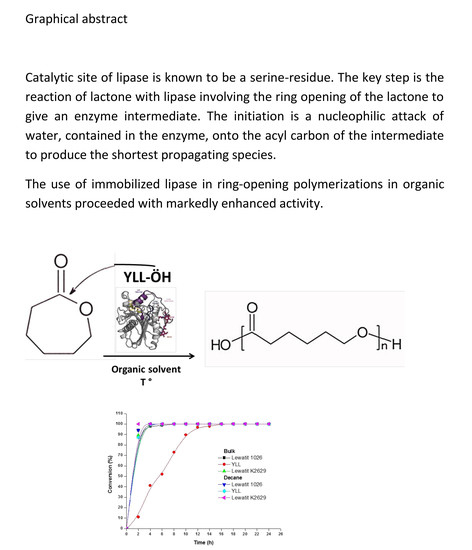
Yarrowia lipolytica Extracellular Lipase Lip2 as Biocatalyst for the Ring-Opening Polymerization of ε-Caprolactone
Abstract
:1. Introduction
Yarrowia lipolytica Lipases as Biocatalysts
2. Results and Discussion
2.1. Lipase Isolation and Immobilization
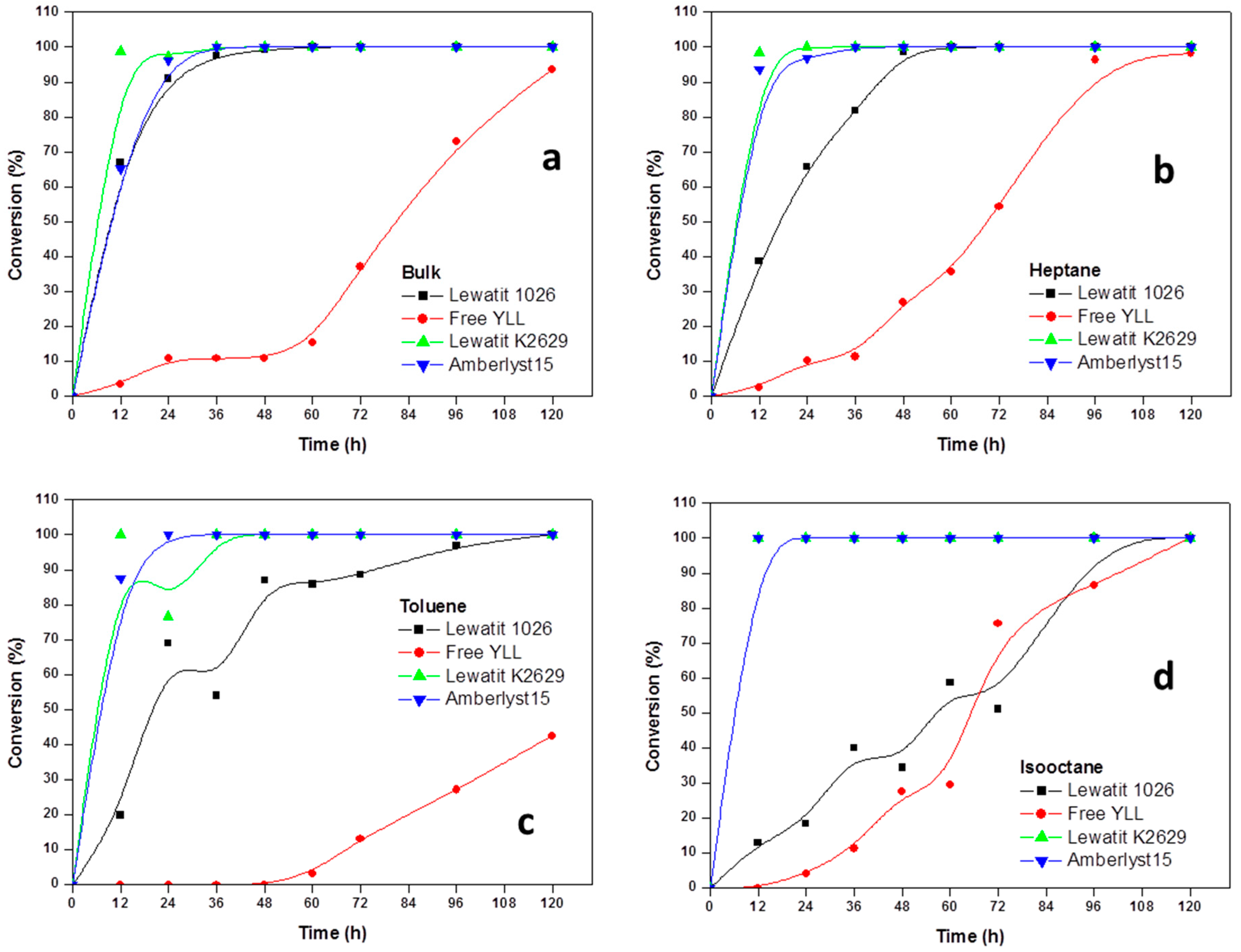
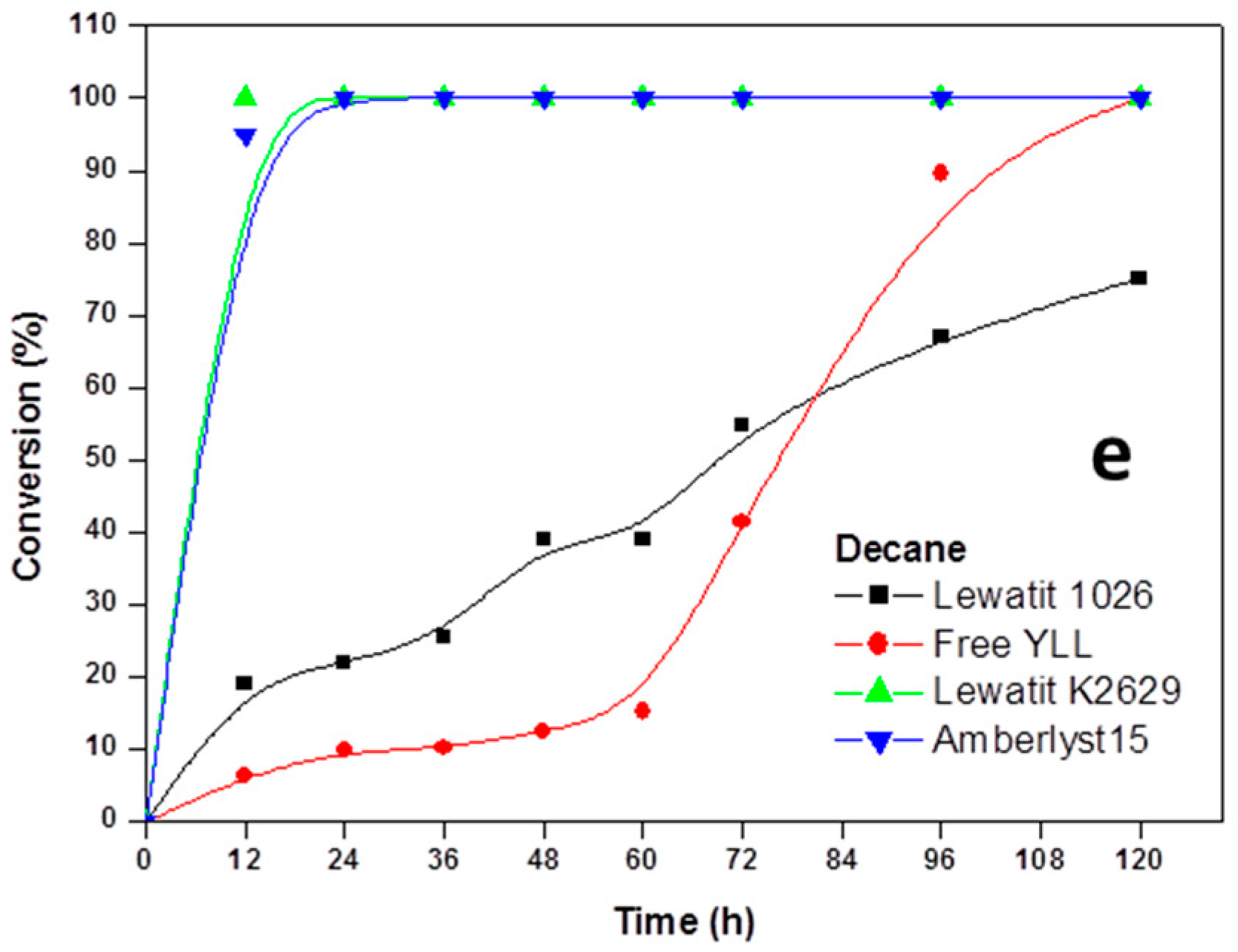
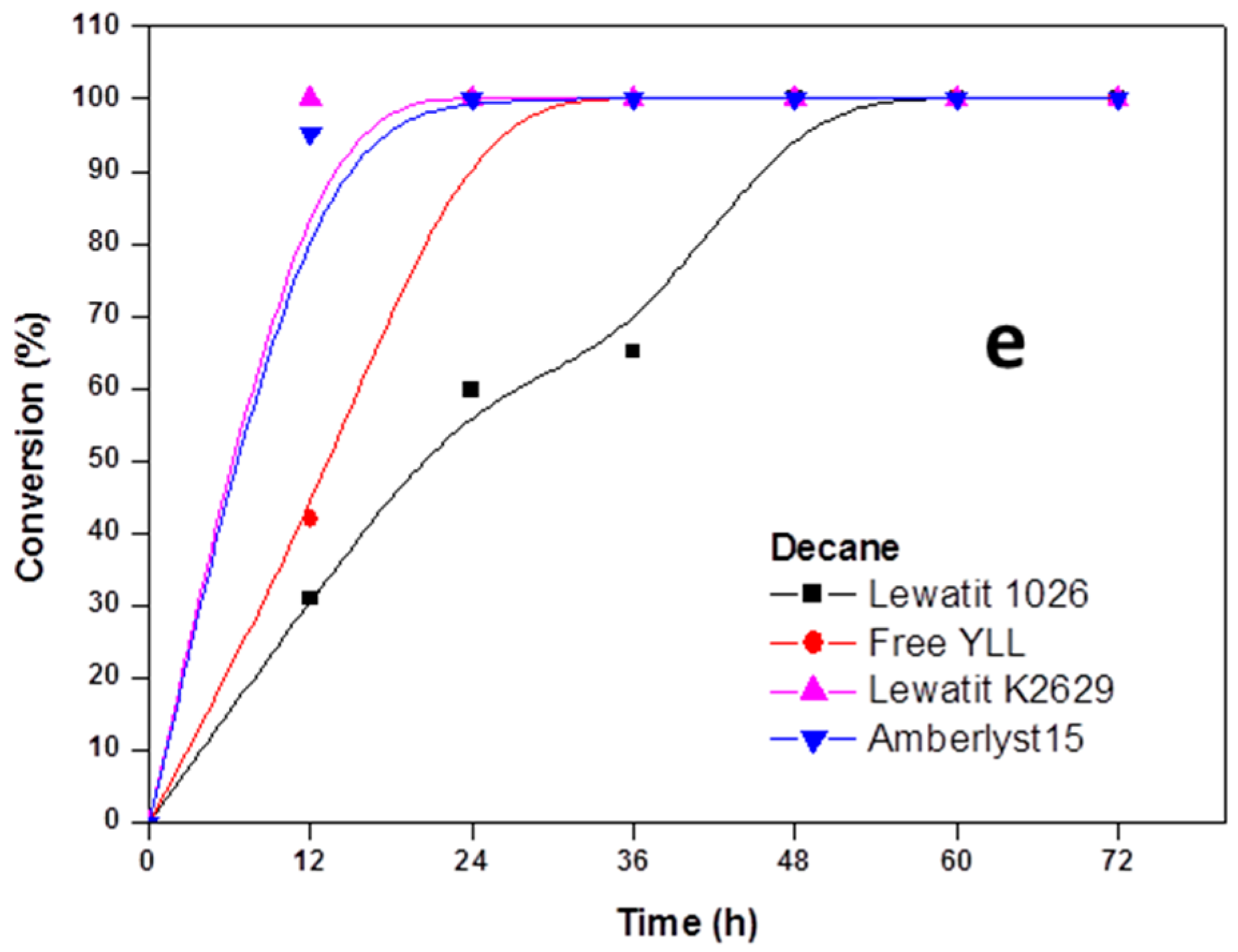
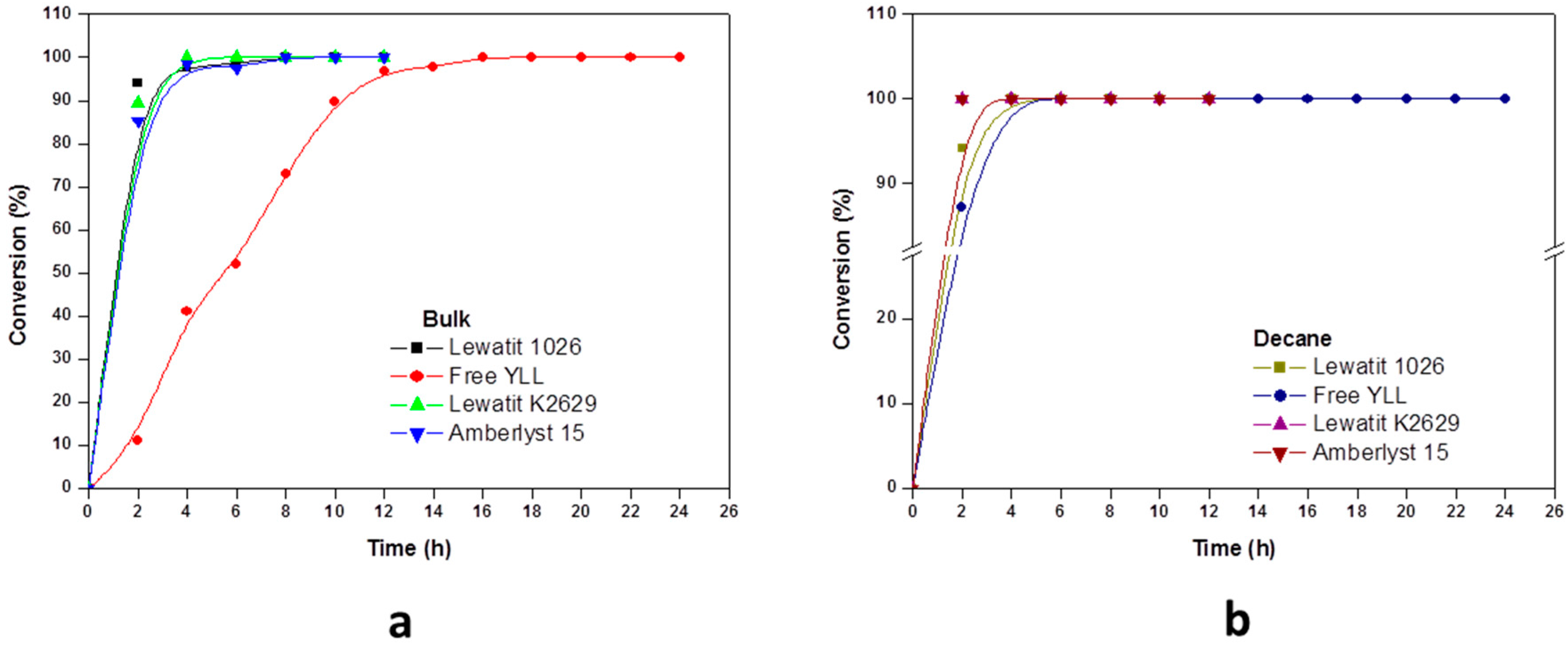
2.2. Enzymatic Synthesis of Poly (ε-Caprolactone) Using Immobilized Y. lipolytica Lipase
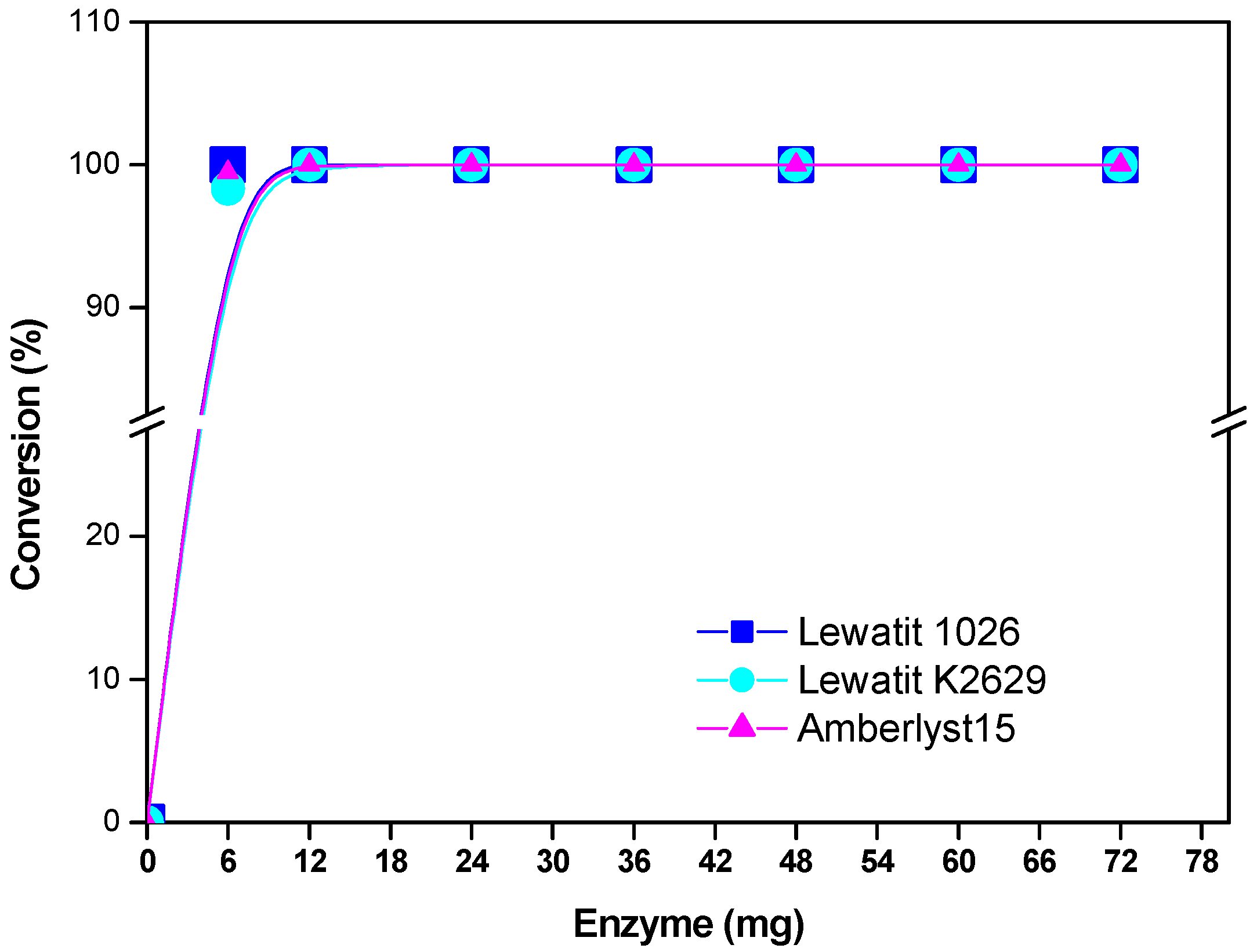
2.3. Enzyme Concentration
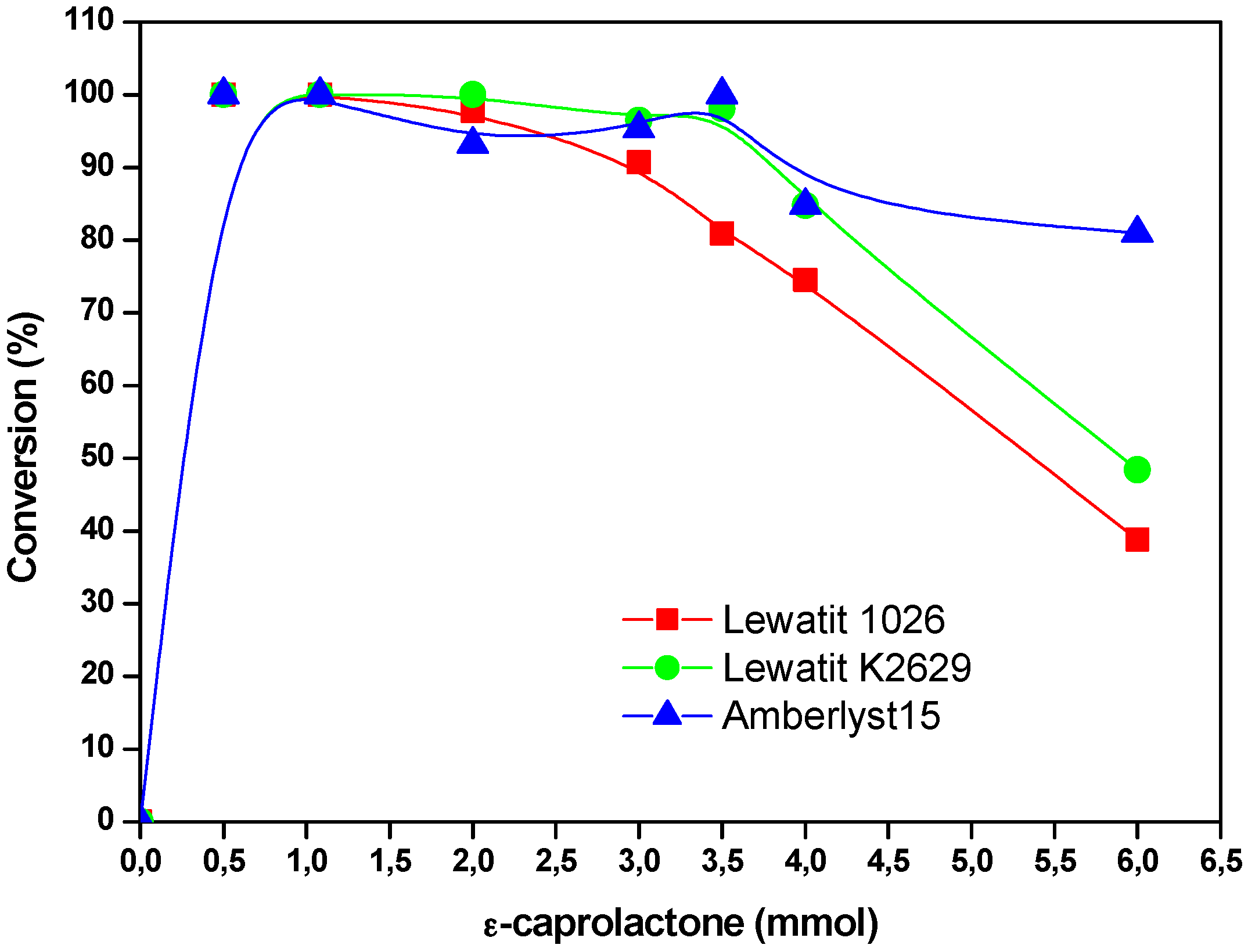
2.4. The Effect of the Substrate Concentration
3. Materials and Methods
3.1. Organism
3.2. Protein Determination
3.3. Lipase Activity
3.4. Lipase Isolation and Immobilization
3.5. Synthesis of Poly (ε-Caprolactone) Using Immobilized Y. lipolytica Lipase
4. Conclusions
