Interaction of Arylidenechromanone/Flavanone Derivatives with Biological Macromolecules Studied as Human Serum Albumin Binding, Cytotoxic Effect, Biocompatibility Towards Red Blood Cells
Abstract
:1. Introduction
2. Results and Discussion
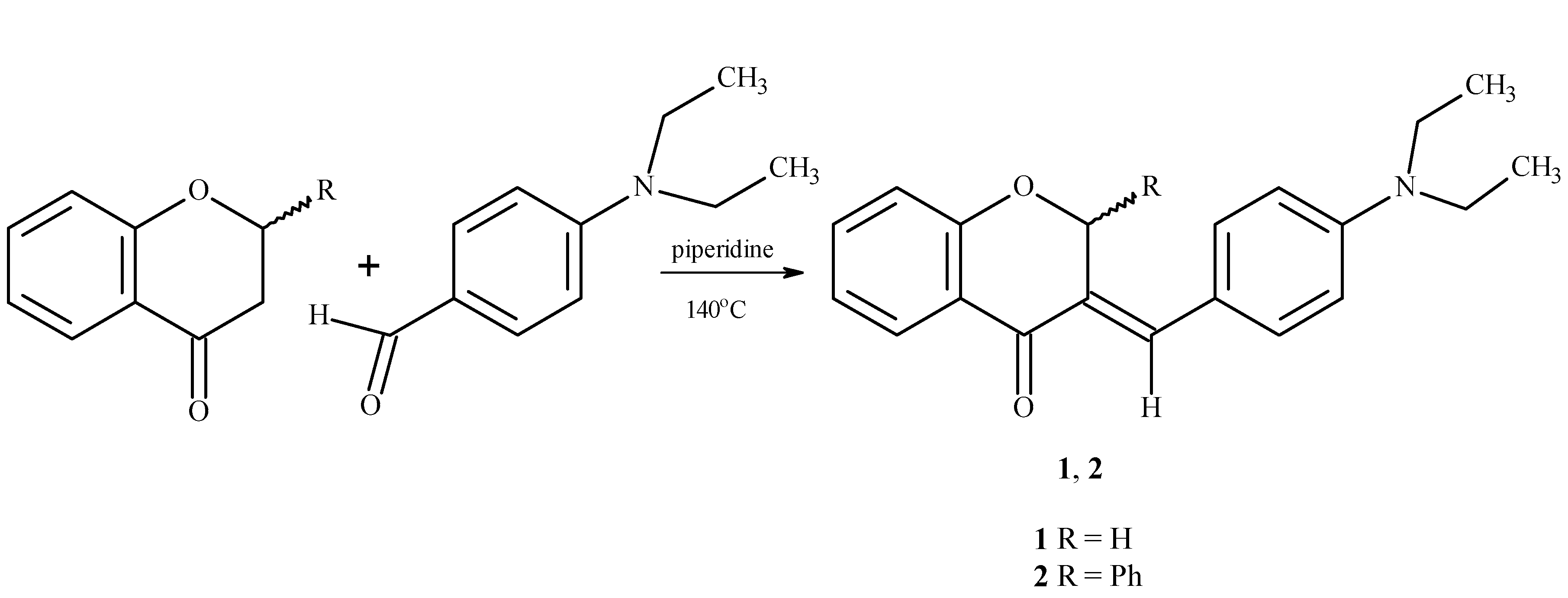
2.1. Chemistry
2.2. Molecular Structures
2.3. Biological Assays
2.3.1. Cytotoxic Activity
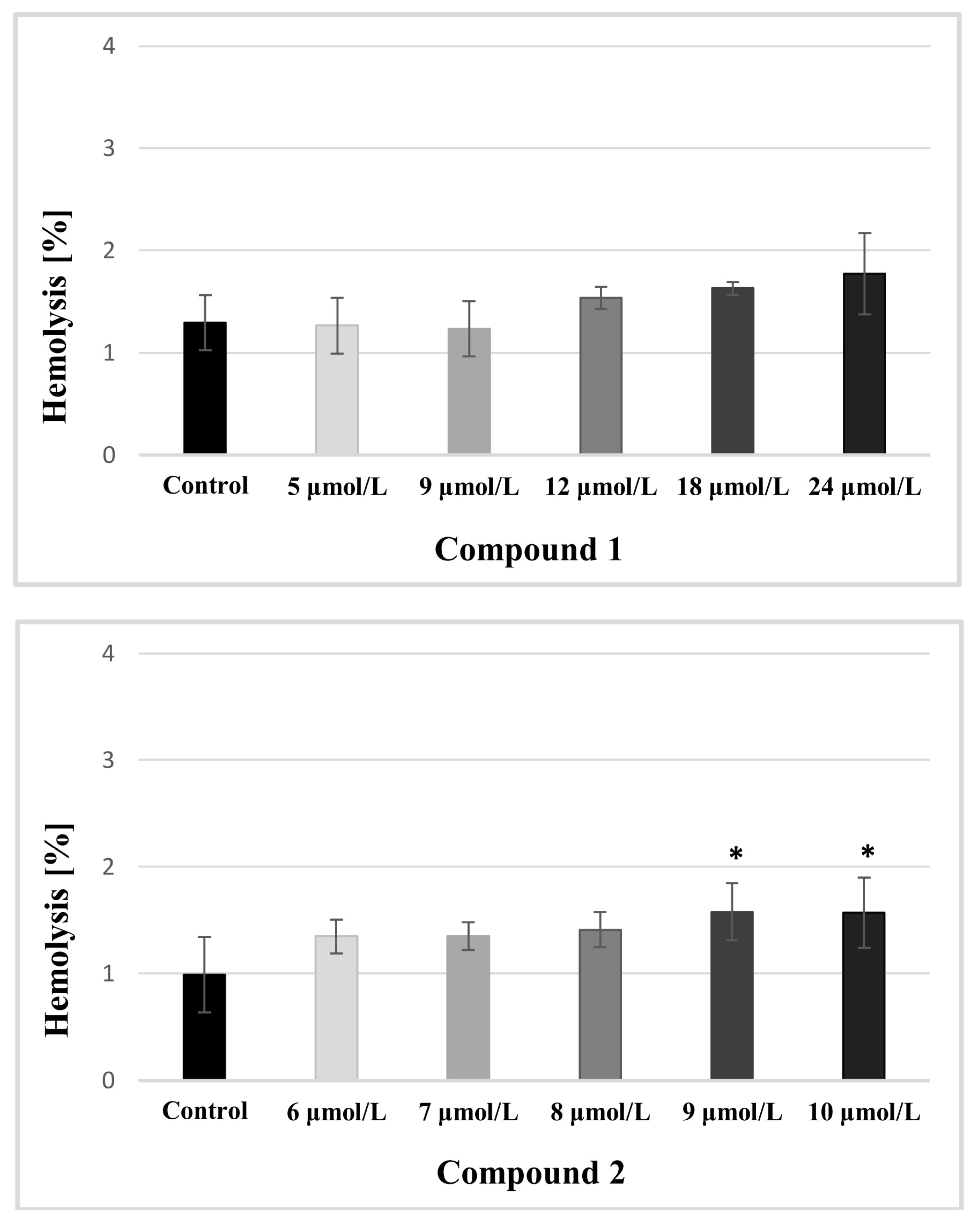
2.3.2. Rbcs Lysis Assay
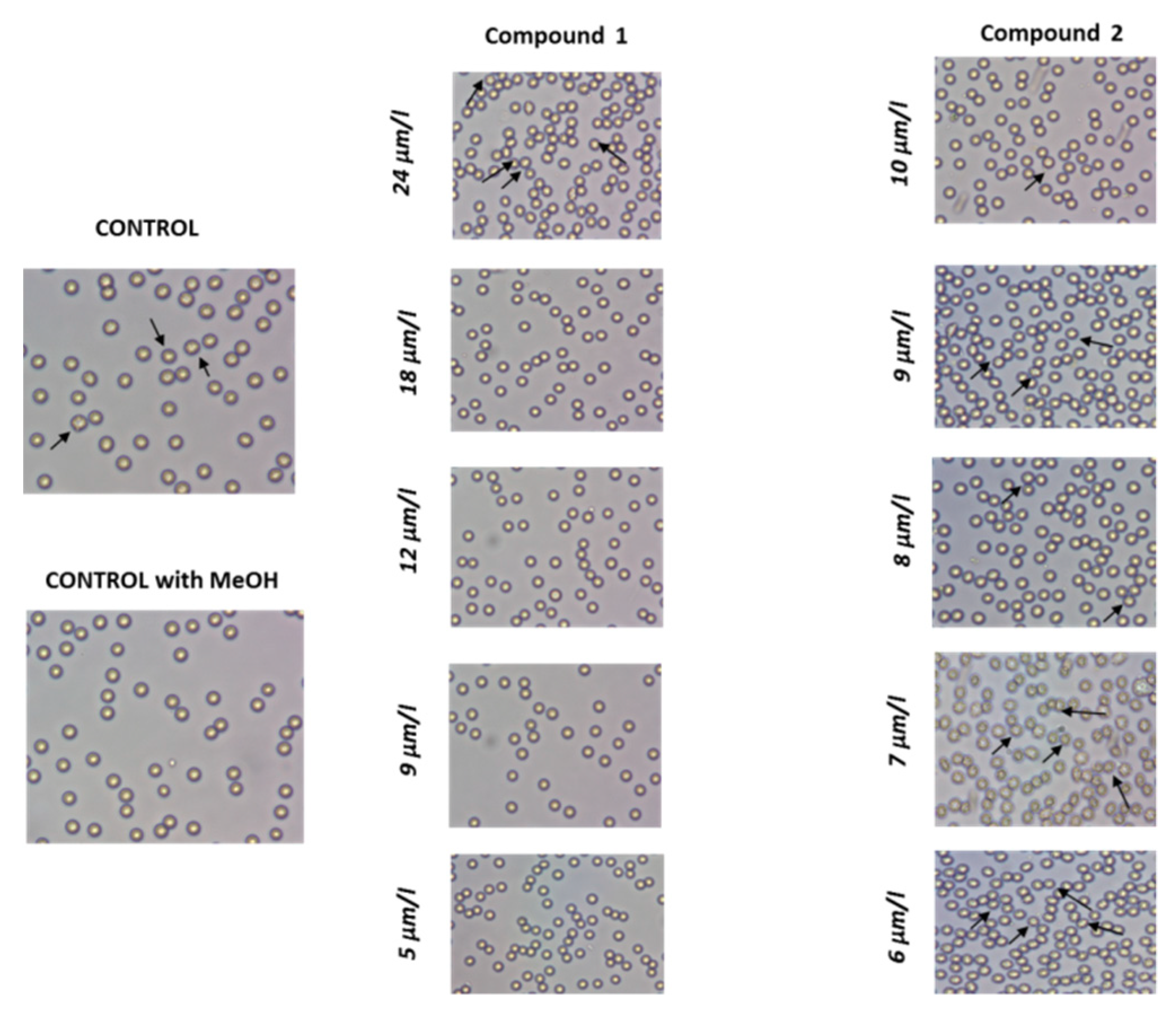
2.3.3. RBCs Morphology
2.4. Computational Studies
3. Materials and Methods
3.1. Synthesis of (E)-3-(4-N,N-diethylaminobenzylidene)chroman-4-one (1)
3.2. Synthesis of (E)-3-(4-N,N-diethylaminobenzylidene)-2-phenylchroman-4-one (2)
3.3. Determination of Lipophilicity of Flavone Derivatives Using RP-TLC Method
3.4. X-ray Diffraction Experiment
3.5. Cells Cultures and Cytotoxicity Assay by MTT
3.6. Red Blood Cells Lysis Assay
3.7. RBCs Morphology
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Czaplińska, M.; Czepas, J.; Gwoździński, K. Budowa, właściwości przeciwutleniające i przeciwnowotworowe flawonoidów. Post. Bioch. 2012, 58, 235–242. [Google Scholar]
- Nijveldt, R.J.; Van Nood, E.; Van Hoorn, D.E.; Boelens, P.G.; Van Norren, K.; Van Leeuwen, P.A. Flavonoids: A review of propable mechanisms of action and potential applications. Am. J. Clin. Nutr. 2001, 74, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Ravishankar, D.; Rajora, A.K.; Greco, F.; Osborn, H.M. Flavonoids as prospective compounds for anti-cancer therapy. Int. J. Biochem. Cell. Biol. 2013, 45, 2821–2831. [Google Scholar] [CrossRef] [PubMed]
