Design and Fabrication of Organ-on-Chips: Promises and Challenges
Abstract
:1. Introduction
2. Conceptual Design of OOC
2.1. Geometry and Dimensions
2.2. Flow Control in OOCs
2.3. Clog Avoidance in OOCs
2.4. Monitoring and Detection
3. Fabrication Materials
3.1. Materials Used in Chip Production
3.1.1. Polydimethylsiloxane (PDMS)
3.1.2. Glass
3.1.3. Thermoplastics
3.2. Other Materials Used in OOC Technology
3.2.1. Hydrogels
3.2.2. Silicon
3.2.3. Metals
3.2.4. Membranes
4. Fabrication Methods
4.1. Soft Lithography
4.2. Hot Embossing
4.3. Injection Molding
4.4. 3D Printing
5. Creative Methods
6. Applications of OOCs
6.1. OOC Technologies
6.1.1. Single Organ Models
- -
- BBB-on-Chip: Drugs are not effective in the CNS unless they pass through the highly selective brain microvascular endothelial cells [234]. The blood-brain barrier (BBB) is a combination of Brain Microvascular Endothelial Cells (BMECs) in the capillaries and the surrounding cells in the CNS, which consists of pericytes and astrocytes [235]. Although the BBB blocks numerous drug compounds from entering the brain, it protects the CNS and brain from pathogens [236]. It is expected that BBB-on-chip models based on human cells will be increasingly used in drug-discovery and drug-delivery research on the brain [237,238,239] as the in vivo expression of many solute carriers and efflux transporters varies widely between human and animal systems due to differences between species [59,240]. In the development of BBB-on-chip models for drug screening, the following aspects are crucial: (i) two compartments recapitulating blood and brain parts separated by a porous membrane provide the possibility to sample both brain and blood channels for permeability assays and to directly control and manipulate both the brain and the blood compartments simultaneously [49,50]; (ii) brain endothelial cells mimicking physiological functions, forming a high barrier integrity and expressing efflux pumps, which requires precise control of shear walls (viscosity and flow) to maintain polarity [241]. The literature provides some good examples of BBB-on-chip models that meet both of these criteria [49,50,242]. For example, Park et al. used a sandwiched double channel separated with a porous microfluidic chip to model the BBB within [49]. They used a unique, developmentally inspired iPS differentiation protocol to obtain brain endothelial cells seeded into the bottom channel of the chip to mimic the brain vasculature. Primary astrocytes and pericytes were seeded in the upper channel to mimic brain parenchyma. They demonstrated effective levels of barrier function for up to two weeks and the validation of delivery systems that transport drugs and therapeutic antibodies through the human BBB. Recently, Liang and Yoon used a well-based design of the BBB-on-chip with integrated sensors for sensing TEER, which was shown to be more effective compared to previous designs [242].
- -
- Lung-on-Chip: The interaction between the flow of air during inhalation and exhalation and the blood capillaries of the lungs is an important phenomenon to observe. One tangible reason for its importance is pandemic diseases, such as COVID-19 and influenza, as this is where viral or bacterial infections begin; therefore, physiologically relevant lung models can be used to develop effective drugs and treatments to protect the entire body [243,244]. Transparent, flexible, and low-cost OOCs are one of the best options to perform this type of research and investigate lung issues such as disease etiology and drug screening [40]. In most lung-on-chip designs, there are two channels separated by a porous membrane to recapitulate the microphysiological environment of the lung [76]. 1. Air channel: Lung epithelial cells are cultured in air without flowing media; they are nourished via the adjacent channel. 2. Blood channel: Lung endothelial cells are cultured here under flow conditions to recapitulate blood capillaries. Cyclic respiratory motion is another factor that must be considered when developing a physiologically relevant lung-on-a-chip model to recapitulate breathing motions at an exact rhythm, rate, and magnitude, which has been shown to have drastic effects on tissue function [34]. Vacuum chambers are the solution presented to exert a cyclic suction when combined with an elastic material to mimic this biomechanical motion [36]. Huang et al. presented a new design of a lung-on-chip, integrating gelatin hydrogel into a PDMS structure that can be subjected to cyclic stress to recapitulate the breathing motions [245]. This improves the similarity to the real organ as the mechanical properties as well as the stiffness are close to that of the human lung, and the results also better match the in vivo environment. In addition, Si et al. recently investigated the use of the lung-on-chip to model viral infections and rapidly screen therapeutic candidates [83]. They proposed a human lung bronchial airway modeled on-chip with lung epithelial cells and pulmonary endothelium. The chips were tested with a virus (coronavirus 2 (SARS-CoV-2)), and the best therapeutic was introduced accordingly.
- -
- Liver-on-Chip: Drug-induced toxicity is a critical factor in drug development models, and the liver is the organ most vulnerable to potential hazards. Regardless of the research conducted to treat liver disease, studying this organ reduces the number of drug failures. OOCs are considered the best approach for studying the liver because animal studies are expensive, time-consuming, and in some cases inaccurate [246]. Cellular components and biomechanical factors are some of the critical parameters for proper functioning of liver chips. There are several cell types in the liver that maintain the physiological functions, including Kupffer, stellate, and endothelial cells; thus, co-culturing approaches are recommended. Geometry and flow are the most important biomechanical aspects in developing a liver-on-chip. Moreover, liver microvessels are sinusoidal and have mainly rectangular cross-sections [247]. Therefore, when designing vascular sinusoids, aspect ratios and velocities must be accurately calculated to maintain a laminar flow regime (Re <1), which directly affects the compensation of the concentration gradients [43,248]. In this regard, Deng et al. performed a study on liver-on-chips to evaluate hepatoprotective activity [249]. They used a sinusoidal, single microchannel (PDMS-glass) chip seeded with four different hepatic cell lines and perfused with laminar flow. Their observations were promising as they recorded different mechanisms of hepatoprotectants. Kim et al. also used a PDMS-glass chip with straight microchannels and a porous membrane to study liver-on-chips in order to study the metastasis of breast-cancer-derived extracellular vesicles to the liver [246].