5. Future Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Barth, G.; Gaillardin, C. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Bicrobiol. Rev. 1997, 19, 219–237. [Google Scholar] [CrossRef]
- Gellissen, G.; Kunze, G.; Gaillardin, C.; Cregg, J.M.; Berardi, E.; Veenhuis, M.; van der Klei, I. New yeast expression platforms based on methylotrophic Hansenula polymorpha and Pichia pastoris and on dimorphic Arxula adeninivorans and Yarrowia lipolytica—A comparison. FEMS Yeast Res. 2005, 5, 1079–1096. [Google Scholar] [CrossRef] [PubMed]
- Beckerich, J.M.; Baudevin, A.B.; Gaillardin, C. Yarrowia lipolytica: A model organism for protein secretion studies. Int. Microbiol. 1998, 1, 123–130. [Google Scholar] [PubMed]
- Fickers, P.; Marty, A.; Nicaud, J.M. The lipases from Yarrowia lipolytica: Genetics, production, regulation, biochemical characterization and biotechnological applications. Biotechnol. Adv. 2011, 29, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, N.N. Applications of lipase. JAOCS 1997, 74, 621–634. [Google Scholar] [CrossRef]
- Peters, I.I.; Nelson, F.E. Preliminary characterization of the lipase of Mycotorula lipolytica. J. Bacteriol. 1948, 55, 593–600. [Google Scholar] [PubMed]
- Peters, I.I.; Nelson, F.E. Factors influencing the production of lipase by Mycotorula lipolytica. J. Bacteriol. 1948, 55, 581–591. [Google Scholar] [PubMed]
- Ota, Y.; Gomi, K.; Kato, S.; Sugiura, T.; Minoda, Y. Purification and some properties of cell-bound lipase from Saccharomycopsis lipolytica. Agric. Biol. Chem. 1982, 46, 2885–2893. [Google Scholar] [CrossRef]
- Ota, Y.; Oikawa, S.; Morimote, Y.; Minoda, Y. Nutritional factors causing mycelial development of Saccharomycopsis lipolytica. Agric. Biol. Chem. 1984, 48, 1933–1940. [Google Scholar]
- Uyama, H.; Kobayashi, S. Enzymatic Ring-Opening Polymerization of Lactones Catalyzed by Lipase. Chem. Lett. 1993, 22, 1149–1150. [Google Scholar] [CrossRef]
- Uyama, H.; Takeya, K.; Kobayashi, S. Synthesis of Polyesters by Enzymatic Ring-Opening Copolymerizatio Using Lipase Catalyst. Proc. Jpn. Acad. Ser. B 1993, 69, 203–207. [Google Scholar] [CrossRef]