- Raffa, D.; Maggio, B.; Raimondi, M.V.; Plescia, F.; Daidone, G. Recent discoveries of anticancer flavonoids. Eur. J. Med. Chem. 2017, 142, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Perez-Vizcaino, F.; Fraga, C.G. Research trends in flavonoids and health. Arch. Biochem. Biophys. 2018, 646, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Sharmila, G.; Bhat, F.A.; Arunkumar, R.; Elumalai, P.; Singh, P.R.; Senthilkumar, K.; Arunakaran, J. Chemopreventive effect of quercetin, a natural dietary flavonoid on prostate cancer in in vivo model. J. Clin. Nutr. 2014, 33, 718–726. [Google Scholar] [CrossRef] [PubMed]
- Demirayak, S.; Yurttas, L.; Gundogdu-Karaburun, N.; Karaburun, A.C.; Kayagil, I. New chroman-4-one/thiochroman-4-one derivatives as potential anticancer agents. Saudi Pharm. J. 2017, 25, 1063–1072. [Google Scholar] [CrossRef]
- Perkin, W.H.; Ray, J.N.; Robinson, R. Experiments on the synthesis of brazilin and hœmatoxylin and their derivatives. Part I. Veratrylidene-7-methoxychromanone and an account of a new synthesis of some benzopyrylium salts. J. Chem. Soc. 1926, 129, 941–953. [Google Scholar] [CrossRef]
- Levai, A.; Schag, J.B. Synthesis of 3-benzylidenechroman-4-ones and -1-thiochroman-4-ones. Pharmazie. 1979, 34, 748–749. [Google Scholar]
- Levai, A.; Dinya, Z.; Schag, J.B.; Toth, G.; Szollosy, A. Synthesis of 3-benzyl-4-chromones and 3-benzyl-1-thio-4-chromones. Chem. Inf. 1981, 36, 465. [Google Scholar]
