Syntheses of Novel 4-Substituted N-(5-amino-1H-1,2,4-triazol-3-yl)pyridine-3-sulfonamide Derivatives with Potential Antifungal Activity
Abstract
:1. Introduction
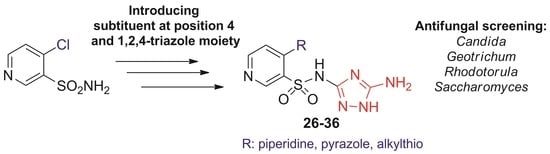
2. Results and Discussion
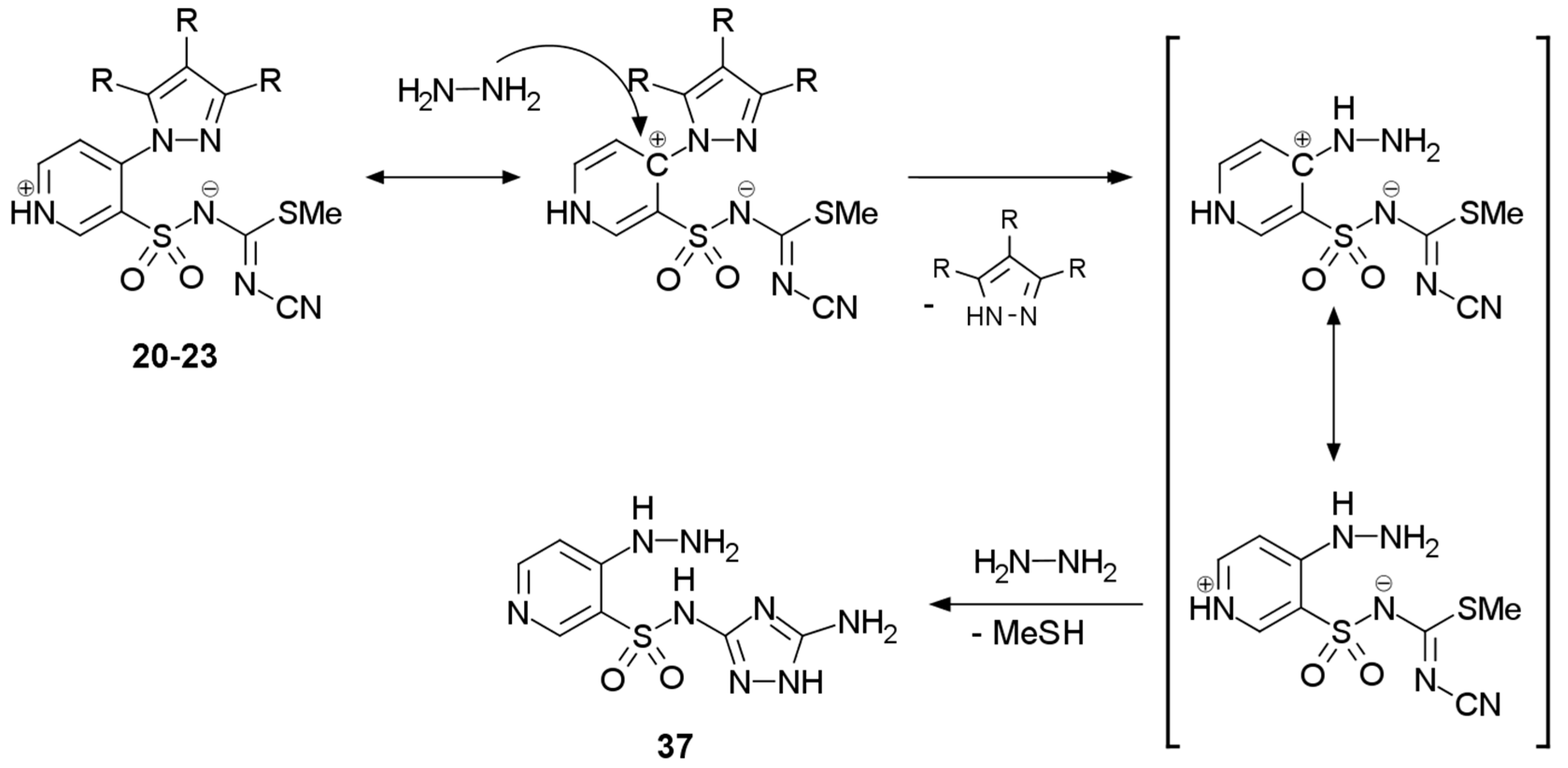
2.1. Chemistry
2.2. Antifungal Activity
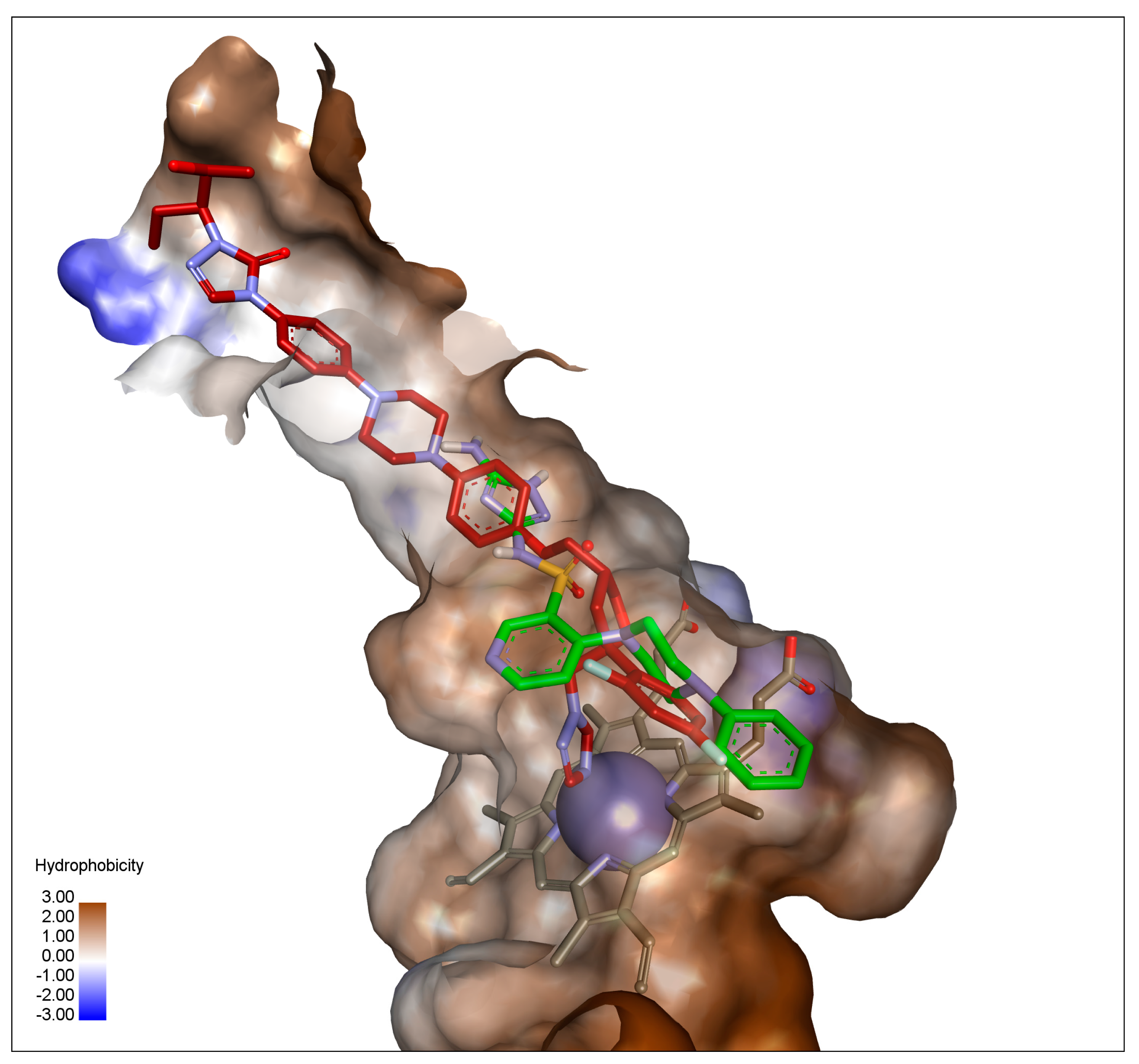
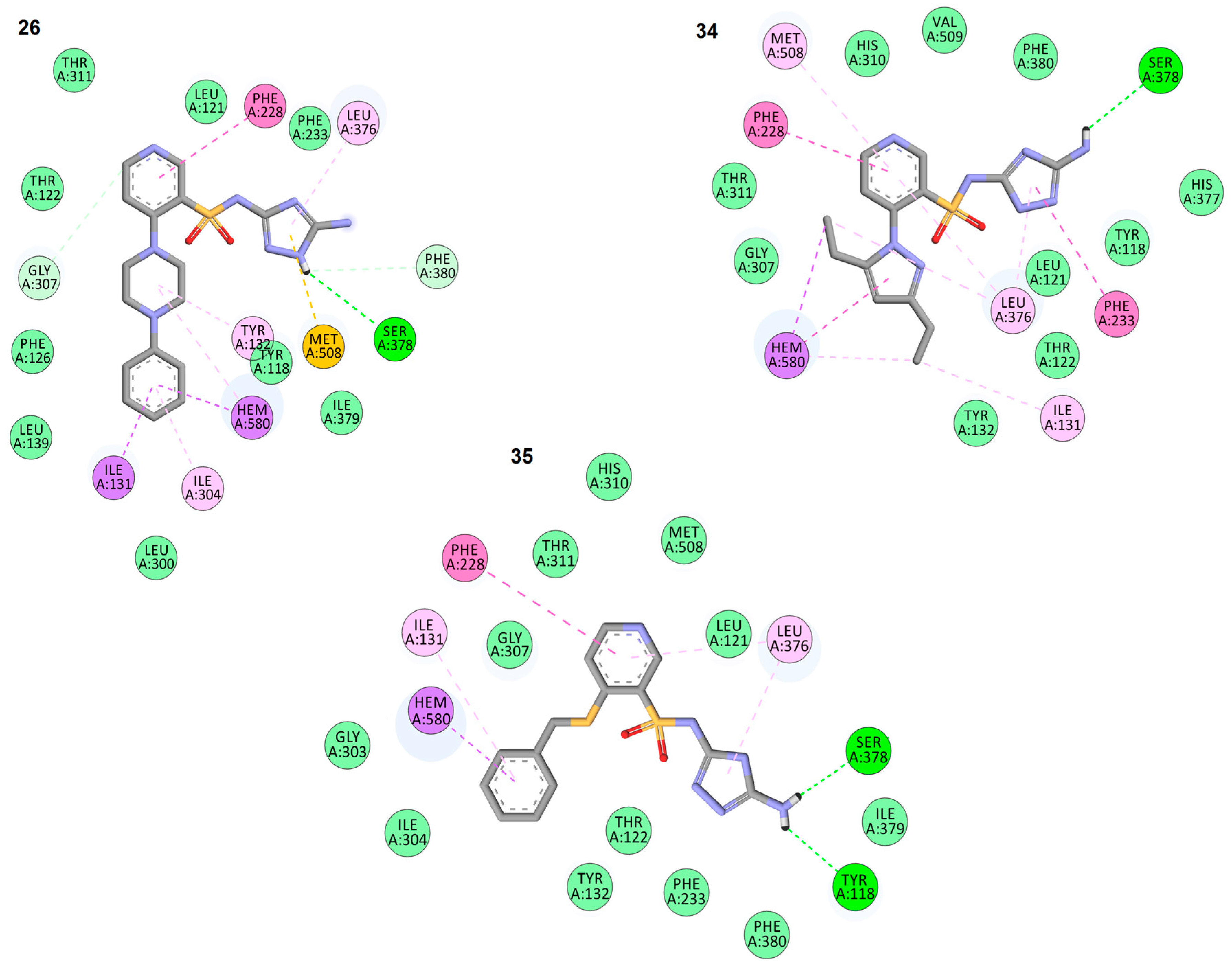
2.3. In Silico Docking Studies
2.4. Anticancer Activity
3. Materials and Methods
3.1. General Information
3.2. Synthesis
3.2.1. Procedure for the Preparation of Methyl N′-Cyano-N-{[4-(piperazin-1-yl)pyridin-3-yl]sulfonyl}carbamimidothioates 15–19
3.2.2. Procedure for the Preparation of Methyl N-(Pyridin-3-yl)sulfonyl-N′-cyanocarbamimidothioate 20–25
3.2.3. Procedure for the Preparation of N-(5-Amino-1H-1,2,4-triazol-3-yl)-4-(piperazin-1-yl)pyridine-3-sulfonamides 26–30
3.2.4. Procedure for the Preparation of N-(5-Amino-1H-1,2,4-triazol-3-yl)-4-R-pyridine-3-sulfonamide 31–36
3.2.5. Procedure for the Preparation of N-(5-Amino-1H-1,2,4-triazol-3-yl)-4-hydrazinylpyridine-3-sulfonamide 37
3.3. Antifungal Evaluation
3.4. Molecular Docking
3.5. Antitumor Evaluation
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Swain, S.S.; Paidesetty, S.K.; Padhy, R.N. Antibacterial activity, computational analysis and host toxicity study of thymol-sulfonamide conjugates. Biomed. Pharmacother. 2017, 88, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. Anti-infective carbonic anhydrase inhibitors: A patent and literature review. Expert Opin. Ther. Pat. 2013, 23, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Rendell, M. The role of sulphonylureas in the management of type 2 diabetes mellitus. Drugs 2004, 64, 1339–1358. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Diuretics: From classical carbonic anhydrase inhibitors to novel applications of the sulfonamides. Curr. Pharm. Des. 2008, 14, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Kaur, I.P.; Smitha, R.; Aggarwal, D.; Kapil, M. Acetazolamide: Future perspective in topical glaucoma therapeutics. Int. J. Pharm. 2002, 248, 1–14. [Google Scholar] [CrossRef]