- Knani, D.; Gutman, A.L.; Kohn, D.H. Enzymatic Polyesterification in Organic Media -Enzyme-Catalyzed Synthesis of Linear Polyesters. I. Condensation Polymerization of Linear Hydroxyesters. II. Ring-Opening Polymerization of ε-Caprolactone. J. Polym. Sci. Part A Polym. Chem. 1993, 31, 1221–1232. [Google Scholar] [CrossRef]
- Foresti, M.L.; Ferreira, M.L. Synthesis of Polycaprolactone Using Free/Supported Enzymatic and Non-Enzymatic Catalysts. Macromol. Rapid Commun. 2004, 25, 2025–2028. [Google Scholar] [CrossRef]
- Kobayashi, S. Enzymatic Polymerization: A New Method of Polymer Synthesis. J. Polym. Sci. Part A Polym. Chem. 1999, 37, 3041–3056. [Google Scholar] [CrossRef]
- Kobayashi, S.; Uyama, H. Biocatalytical Routes to Polymers. In Material Science and Technology-Synthesis of Polymers; Schlueter, A.D., Ed.; Wiley-VCH: Weinheim, Germany, 1999; Volume 54, pp. 549–569. [Google Scholar]
- Kobayashi, S.; Uyama, H.; Ohmae, M. Enzymatic Polymerization for Precision Polymer Synthesis. Bull. Chem. Soc. Jpn. 2001, 74, 613–635. [Google Scholar] [CrossRef]