- Kupcewicz, B.; Balcerowska-Czerniak, G.; Małecka, M.; Paneth, P.; Krajewska, U.; Rozalski, M. Structure-cytotoxic activity relationship of 3-arylideneflavanone and chromanone (E, Z isomers) and 3-arylflavones. Bioorganic Med. Chem. Letters. 2013, 23, 4102–4106. [Google Scholar] [CrossRef] [PubMed]
- Budzisz, E.; Paneth, P.; Geromino, I.; Muzioł, T.; Rozalski, M.; Krajewska, U.; Pipiak, P.; Ponczek, M.B.; Malecka, M.; Kupcewicz, B. The cytotoxic effect of spiroflavanone derivatives, their binding ability to human serum albumin (HSA) and a DFT study on the mechanism of their synthesis. J. Mol. Struct. 1137, 267–276. [Google Scholar] [CrossRef]
- Teng, G.G. Negative lase sensitive lithographic printing plate having specific photosensitive composition. U.S. Patent 7,524,615, 14 February 2008. [Google Scholar]
- Pijewska, L.; Kamecki, J.; Perka-Karolczak, W. 3-Arylideneflavanones. Part 2 Reaction with diazomethane. Pharmazie 1993, 48, 254–257. [Google Scholar]
- Fasano, M.; Curry, S.; Terreno, E.; Galliano, M.; Narciso, G.P.; Notari, S.; Ascenzi, P. The extraordinary ligand binding properties of human serum albumin. IUBMB Life. 2005, 57, 787–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gangwal, R.P.; Damre, M.V.; Das, N.R.; Dhoke, G.V.; Bhadauriya, A.; Varikoti, R.A.; Sangamwar, A.T. Structure based virtual screening to identify selective phosphodiesterase 4B inhibitors. J. Mol. Graph. Model 2015, 57, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Valkonen, A.; Laihia, K.; Kolehmainen, E.; Kauppinen, R.; Perjesi, P. Structural studies of seven homoisoflavonoids, six thiohomoisoflavonoids and four structurally related compounds. Struct. Chem. 2012, 23, 209–217. [Google Scholar] [CrossRef]