- -
- Kidney-on-Chip: The kidney is an organ that balances the body’s fluid and filters the blood. The process of waste removal is an important feature that is closely related to drug composition and toxicity and needs to be monitored accurately [250]. Apart from drugs, there are other conditions that affect the filtration process such as urinary stone disease leading to inflammation, which needs to be thoroughly investigated [251]. Nephrons are small functional units in the kidney that are responsible for purifying the blood [252]. The kidney is composed of various parts, including the glomerulus [253], the proximal [254], and the distal tubule [255], which have been studied individually on a chip. A typical kidney-on-chip has two channels where the urinary lumens are in contact with the interstitial flow. Ultra-filtering is a key consideration in the design of the chip and is tightly controlled by the shear stresses exerted on the cells, which are low (~0.2 dyn/cm2) compared to other organs [256].
- -
- Gut-on-Chip: The gut is a multifunctional organ where orally ingested drugs and nutrients are digested, transported, and absorbed. Therefore, it is an important factor in drug efficacy which must be in concert with the barrier function that blocks certain compounds to protect the body [257]. However, the gut is quite a complex physiological environment as other microbial symbionts also work to promote intestinal health [258]. Studying the gut is a step forward in improving the body’s immune function, and OOCs are providing a superior alternative to other approaches such as in vivo animal studies, which have often failed in the transition of the data to the clinic [259]. The relevant literature distinguishes between two types of gut-on-chips: intestine-on-chip [43,260,261] and colon-on-chip [262,263]. The most commonly used gut-on-chip models have typically two channels connected with a porous media; one is intestinal epithelial and the other is vascular endothelial. Accurate barrier function should be achieved for better results in the stage of designing and fabrication. Further cyclic strains and anaerobic environments are sometimes applied in the corresponding research [264].
- -
- Heart-on-Chip: Heart disease ranks first among potentially fatal diseases worldwide [265]. For this reason, effective and inexpensive drugs for its treatment are especially important to save the lives of many people. Three-dimensional, bioengineered OOCs of the heart are used effectively for drug testing because they can recapitulate the physiological mechanisms and cell interactions associated with the biomechanical factors [266]. “Cardiac motion” (Cardiac motion: the heart’s cyclic motion with a 0.6–2 Hz frequency as a result of the heart beating (40–120 beats per minute) [267]) is due to highly polarized and contractile cells called cardiomyocytes, and their function is directly related to flow rate, calcium ion concentration, and electrical stimuli [268]. Thus, providing cardiac physiology on a chip requires precise design to perform the mechanical, electrical, and chemical functions [269]. In addition, the design of cardiac chip models must take into account the ability of the chip to perform contractility techniques such as muscular thin films and to acquire electrophysiological and morphological data [270]. For example, Liu et al. used a double-channel microfluidic device made of PDMS to model a human heart-on-chip [86]. Their heart-on-chip was lined with vein endothelial cells, induced pluripotent stem cells, and fibroblasts (gingival fibroblasts). The model is expected to be a functional tool for pharmacological studies and personalized medicine.
- -
- Bone-on-Chip: Bones are living tissues that both serve as the structure of the body and produce the major blood cells [271]. It has three main tissues (compact, cancellous, and subchondral), in which different types of bone cells (osteoblasts, osteoclasts, osteocytes, and hematopoietic cells) maintain bone metabolism and blood cell production [272]. Cancellous bone tissue consists of a spongy substance called marrow, which is responsible for blood production in the middle of the bone. Chou et al. have introduced an in vitro model of “bone marrow” (Bone marrow: a sponge-like tissue inside the bone which produces diverse materials, including stem and blood cells [273]) using microfluidics to study toxicities and dysfunction caused by factors such as drugs and radiation. Their chip consists of two channels representing the vasculature and hematopoietic system separated by a porous membrane. They obtained promising results for studying responses to drugs and also to radiation [274]. In another study, Bahmaee et al. presented a new study consisting of a microfluidic device (bioreactor) and a scaffold chamber with a hexagonal pillar pattern to study osteogenesis-on-chip [275]. They claimed that their device is a new and effective platform for testing bone drugs compared to the usual approaches in this field. Additionally, there are related research trends using bone-on-chips to study bone metastasis and metastasis colonization for the purpose of cancer treatment and prevention [276].
- -
- Other Organs: The developing OOC models include different parts and bring revolutionary breakthroughs compared to the previous trend. The skin is the first external organ that protects the body and is very likely to be affected by chemical substances, pollutants, and Ultraviolet Light (UV); thus, conducting research to protect, prevent, or cure corresponding diseases is very important. Previously, optically visible skin layers were studied on chips to mimic the interactions between layers and to investigate the biology behind them [277]. Wufuer et al. designed a three-layer chip representing epidermal, dermal, and endothelial cells to recapitulate the dense skin barrier [278]. They were able to study the drugs and concluded that the chips were suitable for modeling inflammatory skin diseases. In recent studies, skin-on-chip modelers have been looking for new approaches to add hair follicles, sweat glands, and pigmentation for more advanced research [279].
6.1.2. Multi Organ Models
7. Discussion
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Harrison, R. Biology and medicine. Observations on the living developing nerve fiber. Sci. Proc. 1907, 98. [Google Scholar]
- Jedrzejczak-Silicka, M. History of Cell Culture. In New Insights into Cell Culture Technology; Gowder, S.J.T., Ed.; IntechOpen: London, UK, 2017. [Google Scholar]
- PromoCell Scaling up from 2D monolayers to complex 3D cell cultures. Available online: https://www.promocell.com/in-the-lab/scaling-up-from-2d-to-complex-3d-cell-culture/ (accessed on 26 January 2021).