- Kobayashi, S.; Uyama, H.; Kimura, S. Enzymatic Polymerization. Chem. Rev. 2001, 101, 3793–3818. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Uyama, H. In vitro Polyester Synthesis via Enzymatic Polymerization. Curr. Org. Chem. 2002, 6, 209–222. [Google Scholar] [CrossRef]
- Kobayashi, S.; Uyama, H. Enzymatic Polymerization to Polyesters. In Handbook of Biopolymers; Polyesters, I., Doi, Y., Steinbüchel, A., Eds.; Wiley-VCH: Weinheim, Germany, 2002; pp. 373–400. [Google Scholar]
- Kobayashi, S.; Uyama, H. Enzymatic Polymerization. In Encyclopedia of Polymer Science and Technology, 3rd ed.; Kroschwitz, J.I., Ed.; Wiley: New York, NY, USA, 2003; pp. 328–364. [Google Scholar]
- Gross, R.A.; Kumar, A.; Kalra, B. Polymer Synthesis by In Vitro Enzyme Catalysis. Chem. Rev. 2001, 101, 2097–2124. [Google Scholar] [CrossRef] [PubMed]
- Albertsson, A.C.; Varma, I.K. Recent Developments in Ring Opening Polymerization of Lactones for Biomedical Applications. Biomacromolecules 2003, 4, 1466–1486. [Google Scholar] [CrossRef] [PubMed]