- Cremer, D.; Pople, J.A. General definition of ring puckering coordinates. J. Amer. Chem. Soc. 1975, 97, 1354–1358. [Google Scholar] [CrossRef]
- Bhattacharya, A.A.; Grune, T.; Curry, S. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin. J. Mol. Biol. 2000, 303, 721–732. [Google Scholar] [CrossRef]
- Duax, W.L.; Norton, D.A. Atlas of Steroid Structure; Springer Science & Business Media: New York, NY, USA, 2012; pp. 3273–3274. [Google Scholar]
- Kupcewicz, B.; Małecka, M.; Zapadka, M.; Krajewska, U.; Różalski, M.; Budzisz, E. Quantitative relationships between structure and cytotoxic activity of flavonoid derivatives. An application of Hirshfeld surface derived descriptors. Bioorg. Med. Chem. Lett. 2016, 26, 3336–3341. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Perjesi, P.; Umashankar, D.; De Clercg, E.; Balzarini, J.; Kawase, M.; Sakagami, H.; Stables, J.; Lorand, T.; Rozmer, Z.; Dimmock, J. Design, synthesis and antiproliferative activity of some 3-benzylidene-2,3-dihydro-1-benzopyran-4-ones which display selective toxicity for malignant cells. Eur. J. Med. Chem. 2008, 43, 839–845. [Google Scholar] [CrossRef] [Green Version]
- Fischer, D.; Li, Y.; Ahlemeyer, B.; Krieglestein, J. In vitro cytotoxicity testing of polycations: Influence of polymer structure on cell viability and hemolysis. Biomaterials 2003, 24, 1121–1131. [Google Scholar] [CrossRef]