- Hudu, S.A.; Alshrari, A.S.; Syahida, A.; Sekawi, Z. Cell culture, technology: Enhancing the culture of diagnosing human diseases. J. Clin. Diagnostic Res. 2016, 10, DE01–DE05. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saez, A.; Anon, E.; Ghibaudo, M.; du Roure, O.; Di Meglio, J.-M.; Hersen, P.; Silberzan, P.; Buguin, A.; Ladoux, B. Traction forces exerted by epithelial cell sheets. J. Phys. Condens. Matter 2010, 22, 194119. [Google Scholar] [CrossRef] [PubMed]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different. Arch. Med. Sci. 2016, 14, 910–919. [Google Scholar]
- Fennema, E.; Rivron, N.; Rouwkema, J.; van Blitterswijk, C.; De Boer, J. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol. 2013, 31, 108–115. [Google Scholar] [CrossRef]
- Mehta, G.; Hsiao, A.Y.; Ingram, M.; Luker, G.D.; Takayama, S. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J. Control. Release 2012, 164, 192–204. [Google Scholar] [CrossRef] [Green Version]
- Laschke, M.W.; Menger, M.D. Life is 3D: Boosting Spheroid Function for Tissue Engineering. Trends Biotechnol. 2017, 35, 133–144. [Google Scholar] [CrossRef]
- Murphy, K.C.; Whitehead, J.; Zhou, D.; Ho, S.S.; Leach, J.K. Engineering fibrin hydrogels to promote the wound healing potential of mesenchymal stem cell spheroids. Acta Biomater. 2017, 64, 176–186. [Google Scholar] [CrossRef]
- Ryu, N.E.; Lee, S.H.; Park, H. Spheroid Culture System Methods and Applications for Mesenchymal Stem Cells. Cells 2019, 8, 1620. [Google Scholar] [CrossRef] [Green Version]
- Białkowska, K.; Komorowski, P.; Bryszewska, M.; Miłowska, K. Spheroids as a type of three-dimensional cell cultures—examples of methods of preparation and the most important application. Int. J. Mol. Sci. 2020, 21, 6225. [Google Scholar] [CrossRef]
- Lehmann, R.; Lee, C.M.; Shugart, E.C.; Benedetti, M.; Charo, R.A.; Gartner, Z.; Hogan, B.; Knoblich, J.; Nelson, C.M.; Wilson, K.M. Human organoids: A new dimension in cell biology. Mol. Biol. Cell 2019, 30, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Jiao, Y.; Qin, S.; Zhao, W.; Chu, Q.; Wu, K. Organoid technology in disease modelling, drug development, personalized treatment and regeneration medicine. Exp. Hematol. Oncol. 2018, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Koo, B.K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Roux, E.; Bougaran, P.; Dufourcq, P.; Couffinhal, T. Fluid Shear Stress Sensing by the Endothelial Layer. Front. Physiol. 2020, 11, 861. [Google Scholar] [CrossRef] [PubMed]
- Hofer, M.; Lutolf, M.P. Engineering organoids. Nat. Rev. Mater. 2021, 6, 402–420. [Google Scholar] [CrossRef] [PubMed]
- Clinton, J.; McWilliams-Koeppen, P. Initiation, Expansion, and Cryopreservation of Human Primary Tissue-Derived Normal and Diseased Organoids in Embedded Three-Dimensional Culture. Curr. Protoc. Cell Biol. 2019, 82. [Google Scholar] [CrossRef] [Green Version]
- Jang, J.; Yi, H.G.; Cho, D.W. 3D Printed Tissue Models: Present and Future. ACS Biomater. Sci. Eng. 2016, 2, 1722–1731. [Google Scholar] [CrossRef]
- Dey, M.; Ozbolat, I.T. 3D bioprinting of cells, tissues and organs. Sci. Rep. 2020, 10, 14023. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, Y.; Spitz, S.; Turjeman, K.; Selinger, F.; Barenholz, Y.; Ertl, P.; Benny, O.; Bavli, D. Breaking the Third Wall: Implementing 3D-Printing Techniques to Expand the Complexity and Abilities of Multi-Organ-on-a-Chip Devices. Micromachines 2021, 12, 627. [Google Scholar] [CrossRef] [PubMed]
- Kirby, B.J. Micro- and Nanoscale Fluid Mechanics; Cambridge University Press: Cambridge, UK, 2010; ISBN 9783540773405. [Google Scholar]
- Casquillas, G.V.; Houssin, T.; Durieux, L. Microfluidics and Microfluidic Devices: A review. Available online: https://www.elveflow.com/microfluidic-reviews/general-microfluidics/microfluidics-and-microfluidic-device-a-review/ (accessed on 15 October 2021).
- Li, H.-F.; Lin, J.-M. Applications of microfluidic systems in environmental analysis. Anal. Bioanal. Chem. 2009, 393, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Aryasomayajula, A.; Bayat, P.; Rezai, P.; Selvaganapathy, P.R. Microfluidic Devices and Their Applications; Springer: Berlin/Heidelberg, Germany, 2017; ISBN 9783319493473. [Google Scholar]
- Liu, Y.; Sun, L.; Zhang, H.; Shang, L.; Zhao, Y. Microfluidics for Drug Development: From Synthesis to Evaluation. Chem. Rev. 2021, 121, 7468–7529. [Google Scholar] [CrossRef]
- Fiorini, G.S.; Chiu, D.T. Disposable microfluidic devices: Fabrication, function, and application. Biotechniques 2005, 38, 429–446. [Google Scholar] [CrossRef] [Green Version]
- Beißner, N.; Lorenz, T.; Reichl, S. Organ on Chip. In Microsystems for Pharmatechnology: Manipulation of Fluids, Particles, Droplets, and Cells; Dietzel, A., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 299–339. ISBN 978-3-319-26920-7. [Google Scholar]
- Mosig, A.S. Organ-on-chip models: New opportunities for biomedical research. Future Sci. 2016, 3, 2–4. [Google Scholar] [CrossRef] [Green Version]
- Damiati, S.; Kompella, U.B.; Damiati, S.A.; Kodzius, R. Microfluidic devices for drug delivery systems and drug screening. Genes 2018, 9, 103. [Google Scholar] [CrossRef] [Green Version]
- Ingber, D.E. Developmentally inspired human ‘organs on chips’. Development 2018, 145, 10–13. [Google Scholar] [CrossRef] [Green Version]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Ingber, D.E. From mechanobiology to developmentally inspired engineering. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Wikswo, J.P.; Block, F.E., III; Cliffel, D.E.; Goodwin, C.R.; Marasco, C.C.; Markov, D.A.; McLean, D.L.; McLean, J.A.; McKenzie, J.R.; Reiserer, R.S.; et al. Engineering Challenges for Instrumenting and Controlling Integrated Organ-on-Chip Systems. IEEE Trans. Biomed. Eng. 2013, 60, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, Q.; Zourob, M. Organ-on-a-chip engineering: Toward bridging the gap between lab and industry. Biomicrofluidics 2020, 14. [Google Scholar] [CrossRef] [PubMed]
