y.B.; Lin, X.; Shi, K.C.; Cao, X.; Zhou, M. In vitro and in vivo anti-hepatitis B virus activities of the lignin nirtetralin B isolated from Phyllanthus niruri L. J. Ethnopharmacol. 2014, 157, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.X.; Li, X.R.; Wang, K.W.; Zheng, Z.W.; Zhou, M. Lignans with anti-hepatitis B virus activities from Phyllanthus niruri L. Phytother. Res. 2012, 26, 964–968. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wei, W.X.; Li, Y.B.; Liu, X.; Cao, X.J.; Lei, K.C.; Zhou, M. Design, synthesis, biological evaluation and molecular docking studies of phenylpropanoid derivatives as potent anti-hepatitis B virus agents. Eur. J. Med. Chem. 2015, 95, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, Y.B.; Wei, W.X.; Wei, J.C. Synthesis and biological evaluation of phenylpropanoid derivatives. Med. Chem. Res. 2016, 25, 1074–1086. [Google Scholar] [CrossRef]
- Liu, S.; Li, Y.B.; Wei, W.X.; Wang, K.W.; Wang, L.S.; Wang, J.Y. Design, Synthesis, Molecular Docking Studies and Anti-HBV Activity of Phenylpropanoid Derivatives. Chem. Biol. Interact. 2016, 251, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Imoto, H.; Sugiyama, Y.; Kimura, H.; Momose, Y. Studies on Non-Thiazolidinedion Antidiabetic Agents. Novel Oxyiminoalkanoic Acid derivatives as Potent Glucose and LiPid Lowering Agents. Chem. Pharm. Bull. 2003, 51, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.W.; Tan, J.; Zhou, M. Anti-Hepatitis B Virus Activitive N-Phenyl-4-phenyl Butylamidoxime and Its Derivatives. CN106518727A, 2 November 2016. [Google Scholar]
- Howe, N.J.; Blades, K.; Lamont, G.M. Synthesis of the Novel Tetrahydropyrazolo[3,4-c]pyridin-5-one Scaffold. Synlett 2015, 26, 228–232. [Google Scholar] [CrossRef]
- Alfonsi, M.; Arcadi, A.; Chiarini, M. Sequential Rhodium-Catalyzed Stereo- and Regioselective Addition of Organoboron Derivatives to the Alkyl 4-Hydroxy-2-Alkynoates/Lactonizaction Reaction. J. Med. Chem. 2007, 72, 9510–9517. [Google Scholar] [CrossRef] [PubMed]
- Mahmoodi, N.O.; Malekroudi, R.Y. Synthesis of Chiral 5-Aryltetrahydrofuran-2-ones via Yeast Bioreduction of γ-Keto Acids and Their Esters. Rus. J. Med. Chem. 2006, 42, 365–368. [Google Scholar] [CrossRef]