- Stasiuk, M.; Kijanka, G.; Kozubek, A. Zmiany kształtu erytrocytów i czynniki je wywołujące. Post. biochemii. 2009, 55, 425–433. [Google Scholar]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Cryst. 2009, 65, 148–155. [Google Scholar] [CrossRef] [Green Version]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; van de Streek, J.; Wood, P.A. New Features for the Visualization and Investigation of Crystal Structures. J. App. Cryst. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Hansen, M.B.; Nielsen, S.E.; Berg, K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J. Immunol. Methods. 1989, 119, 203–210. [Google Scholar] [CrossRef]
- Markowicz-Piasecka, M.; Sikora, J.; Mateusiak, Ł.; Mikiciuk-Olasik, E.; Huttunen, K.M. New prodrugs of metformin do not influence the overall haemostasis potential and integrity of the erythrocyte membrane. Eur. J. Pharm. 2017, 811, 208–221. [Google Scholar] [CrossRef]
- Markowicz-Piasecka, M.; Mikiciuk-Olasik, E.; SIkora, J. Stability of erythrocyte membrane and overall hemostasis potential—A biocompatibility study of mebrofenin and other iminodiacetic acid derivatives. J. Pharm. Rep. 2015, 67, 1230–1239. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds (E)-3-(4-N,N-diethylaminobenzylidene)chroman-4-one (1) and (E)-3-(4-N,N-diethylaminobenzylidene)-2-phenylchroman-4-one (2) are available from the author Angelika A. Adamus-Grabicka. |

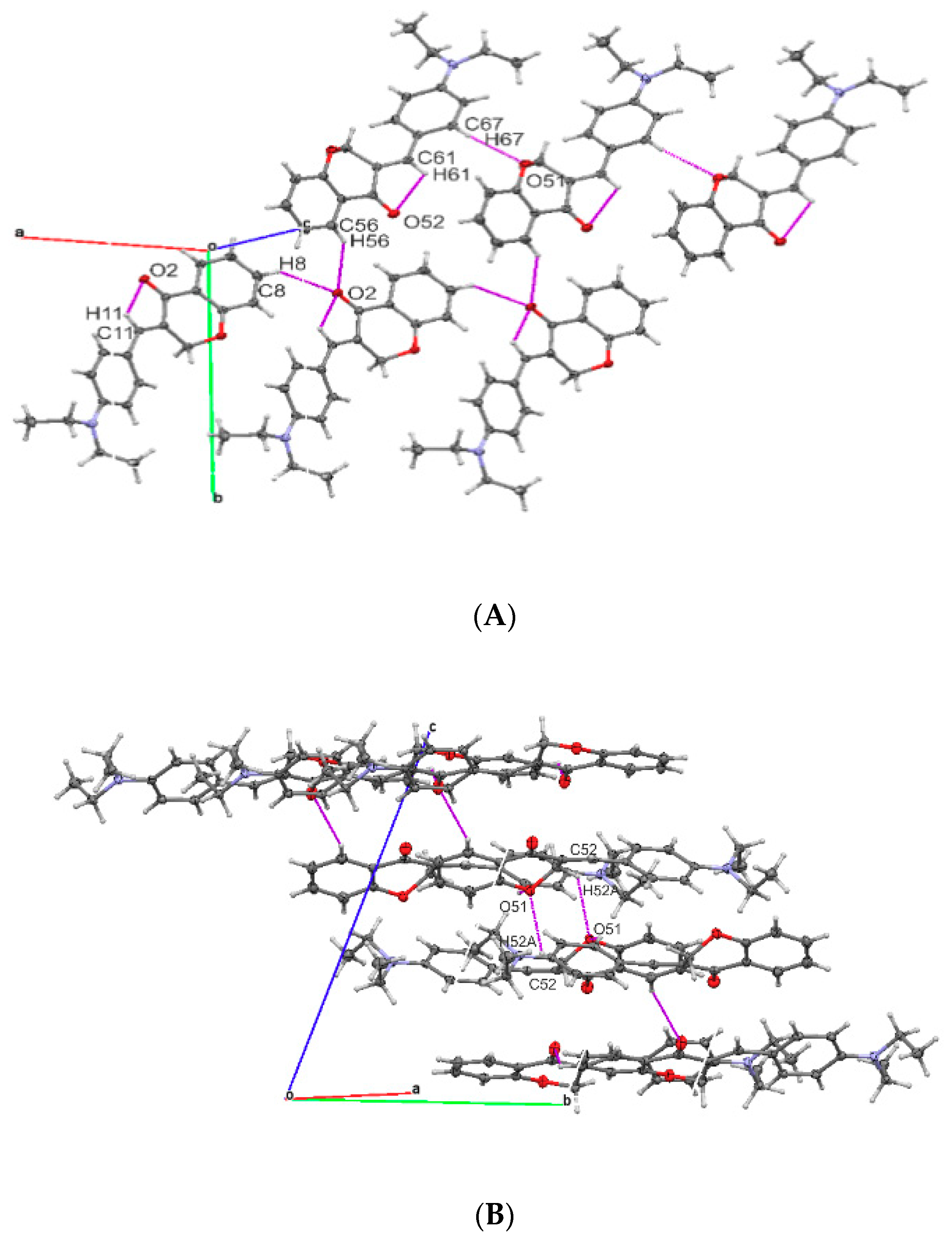
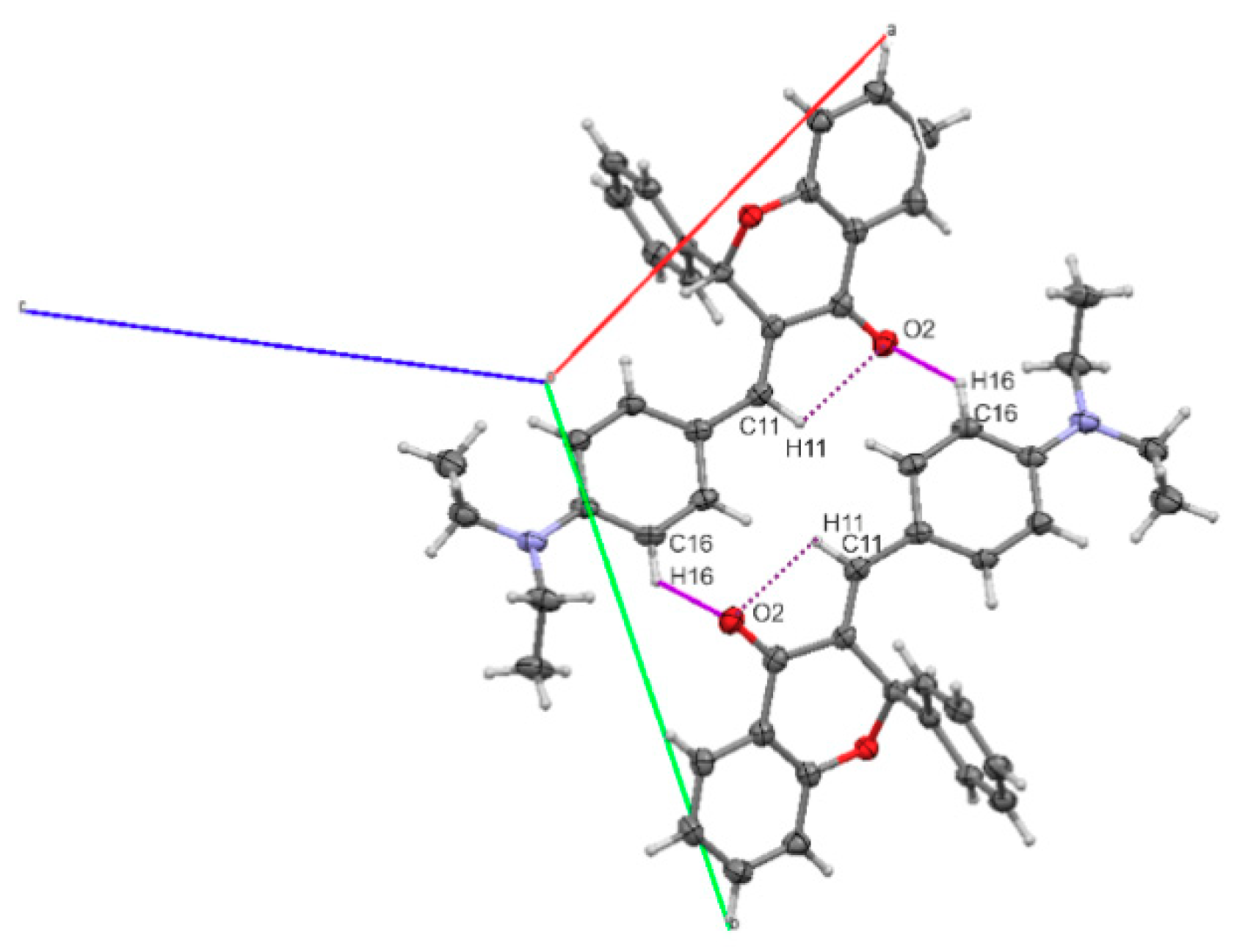