- Angius, F.; Piras, E.; Uda, S.; Madeddu, C.; Serpe, R.; Bigi, R.; Chen, W.; Dittmer, D.P.; Pompei, R.; Ingianni, A. Antimicrobial sulfonamides clear latent Kaposi sarcoma herpesvirus infection and impair MDM2 p53 complex formation. J. Antibiot. (Tokyo) 2017, 70, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Yadav, M.; Srivastava, R.; Singh, N.; Kaur, R.; Gupta, S.K.; Singh, R.K. Design and anti-HIV activity of arylsulphonamides as non-nucleoside reverse transcriptase inhibitors. Med. Chem. Res. 2016, 25, 2842–2859. [Google Scholar] [CrossRef]
- Barone, M.; Pannuzzo, G.; Santagati, A.; Catalfo, A.; De Guidi, G.; Cardile, V. Molecular docking and fluorescence characterization of benzothieno[3,2-d]pyrimidin-4-one sulphonamide thio-derivatives, a novel class of selective cyclooxygenase-2 Inhibitors. Molecules 2014, 19, 6106–6122. [Google Scholar] [CrossRef] [PubMed]
- Nissinen, L.; Ojala, M.; Langen, B.; Dost, R.; Pihlavisto, M.; Käpylä, J.; Marjamäki, A.; Heino, J. Sulfonamide inhibitors of α2β1 integrin reveal the essential role of collagen receptors in in vivo models of inflammation. Pharmacol. Res. Perspect. 2015, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Minoshima, Y.; Sagane, K.; Sugi, N.H.; Mitsuhashi, K.O.; Yamamoto, N.; Kamiyama, H.; Takahashi, K.; Kotake, Y.; Uesugi, M.; et al. Selective degradation of splicing factor CAPERα by anticancer sulfonamides. Nat. Chem. Biol. 2017, 13, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic Anhydrase Inhibition and the Management of Hypoxic Tumors. Metabolites 2017, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Żołnowska, B.; Sławiński, J.; Pogorzelska, A.; Szafrański, K.; Kawiak, A.; Stasiłojć, G.; Belka, M.; Zielińska, J.; Bączek, T. Synthesis, QSAR studies, and metabolic stability of novel 2-alkylthio-4-chloro-N-(5-oxo-4,5-dihydro-1,2,4-triazin-3-yl)benzenesulfonamide derivatives as potential anticancer and apoptosis-inducing agents. Chem. Biol. Drug Des. 2017, 90. [Google Scholar] [CrossRef] [PubMed]