- Starodubtseva, E.V.; Turova, O.V.; Vinogradov, M.G.; Gorshkova, L.S.; Struchkova, M.I. A convenient route to chiral γ-lactones via asymmetric hydrogenation of γ-ketoesters using the RuCl3–BINAP–HCl catalytic system. Tetrahedron 2008, 64, 11713–11717. [Google Scholar] [CrossRef]
- Li, W.H.; Zhang, L.F.; Liu, W. Synthesis and antifungal activity of 2-carboxamide-4-aryl-1,5-benzothiazepines. Chin. J. Org. Chem. 2011, 31, 1252–1257. [Google Scholar]
- Candeloro, V.; Bowie, J.H. 1H-1-benzazepines. The reactions of levulinic acid and β-benzoylpropionic acid with aniline and methoxyanilines. Aust. J. Chem. 1978, 31, 2031–2037. [Google Scholar] [CrossRef]
- Liu, J.X.; Chen, K.Y.; Ren, E.C. Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines. Eur. J. Immunol. 2011, 41, 2097–2106. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.Q.; Huang, Z.M.; Yang, X.B.; Liu, H.Z.; Wu, G.X. In vivo and in vitro antihepatitis B virus activity of total phenolics from Oenanthe javanica. J. Ethnopharmacol. 2008, 118, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Sells, M.A.; Chen, M.L.; Acs, G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. USA 1987, 84, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the all compounds are available from the authors. |

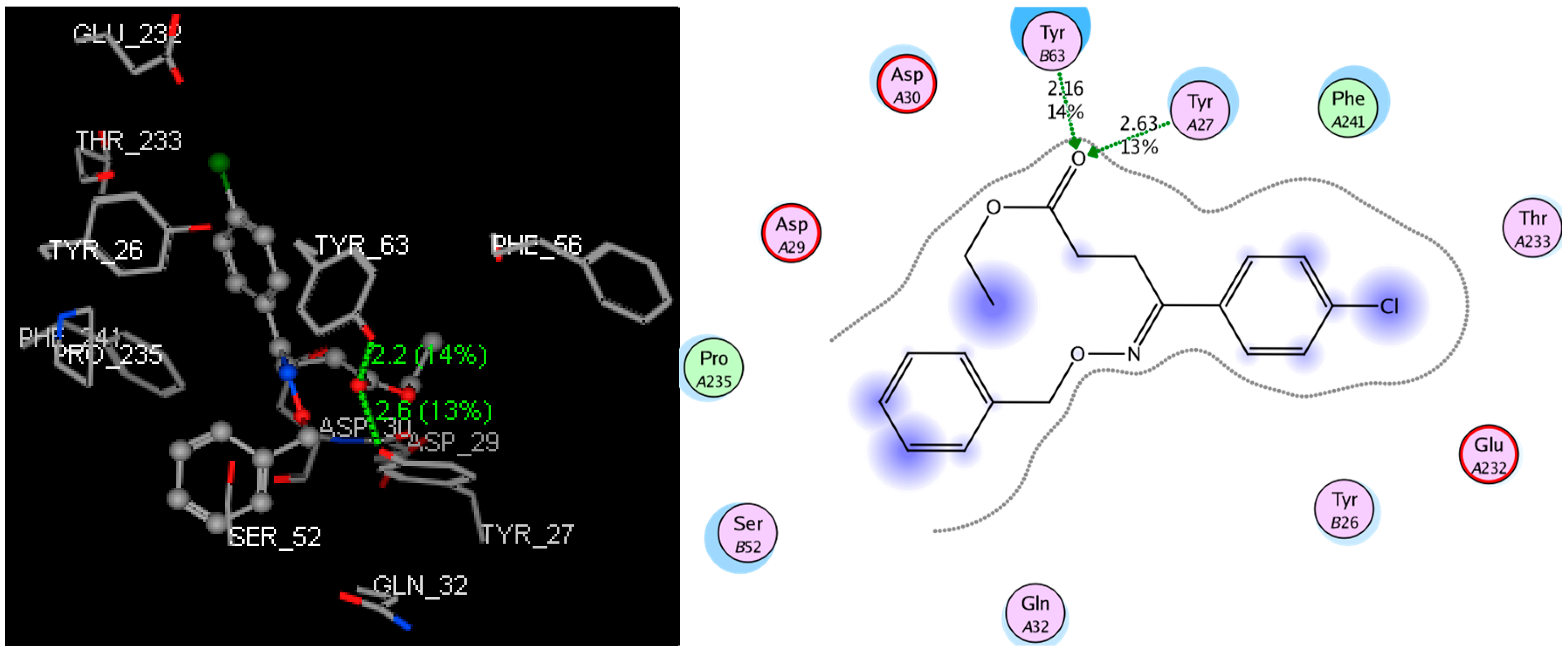



| Compound | S-Score (kcal/mol) | Distance (Å) | Residue | Site of Actions |
|---|---|---|---|---|
| 3a-1 | −9.1113 | 2.72 | Tyr27 | N of -C=N- |
| 3a-2 | −9.7265 | 2.93 | Tyr27 | N of -C=N- |
| 3b-1 | −10.4709 | 2.94 | Tyr26 | N of -C=N- |
| 3b-2 | −11.2203 | 2.53 | Tyr26 | O of -C=O |
| 2.83 | Tyr27 | N of -C=N- | ||
| 3c-1 | −9.1497 | 3.09 | Tyr63 | N of -C=N- |
| 3c-2 | −10.0032 | 2.63 | Tyr27 | O of -C=O |
| 2.16 | Tyr63 | O of -C=O | ||
| 4a-1 | −9.9143 | 3.01 | Tyr26 | N of -C=N- |
| 4a-2 | −11.0692 | 2.94 | Tyr63 | O of -C=O |
| 4b-1 | −10.4782 | 2.82 | Tyr26 | N of -C=N- |
| 4b-2 | −10.8746 | 2.82 | Tyr63 | N of -C=N- |
| 4c-1 | −10.0261 | 3.01 | Tyr27 | N of -C=N- |
| 4c-2 | −10.3516 | 2.81 | Tyr27 | N of -C=N- |
| 2.73 | Tyr63 | N of -C=N- | ||
| 5a-1 | −9.9471 | 2.40 | Tyr26 | N of -C=N- |
| 5a-2 | −10.3332 | 1.96 | Asp30 | H of -N-H |