| D-H | H---A | D---A | D-H---A | |
|---|---|---|---|---|
| C11-H11---O2 | 0.93 | 2.35 | 2.768 (2) | 107 |
| C61-H61---O52 | 0.93 | 2.45 | 2.811 (2) | 103 |
| C8-H8…O2 i | 0.93 | 2.63 | 3.198 (2) | 120 |
| C67-H67---O51 i | 0.93 | 2.59 | 3.409 (2) | 147 |
| C52-H52A…O51 ii | 0.97 | 2.60 | 3.274 (2) | 126 |
| C56-H56…O2 iii | 0.93 | 2.70 | 3.498 (2) | 144 |
| C2-H2B…Cg3 iv | 0.97 | 2.87 | 3.717 (2) | 147 |
| C16-H16…Cg7 v | 0.93 | 2.90 | 3.726 (2) | 148 |
| C21-H21C…Cg2 iv | 0.96 | 2.95 | 3.712 (2) | 137 |
| C59-H59…Cg7 ii | 0.93 | 2.71 | 3.545 (2) | 150 |
| C69-H69B…Cg6 vi | 0.96 | 2.93 | 3.645 (2) | 132 |
| 9 | D-H | H---A | D---A | D-H---A |
|---|---|---|---|---|
| C11-H11---O2 | 0.97 | 2.39 | 2.811 (2) | 105 |
| C16-H16---O2i | 0.93 | 2.64 | 3.271 (2) | 126 |
| C7-H7…Cg4ii | 0.93 | 2.55 | 3.475 (2) | 172 |
| C14-H14…Cg2iii | 0.93 | 2.84 | 3.619 (2) | 142 |
| Compounds | IC50 (µM) | ||
|---|---|---|---|
| HL-60 | NALM-6 | WM-115 | |
| 1 | 11.76 ± 1.97 | 8.69 ± 0.40 | 18.09 ± 3.14 |
| 2 | 8.36 ± 0.63 | 9.08 ± 0.30 | 6.45 ± 0.69 |
| 4-chromanone | 676.7 ± 32.6 | 673.7 ± 22.5 | >1000 |
| 3-benzylideneflavanone [11] | 33.3 ± 3.0 | 29.5 ± 4.7 | 59.4 ± 0.9 |
| Compound | Bioactivity Score | H.A. | n | ΔG° Bind (kcal/mol) | LogP (exp) | miLogP | LE | LELP |
|---|---|---|---|---|---|---|---|---|
| 1 | GPCR ligand −0.15 Ion channel modulator −0.30 Kinase inhibitor −0.30 Nuclear receptor ligand −0.04 Protease inhibitor −0.38 Enzyme inhibitor −0.11 | 23 | 4 | −7.5 | 3.43 | 4.47 | 0.33 | 13.55 |