- Hassell, B.A.; Goyal, G.; Lee, E.; Sontheimer-Phelps, A.; Levy, O.; Chen, C.S.; Ingber, D.E. Human Organ Chip Models Recapitulate Orthotopic Lung Cancer Growth, Therapeutic Responses, and Tumor Dormancy In Vitro. Cell Rep. 2017, 21, 508–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benam, K.H.; Villenave, R.; Lucchesi, C.; Varone, A.; Hubeau, C.; Lee, H.-H.; Alves, S.E.; Salmon, M.; Ferrante, T.C.; Weaver, J.C.; et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 2016, 13, 151–157. [Google Scholar] [CrossRef]
- Benam, K.H.; Mazur, M.; Choe, Y.; Ferrante, T.C.; Novak, R.; Ingber, D.E. Human Lung Small Airway-on-a-Chip Protocol. Methods Mol. Biol. 2017, 1612, 345–365. [Google Scholar] [CrossRef]
- Deng, J.; Wei, W.; Chen, Z.; Lin, B.; Zhao, W.; Luo, Y.; Zhang, X. Engineered liver-on-a-chip platform to mimic liver functions and its biomedical applications: A review. Micromachines 2019, 10, 676. [Google Scholar] [CrossRef] [Green Version]
- Moradi, E.; Jalili-Firoozinezhad, S.; Solati-Hashjin, M. Microfluidic organ-on-a-chip models of human liver tissue. Acta Biomater. 2020, 116, 67–83. [Google Scholar] [CrossRef]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A.; et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 2871. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef]
- Bein, A.; Shin, W.; Jalili-Firoozinezhad, S.; Park, M.H.; Sontheimer-Phelps, A.; Tovaglieri, A.; Chalkiadaki, A.; Kim, H.J.; Ingber, D.E. Microfluidic Organ-on-a-Chip Models of Human Intestine. CMGH 2018, 5, 659–668. [Google Scholar] [CrossRef] [Green Version]
- Bhalerao, A.; Sivandzade, F.; Archie, S.R.; Chowdhury, E.A.; Noorani, B.; Cucullo, L. In vitro modeling of the neurovascular unit: Advances in the field. Fluids Barriers CNS 2020, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Mofazzal Jahromi, M.A.; Abdoli, A.; Rahmanian, M.; Bardania, H.; Bayandori, M.; Moosavi Basri, S.M.; Kalbasi, A.; Aref, A.R.; Karimi, M.; Hamblin, M.R. Microfluidic Brain-on-a-Chip: Perspectives for Mimicking Neural System Disorders. Mol. Neurobiol. 2019, 56, 8489–8512. [Google Scholar] [CrossRef] [PubMed]
- Park, T.E.; Mustafaoglu, N.; Herland, A.; Hasselkus, R.; Mannix, R.; FitzGerald, E.A.; Prantil-Baun, R.; Watters, A.; Henry, O.; Benz, M.; et al. Hypoxia-enhanced Blood-Brain Barrier Chip recapitulates human barrier function and shuttling of drugs and antibodies. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Vatine, G.D.; Barrile, R.; Workman, M.J.; Sances, S.; Barriga, B.K.; Rahnama, M.; Barthakur, S.; Kasendra, M.; Lucchesi, C.; Kerns, J.; et al. Human iPSC-Derived Blood-Brain Barrier Chips Enable Disease Modeling and Personalized Medicine Applications. Cell Stem Cell 2019, 24, 995–1005.e6. [Google Scholar] [CrossRef] [PubMed]
- Linville, R.M.; DeStefano, J.G.; Sklar, M.B.; Xu, Z.; Farrell, A.M.; Bogorad, M.I.; Chu, C.; Walczak, P.; Cheng, L.; Mahairaki, V.; et al. Human iPSC-derived blood-brain barrier microvessels: Validation of barrier function and endothelial cell behavior. Biomaterials 2018, 190–191, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Herland, A.; Van Der Meer, A.D.; FitzGerald, E.A.; Park, T.E.; Sleeboom, J.J.F.; Ingber, D.E. Distinct contributions of astrocytes and pericytes to neuroinflammation identified in a 3D human blood-brain barrier on a chip. PLoS One 2016, 11, e0150360. [Google Scholar] [CrossRef] [Green Version]
- Skardal, A.; Murphy, S.V.; Devarasetty, M.; Mead, I.; Kang, H.W.; Seol, Y.J.; Zhang, Y.S.; Shin, S.R.; Zhao, L.; Aleman, J.; et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Maoz, B.M.; Herland, A.; FitzGerald, E.A.; Grevesse, T.; Vidoudez, C.; Pacheco, A.R.; Sheehy, S.P.; Park, T.-E.; Dauth, S.; Mannix, R.; et al. A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nat. Biotechnol. 2018, 36, 865–874. [Google Scholar] [CrossRef]
- Ma, C.; Peng, Y.; Li, H.; Chen, W. Organ-on-a-Chip: A New Paradigm for Drug Development. Trends Pharmacol. Sci. 2021, 42, 119–133. [Google Scholar] [CrossRef]
- Wilkinson, M. Animal Experimentation: Working Towards a Paradigm Change; Brill: Leiden, The Netherlands, 2019; ISBN 9789004391192. [Google Scholar]
- Van Norman, G.A. Limitations of Animal Studies for Predicting Toxicity in Clinical Trials: Is it Time to Rethink Our Current Approach? JACC Basic Transl. Sci. 2019, 4, 845–854. [Google Scholar] [CrossRef]
- Chan, C.Y.; Huang, P.H.; Guo, F.; Ding, X.; Kapur, V.; Mai, J.D.; Yuen, P.K.; Huang, T.J. Accelerating drug discovery via organs-on-chips. Lab Chip 2013, 13, 4697–4710. [Google Scholar] [CrossRef] [Green Version]
- Ingber, D.E. Is it Time for Reviewer 3 to Request Human Organ Chip Experiments Instead of Animal Validation Studies? Adv. Sci. 2020, 7, 2002030. [Google Scholar] [CrossRef]
- Jiang, B.; Zheng, W.; Zhang, W.; Jiang, X. Organs on microfluidic chips: A mini review. Sci. China Chem. 2014, 57, 356–364. [Google Scholar] [CrossRef]
- Zhang, B.; Korolj, A.; Lai, B.F.L.; Radisic, M. Advances in organ-on-a-chip engineering. Nat. Rev. Mater. 2018, 3, 257–278. [Google Scholar] [CrossRef]
- Eduardo, J.; Villalba-Rodr, A.M.; Aguilar-Aguila-Isa, M.A.; Garc, I.E.; Hern, A.; Ahmed, I.; Sharma, A.; Parra-Sald, R.; Iqbal, H.M.N. Organs-on-a-Chip Module: A Review from the Development and Applications Perspective. Micromachines 2018, 9, 536. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Liu, J.; Wang, X.; Feng, L.; Wu, J.; Zhu, X.; Wen, W.; Gong, X. Organ-on-a-chip: Recent breakthroughs and future prospects. Biomed. Eng. 2020, 19, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quan, Y.; Sun, M.; Tan, Z.; Eijkel, J.C.T.; Van Den Berg, A.; Van Der Meer, A.; Xie, Y. Organ-on-a-chip: The next generation platform for risk assessment of radiobiology. RSC Adv. 2020, 10, 39521–39530. [Google Scholar] [CrossRef]