- Brzozowski, Z.; Sławiński, J.; Sączewski, F.; Innocenti, A.; Supuran, C.T. Carbonic anhydrase inhibitors: Synthesis and inhibition of the human cytosolic isozymes I and II and transmembrane isozymes IX, XII (cancer-associated) and XIV with 4-substituted 3-pyridinesulfonamides. Eur. J. Med. Chem. 2010, 45, 2396–2404. [Google Scholar] [CrossRef] [PubMed]
- Brzozowski, Z.; Sławiński, J.; Innocenti, A.; Supuran, C.T. Carbonic anhydrase inhibitors. Regioselective synthesis of novel 1-substituted 1,4-dihydro-4-oxo-3-pyridinesulfonamides and their inhibition of the human cytosolic isozymes I and II and transmembrane cancer-associated isozymes IX and XII. Eur. J. Med. Chem. 2010, 45, 3656–3661. [Google Scholar] [CrossRef] [PubMed]
- Sławiński, J.; Szafrański, K.; Vullo, D.; Supuran, C.T. Carbonic anhydrase inhibitors. Synthesis of heterocyclic 4-substituted pyridine-3-sulfonamide derivatives and their inhibition of the human cytosolic isozymes I and II and transmembrane tumor-associated isozymes IX and XII. Eur. J. Med. Chem. 2013, 69, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Szafrański, K.; Sławiński, J. Synthesis of Novel 1-(4-Substituted pyridine-3-sulfonyl)-3-phenylureas with Potential Anticancer Activity. Molecules 2015, 20, 12029–12044. [Google Scholar] [CrossRef] [PubMed]
- Perfect, J.R. “Is there an emerging need for new antifungals?”. Expert Opin. Emerg. Drugs 2016, 8214, 129–131. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.D. Changing patterns and trends in systemic fungal infections. J. Antimicrob. Chemother. 2005, 56, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Strushkevich, N.; Usanov, S.A.; Park, H.W. Structural basis of human CYP51 inhibition by antifungal azoles. J. Mol. Biol. 2010, 397, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, D.J.; Hitchcock, C.A.; Carol, M. Current and Emerging Azole Antifungal Agents. Clin. Microbiol. Rev. 1999, 12, 40–79. [Google Scholar] [PubMed]