| 2 | GPCR ligand −0.04 Ion channel modulator −0.32 Kinase inhibitor −0.26 Nuclear receptor ligand 0.03 Protease inhibitor −0.31 Enzyme inhibitor −0.08 | 29 | 1 | −9.1 | 5.69 | 6.05 | 0.31 | 19.52 |
| 1 | 2 | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C20H21NO2 | C26H25NO2 |
| Mr | 307.38 | 383.47 |
| Cell setting, space group | Triclinic, P-1 | Monoclinic, P21/n |
| a, b, c (Å) | 7.732(2),13.998(2), 16.312(2) | 11.764(2), 13.455(2), 12.832(2) |
| α, β, γ (°) | 64.74(2), 87.07(2), 87.58(2) | 90.0, 93.75(2), 90.0 |
| V (Å3) | 1594.1(2) | 2026.8(2) |
| Z | 4 | 4 |
| Dx (Mg m−3) | 1.376 | 1.257 |
| Temperature (K) | 100 | 100 |
| Radiation type (Å) | 0.71073 | 0.71073 |
| μ (mm−1) | 0.082 | 0.079 |
| Crystal form, colour | prism, colourless | block, orange |
| Crystal size (mm) | 0.3 × 0.2 × 0.18 | 0.3 × 0.2 × 0.18 |
| Data collection | ||
| Diffractometer | SuperNova, Atlas detector | SuperNova, Atlas detector |
| No. of measured and unique reflections | 13098, 6603 | 25887, 4193 |
| Observed data with F2 4σ(F2) | 5664 | 3316 |
| Rint | 0.0265 | 0.0305 |
| θmax (°) | 26.49 | 26.49 |
| Overall completeness to θmax [%] | 99.8 | 99.8 |
| Refinement (SHELX) | ||
| Refinement on | F2 | F2 |
| All data | 6603 | 4193 |
| R, wR [I 2 (I)] | 0.0362, 0.0941 | 0.0513, 0.1347 |
| R, wR (all data) | 0.0434, 0.0989 | 0.0686, 0.1453 |
| Goodness of fit | 1.067 | 1.066 |
| No. of parameters | 419 | 268 |