- Varma, I.K.; Albertsson, A.C.; Rajkhowa, R.; Srivastava, R.K. Enzyme Catalyzed Synthesis of Polyesters. Prog. Polym. Sci. 2005, 30, 949–981. [Google Scholar] [CrossRef]
- Barrera-Rivera, K.A.; Flores-Carreón, A.; Martínez-Richa, A. Enzymatic Ring-Opening Polymerization of ε-Caprolactone by a New Lipase from Yarrowia lipolytica. J. Appl. Polym. Sci. 2008, 109, 708–719. [Google Scholar] [CrossRef]
- Bordes, F.; Barbe, S.; Escalier, P.; Mourey, L.; André, L.; Marty, A.; Tranier, S. Exploring the Conformational States and Rearrangements of Yarrowia lipolytica Lipase. Biophys. J. 2010, 99, 2225–2234. [Google Scholar] [CrossRef] [PubMed]
- Bartniki-García, S.; Nickerson, W.J. Induction of yeast-like development in Mucor by carbon dioxide. J. Bacteriol. 1962, 84, 829–840. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Gupta, N.; Rathi, P.; Gupta, R. Simplified para-nitrophenyl palmitate assay for lipases and esterases. Anal. Biochem. 2002, 311, 98–99. [Google Scholar] [CrossRef]