- Yang, J.-W.; Shen, Y.-C.; Lin, K.-C.; Cheng, S.-J.; Chen, S.-L.; Chen, C.-Y.; Kumar, P.V.; Lin, S.-F.; Lu, H.-E.; Chen, G.-Y. Organ-on-a-Chip: Opportunities for Assessing the Toxicity of Particulate Matter. Front. Bioeng. Biotechnol. 2020, 8, 519. [Google Scholar] [CrossRef] [PubMed]
- Dersoir, B.; Salmon, J. Clogging in Micro-Channels: From Colloidal Particle to Clog; Université Rennes: Rennes, France, 2015. [Google Scholar]
- Sarasu, S.; Rama, K. Design and development of organ on chip using microfluidic technology for simulation. In Proceedings of the 2013 International Conference on Optical Imaging Sensor and Security, ICOSS 2013, Coimbatore, India, 2–3 July 2013. [Google Scholar]
- Zheng, F.; Fu, F.; Cheng, Y.; Wang, C.; Zhao, Y.; Gu, Z. Organ-on-a-Chip Systems: Microengineering to Biomimic Living Systems. Small 2016, 12, 2253–2282. [Google Scholar] [CrossRef]
- Tuma, R.F.; Duran, W.N.; Ley, K. Handbook of Physiology: Microcirculation; Academic Press: Cambridge, MA, USA, 2011. [Google Scholar]
- Zakharova, M.; do Carmo, M.A.P.; van der Helm, M.W.; Le-The, H.; de Graaf, M.N.S.; Orlova, V.; van den Berg, A.; van der Meer, A.D.; Broersen, K.; Segerink, L.I. Multiplexed blood–brain barrier organ-on-chip. Lab Chip 2020, 20, 3132–3143. [Google Scholar] [CrossRef]
- Kobayashi, J.; Kikuchi, A.; Aoyagi, T.; Okano, T. Cell sheet tissue engineering: Cell sheet preparation, harvesting/manipulation, and transplantation. J. Biomed. Mater. Res. Part A 2019, 107, 955–967. [Google Scholar] [CrossRef]
- Lee, D.-K.; Kwon, J.Y.; Cho, Y.H. Fabrication of microfluidic channels with various cross-sectional shapes using anisotropic etching of Si and self-alignment. Appl. Phys. A 2019, 125, 1–7. [Google Scholar] [CrossRef]
- Koeppen, B.M.; Stanton, B.A. Berne and Levy Physiology E-Book; Sciences, E.H., Ed.; Elsevier Health Sciences: Amsterdam, The Netherlands, 2017; ISBN 9780323523400. [Google Scholar]
- Wang, X.; Phan, D.T.T.; George, S.C.; Lee, A.P.; Louis, S. An on-chip microfluidic pressure regulator that facilitates reproducible loading of cells and hydrogels into microphysiological system platforms. Lab chip 2017, 16, 868–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaarj, K.; Yoon, J.-Y. Methods of Delivering Mechanical Stimuli to Organ-on-a-Chip. Micromachines 2019, 10, 700. [Google Scholar] [CrossRef] [Green Version]
- Huh, D.; Leslie, D.C.; Matthews, B.D.; Fraser, J.P.; Jurek, S.; Hamilton, G.A.; Thorneloe, K.S.; McAlexander, M.A.; Ingber, D.E. A Human Disease Model of Drug Toxicity–Induced Pulmonary Edema in a Lung-on-a-Chip Microdevice. Sci. Transl. Med. 2012, 4, 159ra147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, A.; Barrile, R.; van der Meer, A.; Mammoto, A.; Mammoto, T.; De Ceunynck, K.; Aisiku, O.; Otieno, M.; Louden, C.; Hamilton, G.; et al. Primary Human Lung Alveolus-on-a-chip Model of Intravascular Thrombosis for Assessment of Therapeutics. Clin. Pharmacol. Ther. 2018, 103, 332–340. [Google Scholar] [CrossRef]
- Deosarkar, S.P.; Prabhakarpandian, B.; Wang, B.; Sheffield, J.B.; Krynska, B.; Kiani, M.F. A Novel Dynamic Neonatal Blood-Brain Barrier on a Chip. PLoS One 2015, 10, e0142725. [Google Scholar] [CrossRef] [Green Version]
- Ahn, S.I.; Sei, Y.J.; Park, H.-J.; Kim, J.; Ryu, Y.; Choi, J.J.; Sung, H.-J.; MacDonald, T.J.; Levey, A.I.; Kim, Y. Microengineered human blood–brain barrier platform for understanding nanoparticle transport mechanisms. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Griep, L.M.; Wolbers, F.; de Wagenaar, B.; ter Braak, P.M.; Weksler, B.B.; Romero, I.A.; Couraud, P.O.; Vermes, I.; van der Meer, A.D.; van den Berg, A. BBB ON CHIP: Microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed. Microdevices 2012, 15, 145–150. [Google Scholar] [CrossRef]
- Shrestha, J.; Ghadiri, M.; Shanmugavel, M.; Razavi Bazaz, S.; Vasilescu, S.; Ding, L.; Ebrahimi Warkiani, M. A rapidly prototyped lung-on-a-chip model using 3D-printed molds. Organs Chip 2019, 1, 100001. [Google Scholar] [CrossRef]
- Thacker, V.V.; Dhar, N.; Sharma, K.; Barrile, R.; Karalis, K.; McKinney, J.D. A lung-on-chip model of early m. Tuberculosis infection reveals an essential role for alveolar epithelial cells in controlling bacterial growth. eLife 2020, 9, 1–73. [Google Scholar] [CrossRef] [PubMed]
- Si, L.; Bai, H.; Rodas, M.; Cao, W.; Oh, C.Y.; Jiang, A.; Moller, R.; Hoagland, D.; Oishi, K.; Horiuchi, S.; et al. A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat. Biomed. Eng. 2021, 5, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Villenave, R.; Wales, S.Q.; Hamkins-Indik, T.; Papafragkou, E.; Weaver, J.C.; Ferrante, T.C.; Bahinski, A.; Elkins, C.A.; Kulka, M.; Ingber, D.E. Human Gut-On-A-Chip Supports Polarized Infection of Coxsackie B1 Virus In Vitro. PLoS One 2017, 12, e0169412. [Google Scholar] [CrossRef]
- Liu, H.; Bolonduro, O.A.; Hu, N.; Ju, J.; Rao, A.A.; Duffy, B.M.; Huang, Z.; Black, L.D.; Timko, B.P. Heart-on-a-Chip Model with Integrated Extra- and Intracellular Bioelectronics for Monitoring Cardiac Electrophysiology under Acute Hypoxia. Nano Lett. 2020, 20, 2585–2593. [Google Scholar] [CrossRef]
- Pennathur, S. Flow control in microfluidics: Are the workhorse flows adequate? Lab Chip 2008, 8, 383–387. [Google Scholar] [CrossRef]
- Unger, M.A.; Chou, H.-P.; Thorsen, T.; Scherer, A.; Quake, S.R. Monolithic Microfabricated Valves and Pumps by Multilayer Soft Lithography. Science 2000, 288, 113–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- PERISTALTIC PUMPS—Flow rate pumps. Available online: https://www.drifton.eu/shop/9-flow-rate-tube-pumps/ (accessed on 19 March 2021).