| ρmax, ρmin (eÅ−3) | 0.249, −0.212 | 0.241, −0.265 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adamus-Grabicka, A.A.; Markowicz-Piasecka, M.; Ponczek, M.B.; Kusz, J.; Małecka, M.; Krajewska, U.; Budzisz, E. Interaction of Arylidenechromanone/Flavanone Derivatives with Biological Macromolecules Studied as Human Serum Albumin Binding, Cytotoxic Effect, Biocompatibility Towards Red Blood Cells. Molecules 2018, 23, 3172. https://doi.org/10.3390/molecules23123172
Adamus-Grabicka AA, Markowicz-Piasecka M, Ponczek MB, Kusz J, Małecka M, Krajewska U, Budzisz E. Interaction of Arylidenechromanone/Flavanone Derivatives with Biological Macromolecules Studied as Human Serum Albumin Binding, Cytotoxic Effect, Biocompatibility Towards Red Blood Cells. Molecules. 2018; 23(12):3172. https://doi.org/10.3390/molecules23123172
Chicago/Turabian StyleAdamus-Grabicka, Angelika A., Magdalena Markowicz-Piasecka, Michał B. Ponczek, Joachim Kusz, Magdalena Małecka, Urszula Krajewska, and Elzbieta Budzisz. 2018. "Interaction of Arylidenechromanone/Flavanone Derivatives with Biological Macromolecules Studied as Human Serum Albumin Binding, Cytotoxic Effect, Biocompatibility Towards Red Blood Cells" Molecules 23, no. 12: 3172. https://doi.org/10.3390/molecules23123172
APA StyleAdamus-Grabicka, A. A., Markowicz-Piasecka, M., Ponczek, M. B., Kusz, J., Małecka, M., Krajewska, U., & Budzisz, E. (2018). Interaction of Arylidenechromanone/Flavanone Derivatives with Biological Macromolecules Studied as Human Serum Albumin Binding, Cytotoxic Effect, Biocompatibility Towards Red Blood Cells. Molecules, 23(12), 3172. https://doi.org/10.3390/molecules23123172