- Kharb, R.; Sharma, P.C.; Yar, M.S. Pharmacological significance of triazole scaffold. J. Enzym. Inhib. Med. Chem. 2011, 26, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, C.; Tang, S.; Wu, Q.; Hu, H.; Zhao, Q.; Zou, Y. Synthesis, In Vitro Biological Evaluation, and Molecular Docking of New Triazoles as Potent Antifungal Agents. Arch. Pharm. 2016, 349, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Zayed, M.; El-Messery, S.; Al-Agamy, M.; Abdel-Rahman, H. Design, Synthesis, Antimicrobial Evaluation and Molecular Modeling Study of 1,2,4-Triazole-Based 4-Thiazolidinones. Molecules 2016, 21, 568. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.K.; Ahamd, A.; Pandey, O.P.; Nizamuddin, K. Polyethylene Glycol Mediated, One-Pot, Three-Component Synthetic Protocol for Novel 3-[3-Substituted-5-mercapto-1,2,4-triazol-4-yl]-spiro-(indan-1′,2-thiazolidin)-4-ones as New Class of Potential Antimicrobial and Antitubercular Agents. J. Heterocycl. Chem. 2014, 51, 1233–1239. [Google Scholar] [CrossRef]
- Zoumpoulakis, P.; Camoutsis, C.; Pairas, G.; Soković, M.; Glamočlija, J.; Potamitis, C.; Pitsas, A. Synthesis of novel sulfonamide-1,2,4-triazoles, 1,3,4-thiadiazoles and 1,3,4-oxadiazoles, as potential antibacterial and antifungal agents. Biological evaluation and conformational analysis studies. Bioorg. Med. Chem. 2012, 20, 1569–1583. [Google Scholar] [CrossRef] [PubMed]
- Somagond, S.M.; Kamble, R.R.; Kattimani, P.P.; Joshi, S.D.; Dixit, S.R. Design, synthesis, docking and in vitro antifungal study of 1,2,4-triazole hybrids of 2-(aryloxy)quinolines. Heterocycl. Commun. 2017, 23, 317–324. [Google Scholar] [CrossRef]