- Harzevili, F.D. Biotechnological Applications of the Yeast Yarrowia lipolytica; Springer Briefs in Microbiology; Springer: Cham, Switzerland, 2014. [Google Scholar]
Sample Availability: Samples of the compounds are available from the authors. |

| Resin | Protein Content (mg/g) | Protein Adsorption (%) | Protein Activity (U/g) * |
|---|---|---|---|
| Lewatit VPOC 1026 | 0.14 | 87 | 47 |
| Lewatit VPOC K2629 | 0.14 | 92 | 805 |
| Lewatit CNP-105 | 0.14 | 88 | 291 |
| Lewatit 1064 MD PH | 0.10 | 42 | 137 |
| Lewatit VPOC 1163 | 0.10 | 52 | 86 |
| Lewatit VPOC 1065 weakly basic | 0.11 | 69 | 19 |
| Lewatit MP62 free base | 0.10 | 50 | 81 |
| Lewatit monoplus TP214 | 0.14 | 88 | 23 |
| Lewatit VPOC K3433 | 0.02 | 18 | 3 |
| Lewatit SP112 | 0.10 | 51 | 144 |
| Amberlite XAD16 | 0.11 | 69 | 31 |
| Amberlite XAD7HP | 0.15 | 96 | 35 |
| Amberlite XAD1180 | 0.15 | 95 | 5 |
| Amberlite XAD4 | 0.10 | 64 | 72 |