- Syringe Pumps from LONGER and SHENCHEN. Available online: https://www.drifton.eu/shop/24-syringe-pumps/ (accessed on 19 March 2021).
- Brask, A. Electroosmotic Micropumps; Technical University of Denmark: Lyngby, Denmark, 2005. [Google Scholar]
- Nguyen, N.-T.; Wereley, S.T.; Shaegh, S.A.M. Fundamentals and Applications of Microfluidics; Artech house: Norwood, MA, USA, 2019. [Google Scholar]
- Toh, A.G.G.; Ng, S.H.; Wang, Z. Fabrication and testing of embedded microvalves within PMMA microfluidic devices. Microsyst. Technol. 2009, 15, 1335–1342. [Google Scholar] [CrossRef]
- Siegrist, J.; Gorkin, R.; Clime, L.; Roy, E.; Peytavi, R.; Kido, H.; Bergeron, M.; Veres, T.; Madou, M. Serial siphon valving for centrifugal microfluidic platforms. Microfluid. Nanofluidics 2010, 9, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Guo, L. Modeling of capillary burst valve and siphon with hydrophilic cover for centrifugal microfluidic systems. In Advanced Materials Research; Trans Tech Publications Ltd.: Bäch, Switzerland, 2013; Volume 631–632, pp. 858–863. [Google Scholar]
- Jiang, X.; Lillehoj, P.B. Pneumatic microvalves fabricated by multi-material 3D printing. In Proceedings of the 2017 IEEE 12th International Conference on Nano/Micro Engineered and Molecular Systems (NEMS), Los Angeles, CA, USA, 9–12 April 2017; pp. 38–41. [Google Scholar]
- Woolf, M.S.; Dignan, L.M.; Lewis, H.M.; Tomley, C.J.; Nauman, A.Q.; Landers, J.P. Optically-controlled closable microvalves for polymeric centrifugal microfluidic devices. Lab Chip 2020, 20, 1426–1440. [Google Scholar] [CrossRef]
- Jensen, E.C.; Zeng, Y.; Kim, J.; Mathies, R.A. Microvalve enabled digital microfluidic systems for high-performance biochemical and genetic analysis. JALA—J. Assoc. Lab. Autom. 2010, 15, 455–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dersoir, B.; de Saint Vincent, M.R.; Abkarian, M.; Tabuteau, H. Clogging of a single pore by colloidal particles. Microfluid. Nanofluidics 2015, 19, 953–961. [Google Scholar] [CrossRef]
- Van Der Sman, R.G.M. Simulations of confined suspension flow at multiple length scales. Soft Matter 2009, 5, 4376–4387. [Google Scholar] [CrossRef]
- Pang, L.; Shen, S.; Ma, C.; Ma, T.; Zhang, R.; Tian, C.; Zhao, L.; Liu, W.; Wang, J. Deformability and size-based cancer cell separation using an integrated microfluidic device. Analyst 2015, 140, 7335–7346. [Google Scholar] [CrossRef]
- Dressaire, E.; Alban, S. Clogging of microfluidic systems. Soft Matter 2017, 13, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Sauret, A.; Somszor, K.; Villermaux, E.; Dressaire, E. Growth of clogs in parallel microchannels. Phys. Rev. Fluids 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Duchêne, C.; Filipe, V.; Huille, S.; Lindner, A. Clogging of microfluidic constrictions by monoclonal antibody aggregates: Role of aggregate shape and deformability. Soft Matter 2020, 16, 921–928. [Google Scholar] [CrossRef]
- Yüce, M.; Sert, F.; Torabfam, M.; Parlar, A.; Gürel, B.; Çakır, N.; Dağlıkoca, D.E.; Khan, M.A.; Çapan, Y. Fractionated charge variants of biosimilars: A review of separation methods, structural and functional analysis. Anal. Chim. Acta 2021, 1152. [Google Scholar] [CrossRef]
- Wilson, A. Modeling Clogging in Microfluidic Devices; Tufts University: Medford, MA, USA, 2018. [Google Scholar]
- Charcosset, C. Microfiltration. In Membrane Processes in Biotechnology and Pharmaceutics; Elsevier: Amsterdam, The Netherlands, 2012; pp. 101–141. ISBN 978-0-444-56334-7. [Google Scholar]
- Cheng, Y.; Wang, Y.; Ma, Z.; Wang, W.; Ye, X. A bubble- and clogging-free microfluidic particle separation platform with multi-filtration. Lab Chip 2016, 16, 4517–4526. [Google Scholar] [CrossRef]
- Hourtane, V.; Bodiguel, H.; Colin, A. Dense bubble traffic in microfluidic loops: Selection rules and clogging. Phys. Rev. E 2016, 93, 032607. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Wang, B.; Meng, J.; Zhang, S.; Wang, S. How to Prevent Bubbles in Microfluidic Channels. Langmuir 2021. [Google Scholar] [CrossRef]
- Madou, M.J. Fundamentals of Microfabrication: The Science of Miniaturization, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2018; ISBN 9781482274004. [Google Scholar]
- Kratz, S.R.A.; Höll, G.; Schuller, P.; Ertl, P.; Rothbauer, M. Latest Trends in Biosensing for Microphysiological Organs-on-a-Chip and Body-on-a-Chip Systems. Biosensors 2019, 9, 110. [Google Scholar] [CrossRef] [Green Version]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C. Foundations of MEMS; Prentice Hall: Hoboken, NJ, USA, 2012; ISBN 9780132497367. [Google Scholar]
- Ferrari, E.; Palma, C.; Vesentini, S.; Occhetta, P.; Rasponi, M. Integrating Biosensors in Organs-on-Chip Devices: A Perspective on Current Strategies to Monitor Microphysiological Systems. Biosensors 2020, 10, 110. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, S.; Silverio, V. Introduction to Microfabrication Techniques for Microfluidics Devices; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Baracu, A.M.; Gugoasa, L.A.D. Review—Recent Advances in Microfabrication, Design and Applications of Amperometric Sensors and Biosensors. J. Electrochem. Soc. 2021, 168, 037503. [Google Scholar] [CrossRef]