- Ozdemir, S.B.; Demirbas, N.; Cebeci, Y.U.; Bayrak, H.; Mermer, A.; Ceylan, S.; Demirbas, A. Synthesis and Antimicrobial Activities of Hybrid Heterocyclic Molecules Based on 1-(4-Fluorophenyl)piperazine Skeleton. Lett. Drug Des. Discov. 2017, 14. [Google Scholar] [CrossRef]
- Karaca Gençer, H.; Acar Çevik, U.; Levent, S.; Sağlık, B.N.; Korkut, B.; Özkay, Y.; Ilgın, S.; Öztürk, Y. New Benzimidazole-1,2,4-Triazole Hybrid Compounds: Synthesis, Anticandidal Activity and Cytotoxicity Evaluation. Molecules 2017, 22, 507. [Google Scholar] [CrossRef] [PubMed]
- Mansueto, R.; Perna, F.M.; Salomone, A.; Perrone, S.; Florio, S.; Capriati, V. Efficient Regioselective Synthesis of 3,4,5-Trisubstituted 1,2,4-Triazoles on the Basis of a Lithiation-Trapping Sequence. European J. Org. Chem. 2014, 2014, 6653–6657. [Google Scholar] [CrossRef]
- Katritzky, A.R.; Tymoshenko, D.O.; Chen, K.; Fattah, A.A. Ring and side chain reactivities of 1-([1,3,4]oxadiazol-2-ylmethyl)-1H-benzotriazoles. ARKIVOC 2001, 2001, 101–108. [Google Scholar] [CrossRef]
- Kleschick, W.A.; Dunbar, J.E.; Snider, S.W.; Vinogradoff, A. Regiospecific Synthesis of Arenesulfonamide Derivatives of 3,5-Diamino-1,2,4-triazole. J. Org. Chem. 1988, 53, 3120–3122. [Google Scholar] [CrossRef]
- Wouters, J.; Michaux, C.; Durant, F.; Michel Dogné, J.; Delarge, J.; Masereel, B. Isosterism among analogues of torasemide: Conformational, electronic and lipophilic properties. Eur. J. Med. Chem. 2000, 35, 923–929. [Google Scholar] [CrossRef]
- Hargrove, T.Y.; Friggeri, L.; Wawrzak, Z.; Qi, A.; Hoekstra, W.J.; Schotzinger, R.J.; York, J.D.; Peter Guengerich, F.; Lepesheva, G.I. Structural analyses of Candida albicans sterol 14α-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis. J. Biol. Chem. 2017, 292, 6728–6743. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.K.; Leung, S.S.F.; Guilbert, C.; Jacobson, M.P.; Mckerrow, J.H.; Podust, L.M. Structural characterization of CYP51 from Trypanosoma cruzi and Trypanosoma brucei bound to the antifungal drugs posaconazole and fluconazole. PLoS Negl. Trop. Dis. 2010, 4. [Google Scholar] [CrossRef] [PubMed]