| 2.52 | Tyr63 | N of -C=N- | ||
| 5b-1 | −10.2883 | 2.75 | Tyr63 | N of -C=N- |
| 5b-2 | −12.0185 | 2.66 | Tyr63 | O of -C=O |
| 5c-1 | −9.9406 | 2.50 | Tyr27 | O of -C=O |
| 2.44 | Tyr63 | O of -C=O | ||
| 5c-2 | −11.2609 | 2.92 | Tyr63 | N of -C=N- |
| Compound | TC50 a (μM) | HBsAg b | HBeAg c | ||
|---|---|---|---|---|---|
| IC50 d (μM) | SI e | IC50 d (μM) | SI e | ||
| 3a-1 | >1500 | 292.73 | >5.12 | - f | - |
| 3a-2 | >1500 | - | - | - | - |
| 3b-1 | 1383.74 | - | - | - | - |
| 3b-2 | 429.88 | 86.85 | 4.95 | - | - |
| 3c-1 | 450.67 | 224.82 | 2.00 | - | - |
| 3c-2 | 127.10 | 94.71 | 1.34 | 93.91 | 1.34 |
| 4a-1 | >1500 | 154.50 | >9.71 | - | - |
| 4a-2 | >1500 | - | - | - | - |
| 4b-1 | 347.55 | 127.68 | 2.72 | - | - |
| 4b-2 | 1153.49 | 233.60 | 4.94 | - | - |
| 4c-1 | >1500 | - | - | - | - |
| 4c-2 | >1500 | - | - | - | - |
| 5a-1 | >1500 | 74.92 | >20.02 | 273.87 | >5.48 |
| 5a-2 | >1500 | 156.27 | >9.60 | 220.09 | >6.82 |
| 5b-1 | 473.25 | 207.63 | 2.28 | - | - |
| 5b-2 | >1500 | 101.19 | >14.82 | 191.58 | >7.83 |
| 5c-1 | 1138.45 | 39.93 | 28.51 | 245.96 | 4.63 |
| 5c-1 | >1500 | - | - | - | - |
| 3TC g | 517.40 | 290.73 | 1.78 | 358.59 | 1.44 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, J.; Zhou, M.; Cui, X.; Wei, Z.; Wei, W. Discovery of Oxime Ethers as Hepatitis B Virus (HBV) Inhibitors by Docking, Screening and In Vitro Investigation. Molecules 2018, 23, 637. https://doi.org/10.3390/molecules23030637
Tan J, Zhou M, Cui X, Wei Z, Wei W. Discovery of Oxime Ethers as Hepatitis B Virus (HBV) Inhibitors by Docking, Screening and In Vitro Investigation. Molecules. 2018; 23(3):637. https://doi.org/10.3390/molecules23030637
Chicago/Turabian StyleTan, Jie, Min Zhou, Xinhua Cui, Zhuocai Wei, and Wanxing Wei. 2018. "Discovery of Oxime Ethers as Hepatitis B Virus (HBV) Inhibitors by Docking, Screening and In Vitro Investigation" Molecules 23, no. 3: 637. https://doi.org/10.3390/molecules23030637
APA StyleTan, J., Zhou, M., Cui, X., Wei, Z., & Wei, W. (2018). Discovery of Oxime Ethers as Hepatitis B Virus (HBV) Inhibitors by Docking, Screening and In Vitro Investigation. Molecules, 23(3), 637. https://doi.org/10.3390/molecules23030637