- Kilic, T.; Navaee, F.; Stradolini, F.; Renaud, P.; Carrara, S. Organs-on-chip monitoring: Sensors and other strategies. Microphysiological Syst. 2018, 1, 1. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Aleman, J.; Shin, S.R.; Kilic, T.; Kim, D.; Mousavi Shaegh, S.A.; Massa, S.; Riahi, R.; Chae, S.; Hu, N.; et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. USA 2017, 114, E2293–E2302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajeddin, A.; Mustafaoglu, N.; Yapici, M.K. On-chip measurement of pH using a microcantilever: A biomimetic design approach. In Proceedings of the 2021 Symposium on Design, Test, Integration & Packaging of MEMS and MOEMS (DTIP), Paris, France, 25–27 August 2021. [Google Scholar] [CrossRef]
- Shaegh, S.A.M.; De Ferrari, F.; Zhang, Y.S.; Nabavinia, M.; Mohammad, N.B.; Ryan, J.; Pourmand, A.; Laukaitis, E.; Sadeghian, R.B.; Nadhman, A.; et al. A microfluidic optical platform for real-time monitoring of pH and oxygen in microfluidic bioreactors and organ-on-chip devices. Biomicrofluidics 2016, 10. [Google Scholar] [CrossRef]
- Tanumihardja, E.; Slaats, R.H.; Van Der Meer, A.D.; Passier, R.; Olthuis, W.; Van Den Berg, A. Measuring Both pH and O2with a Single On-Chip Sensor in Cultures of Human Pluripotent Stem Cell-Derived Cardiomyocytes to Track Induced Changes in Cellular Metabolism. ACS Sensors 2021, 6, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.G.; Chen, C. An arduino-based sensor to measure transendothelial electrical resistance. Sens. Actuators A Phys. 2020, 314, 112216. [Google Scholar] [CrossRef]
- Henry, O.Y.F.; Villenave, R.; Cronce, M.; Leineweber, W.; Benz, M.; Ingber, D.E. Organs-on-Chips with integrated electrodes for Trans-Epithelial Electrical Resistance (TEER) measurements of human epithelial barrier function. Lab Chip 2017, 17, 2264. [Google Scholar] [CrossRef]
- Khalid, M.A.U.; Kim, Y.S.; Ali, M.; Lee, B.G.; Cho, Y.J.; Choi, K.H. A lung cancer-on-chip platform with integrated biosensors for physiological monitoring and toxicity assessment. Biochem. Eng. J. 2020, 155, 107469. [Google Scholar] [CrossRef]
- Anderson, J.M.; Itallie, C.M. Van Physiology and Function of the Tight Junction. Cold Spring Harb. Perspect. Biol. 2009, 1, 2584–2585. [Google Scholar] [CrossRef] [PubMed]
- Harris, T.J.C.; Tepass, U. Adherens junctions: From molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 2010, 117, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Einspanier, R.; Schoen, J. Transepithelial electrical resistance (TEER): A functional parameter to monitor the quality of oviduct epithelial cells cultured on filter supports. Histochem. Cell Biol. 2015, 144, 509–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asif, A.; Kim, K.H.; Jabbar, F.; Kim, S.; Choi, K.H. Real-time sensors for live monitoring of disease and drug analysis in microfluidic model of proximal tubule. Microfluid. Nanofluidics 2020, 24, 1–10. [Google Scholar] [CrossRef]
- Moya Lara, A.; Gabriel Buguña, G.; Ramon i Garcia, E.; Aguiló Llobet, J. Integrated sensors for overcoming organ-on-a-chip monitoring challenges. PhD. Thesis, Universitat Autònoma de Barcelona, Departament de Microelectrònica i Sistemes Electrònics, Barcelona, Spain, 2017. [Google Scholar]
- Wang, M.; Duan, B. Materials and Their Biomedical Applications. Encycl. Biomed. Eng. 2019, 1–3, 135–152. [Google Scholar] [CrossRef]
- Torino, S.; Corrado, B.; Iodice, M.; Coppola, G. PDMS-Based Microfluidic Devices for Cell Culture. Inventions 2018, 3, 65. [Google Scholar] [CrossRef] [Green Version]
- Raj M, K.; Chakraborty, S. PDMS microfluidics: A mini review. J. Appl. Polym. Sci. 2019, 137, 48958. [Google Scholar] [CrossRef]
- Winkler, T.E.; Feil, M.; Stronkman, E.F.G.J.; Matthiesen, I.; Herland, A. Low-cost microphysiological systems: Feasibility study of a tape-based barrier-on-chip for small intestine modeling. Lab Chip 2020, 20, 1212–1226. [Google Scholar] [CrossRef]
- Piruska, A.; Nikcevic, I.; Lee, S.H.; Ahn, C.; Heineman, W.R.; Limbach, P.A.; Seliskar, C.J. The autofluorescence of plastic materials and chips measured under laser irradiation. Lab Chip 2005, 5, 1348–1354. [Google Scholar] [CrossRef]
- Campbell, S.B.; Wu, Q.; Yazbeck, J.; Liu, C.; Okhovatian, S.; Radisic, M. Beyond Polydimethylsiloxane: Alternative Materials for Fabrication of Organ-on-a-Chip Devices and Microphysiological Systems. ACS Biomater. Sci. Eng. 2020. [Google Scholar] [CrossRef]
- Zhou, J.; Vera, E.A.; Veolcker, N.H. Recent developments in PDMS surface modification for microfluidic devices. Electrophoresis 2010, 31, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Holczer, E.; Fürjes, P. Effects of embedded surfactants on the surface properties of PDMS; applicability for autonomous microfluidic systems. Microfluid. Nanofluidics 2017, 21, 81. [Google Scholar] [CrossRef] [Green Version]