- Forbes, B.; Sahm, D.; Weissfeld, A. Bailey & Scott’s Diagnostic Microbiology, 12th ed.; Mosby Elsevier: St. Louis, MO, USA, 2007. [Google Scholar]
- Winn, W.; Allen, S.; Janda, W.; Koneman, E.; Procop, G.; Schrekenberger, P.; Woods, G. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology, 6th ed.; Lippincott Williams & Wilkins: Baltimore, MD, USA, 2006. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Boyd, M.R.; Paull, K.D. Some practical considerations and applications of the national cancer institute in vitro anticancer drug discovery screen. Drug Dev. Res. 1995, 34, 91–109. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 2–37 are available from the authors. |


| Compd. | M.W: | Candida albicans (9 strains) | Candida glabrata (5 strains) | Candida guilliermondii (2 strains) | Candida krusei (4 strains) | Candida lusitaniae (3 strains) | Candida parapsilosis (4 strains) | Candida tropicalis (4 strains) | Candida utilis (1 strain) | Geotrichum candidum (2 strains) | Rhodotorula mucilaginosa (2 strains) | Saccharomyces cerevisiae (1 strain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 26 | 400.46 | ≤6.2; 50; 100; 100; 100; *; *; *; * A | *; *; *; *; 100 B | 12.5; 25 | 100; *; *; 100 C | 100; *; 12.5 D | *; *; *; *, E | 100; *; *; 100 F | * | *; * | ≤6.2; 50 | * |
| 27 | 434.90 | *; *; *; *;*; *; *; *; * A | *; *; *; *; * B | *; * | *; *; *; * C | *; *; *, D | *; *; *; *, E | *; *; *; *, F | * | *; * | *; * | * |
| 28 | 418.45 | 12.5; 12.5; 100; 100; *; *; *; *;* A | *; *; *; *; * B | 50; 100 | 100; *; *; * C | 50; 100; 50 D | *; *; *; * E | *; *; *; * F | 100 | *; * | 12.5; 25 | * |
| 29 | 469.35 | *; *; *; *;*; *; *; *; * A | *; *; *; *; * B | *; * | *; *; *; * C | *; *; * D | *; *; *; * E | *; *; *; * F | * | *; * | *; * | * |