| Amberlyst 15 | 0.12 | 74 | 512 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrera-Rivera, K.A.; Martínez-Richa, A. Yarrowia lipolytica Extracellular Lipase Lip2 as Biocatalyst for the Ring-Opening Polymerization of ε-Caprolactone. Molecules 2017, 22, 1917. https://doi.org/10.3390/molecules22111917
Barrera-Rivera KA, Martínez-Richa A. Yarrowia lipolytica Extracellular Lipase Lip2 as Biocatalyst for the Ring-Opening Polymerization of ε-Caprolactone. Molecules. 2017; 22(11):1917. https://doi.org/10.3390/molecules22111917
Chicago/Turabian StyleBarrera-Rivera, Karla A., and Antonio Martínez-Richa. 2017. "Yarrowia lipolytica Extracellular Lipase Lip2 as Biocatalyst for the Ring-Opening Polymerization of ε-Caprolactone" Molecules 22, no. 11: 1917. https://doi.org/10.3390/molecules22111917
APA StyleBarrera-Rivera, K. A., & Martínez-Richa, A. (2017). Yarrowia lipolytica Extracellular Lipase Lip2 as Biocatalyst for the Ring-Opening Polymerization of ε-Caprolactone. Molecules, 22(11), 1917. https://doi.org/10.3390/molecules22111917