| 30 | 430.48 | 12.5; 100; 100; *; *; *; *;*; * A | *; *; *; *;* B | *; * | *; *; *; * C | *; *;* D | *; *; *; * E | *; *; *; * F | * | *; * | 100; * | * |
| 31 | 334.36 | 25; 50; *; *; *; *;*; *; * A | *; *; *; *; * B | *; * | *; *; *; * C | *; *; * D | *; *; *; * E | 25; *; *; 50 F | * | *; * | 100; * | * |
| 32 | 348.38 | 25; 50; 100; *; *; *; *;*; * A | 50; *; *; * B | *; * | 25; *; *; 50 C | *; *; * D | *; *; *; * E | 100 *; *; 100 F | 100 | *; * | 25; 25 | * |
| 33 | 390.46 | *; *; *; *;*; *; *; *; * A | *; *; *; *; * B | *; * | *; *; *; * C | *; *; * D | *; *; *; * E | *; *; *; * F | * | *; * | *; * | * |
| 34 | 362.41 | ≤6.2; ≤6.2; 100; *; *; *; *;*; * A | *; *; *; *; * B | *; * | 100; *; *;* C | 50; 100; 100 D | *; *; *; * E | *; *; *; * F | * | *; * | 100; 200 | * |
| 35 | 362.43 | ≤6.2; 12.5; 50; 100; 100; *; *; *; 50 A | 100; 100; *; *; 100 B | 100; * | 12.5;*; *; 50 C | *; *; * D | *; *; *; * E | *; *; *; * F | * | *; * | 6.2; 100 | * |
| 36 | 329.36 | 12.5; 50; 100; 100; *; *; *; *; 100 A | 25; 100; *; *; 100 B | 50; 100 | 100; *; *; 100 C | 100; *; 100 D | *; *; *; * E | *; *; *; * F | * | *; * | 25; 100 | * |
| Fl | 306.28 | 12.5; 50; #; #; #; #; #; #; # A | 25; 50; #; #; 25 B | 12.5; 50 | 25; 50; 50; 50 C | 12.5; 12.5; 12.5 D | 6.2; 12.5; 25; 3.1 E | 25; 50; #; 6.2 F | 25 | 12.5; 25 | #; # | # |
| Compound Binding Affinity | Molecule Fragment Involved in Interaction | Interacting Residue | Type of Interaction | Interaction Distance [Å] |
|---|---|---|---|---|
| 26 −9.9 (kcal/mol) | 5-amino-1H-1,2,4-triazole | LEU 376 | π-alkyl | 4.93 |
| SER378 | hydrogen bond | 2.67 | ||
| PHE380 | π-donor hydrogen | 4.93 | ||
| MET508 | π-sulfur | 2.67 | ||
| pyridine-3-sulfonamide scaffold | PHE228 | π-π | 5.13 | |
| GLY307 | π-donor hydrogen | 3.56 | ||
| 4-phenylpiperazine substituent | TYR132 | π-alkyl | 5.24 | |
| HEME | π-alkyl | 5.22 | ||
| HEME | π-σ | 3.23 | ||
| ILE304 | π-alkyl | 5.42 | ||
| ILE131 | π-σ | 3.70 | ||
| ILE131 | π-σ | 3.51 | ||
| 34 −9.3 (kcal/mol) | 5-amino-1H-1,2,4-triazole | LEU 376 | 5.06 | |
| SER378 | hydrogen bond | 2.75 (OH) 2.77 (C=O) | ||
| PHE233 | π-π | 2.29 | ||
| pyridine-3-sulfonamide scaffold | PHE228 | π-π | 5.05 | |
| LEU 376 | π-alkyl | 5.41 | ||
| MET508 | π-alkyl | 5.38 | ||
| 3,5-diethyl-1H-pyrazole substituent | LEU 376 | π-alkyl | 4.03 | |
| ILE131 | π-alkyl | 4.66 | ||
| HEME | π-σ | 3.69 | ||
| HEME | π-π | 4.97 | ||
| HEME | π-alkyl | 3.71 | ||
| 35 −9.1 (kcal/mol) | 5-amino-1H-1,2,4-triazole | LEU 376 | π-alkyl | 4.39 |
| SER378 | hydrogen bond | 1.83 | ||
| TYR118 | hydrogen bond | 2.19 | ||
| pyridine-3-sulfonamide scaffold | LEU 376 | π-alkyl | 5.27 | |
| PHE228 | π-π | 5.66 | ||
| benzylthio substituent | ILE131 | π-alkyl | 5.16 | |
| HEME | π-σ | 4.18 | ||
| HEME | π-σ | 3.74 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szafrański, K.; Sławiński, J.; Kędzia, A.; Kwapisz, E. Syntheses of Novel 4-Substituted N-(5-amino-1H-1,2,4-triazol-3-yl)pyridine-3-sulfonamide Derivatives with Potential Antifungal Activity. Molecules 2017, 22, 1926. https://doi.org/10.3390/molecules22111926
Szafrański K, Sławiński J, Kędzia A, Kwapisz E. Syntheses of Novel 4-Substituted N-(5-amino-1H-1,2,4-triazol-3-yl)pyridine-3-sulfonamide Derivatives with Potential Antifungal Activity. Molecules. 2017; 22(11):1926. https://doi.org/10.3390/molecules22111926
Chicago/Turabian StyleSzafrański, Krzysztof, Jarosław Sławiński, Anna Kędzia, and Ewa Kwapisz. 2017. "Syntheses of Novel 4-Substituted N-(5-amino-1H-1,2,4-triazol-3-yl)pyridine-3-sulfonamide Derivatives with Potential Antifungal Activity" Molecules 22, no. 11: 1926. https://doi.org/10.3390/molecules22111926
APA StyleSzafrański, K., Sławiński, J., Kędzia, A., & Kwapisz, E. (2017). Syntheses of Novel 4-Substituted N-(5-amino-1H-1,2,4-triazol-3-yl)pyridine-3-sulfonamide Derivatives with Potential Antifungal Activity. Molecules, 22(11), 1926. https://doi.org/10.3390/molecules22111926