- Roman, G.T.; Culbertson, C.T. Surface engineering of poly(dimethylsiloxane) microfluidic devices using transition metal sol-gel chemistry. Langmuir 2006, 22, 4445–4451. [Google Scholar] [CrossRef] [PubMed]
- Slentz, B.E.; Penner, N.A.; Lugowska, E.; Regnier, F. Nanoliter capillary electrochromatography columns based on collocated monolithic support structures molded in poly(dimethyl siloxane). Electrophoresis 2001, 22, 3736–3743. [Google Scholar] [CrossRef]
- Giri, B. Laboratory Methods in Microfluidics; Elsevier: Amsterdam, The Netherlands, 2017; ISBN 9780128132364. [Google Scholar]
- Iliescu, C.; Taylor, H.; Avram, M.; Miao, J.; Franssila, S. A practical guide for the fabrication of microfluidic devices using glass and silicon. Biomicrofluidics 2012, 6, 16505. [Google Scholar] [CrossRef] [Green Version]
- Stucki, J.D.; Guenat, O.T. A microfluidic bubble trap and oscillator. Lab Chip 2015, 15, 4393–4397. [Google Scholar] [CrossRef] [Green Version]
- Harink, B.; Le Gac, S.; Barata, D.; Van Blitterswijk, C.; Habibovic, P. Microtiter plate-sized standalone chip holder for microenvironmental physiological control in gas-impermeable microfluidic devices. Lab Chip 2014, 14, 1816–1820. [Google Scholar] [CrossRef]
- Haq, M.R.; Kim, Y.K.; Kim, J.; Ju, J.; Kim, S.M. Fabrication of all glass microfluidic device with superior chemical and mechanical resistances by glass molding with vitreous carbon mold. J. Micromech. Microeng. 2019, 29, 075010. [Google Scholar] [CrossRef]
- Rodrigues, R.O.; Lima, R.; Gomes, H.T.; Silva, A.M.T. Polymer microfluidic devices: An overview of fabrication methods. U. Porto J. Eng. 2015, 1, 67–79. [Google Scholar] [CrossRef] [Green Version]
- Hirama, H.; Satoh, T.; Sugiura, S.; Shin, K.; Onuki-Nagasaki, R.; Kanamori, T.; Inoue, T. Glass-based organ-on-a-chip device for restricting small molecular absorption. J. Biosci. Bioeng. 2019, 127, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, R.T.; Bowser, M.T. Micro free-flow electrophoresis: Theory and applications. Anal. Bioanal. Chem. 2009, 394, 187–198. [Google Scholar] [CrossRef] [Green Version]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Tsao, C.W.; Hromada, L.; Liu, J.; Kumar, P.; DeVoe, D.L. Low temperature bonding of PMMA and COC microfluidic substrates using UV/ozone surface treatment. Lab Chip 2007, 7, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Gencturk, E.; Mutlu, S.; Ulgen, K.O. Advances in microfluidic devices made from thermoplastics used in cell biology and analyses. Biomicrofluidics 2017, 11. [Google Scholar] [CrossRef]
- Becker, H.; Gärtner, C. Polymer microfabrication technologies for microfluidic systems. Anal. Bioanal. Chem. 2008, 390, 89–111. [Google Scholar] [CrossRef]
- Schaffarczyk, D.; Knaus, J.; Peeters, G.; Scholl, D.; Schwitalla, A.; Koslowski, C.; Cölfen, H. Polyetheretherketone implant surface functionalization technologies and the need for a transparent quality evaluation system. Polym. Int. 2021, 70, 1002–1015. [Google Scholar] [CrossRef]
- Ahmad, Z.; Kumar, K.D.; Saroop, M.; Preschilla, N.; Biswas, A.; Bellare, J.R.; Bhowmick, A.K. Highly transparent thermoplastic elastomer from isotactic polypropylene and styrene/ethylene-butylene/styrene triblock copolymer: Structure-property correlations. Polym. Eng. Sci. 2010, 50, 331–341. [Google Scholar] [CrossRef]
- Becker, H.; Gärtner, C. Polymer microfabrication methods for microfluidic analytical applications. Electrophoresis 2000, 21, 12–26. [Google Scholar] [CrossRef]
- Ren, K.; Zhou, J.; Wu, H. Materials for microfluidic chip fabrication. Acc. Chem. Res. 2013, 46, 2396–2406. [Google Scholar] [CrossRef]
- Trinh, K.T.L.; Thai, D.A.; Chae, W.R.; Lee, N.Y. Rapid Fabrication of Poly(methyl methacrylate) Devices for Lab-ona-Chip Applications Using Acetic Acid and UV Treatment. ACS Omega 2020, 5, 17396–17404. [Google Scholar] [CrossRef]
- Wong, J.F.; Mohan, M.D.; Young, E.W.K.; Simmons, C.A. Integrated electrochemical measurement of endothelial permeability in a 3D hydrogel-based microfluidic vascular model. Biosens. Bioelectron. 2020, 147. [Google Scholar] [CrossRef]
- Goy, C.B.; Chaile, R.E.; Madrid, R.E. Microfluidics and hydrogel: A powerful combination. React. Funct. Polym. 2019, 145, 104314. [Google Scholar] [CrossRef]
- Ahmed, E.M. Hydrogel: Preparation, characterization, and applications: A review. J. Adv. Res. 2015, 6, 105–121. [Google Scholar] [CrossRef] [Green Version]
- Sung, J.H.; Yu, J.; Luo, D.; Shuler, M.L.; March, J.C. Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip 2011, 11, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Tenje, M.; Cantoni, F.; Porras Hernández, A.M.; Searle, S.S.; Johansson, S.; Barbe, L.; Antfolk, M.; Pohlit, H. A practical guide to microfabrication and patterning of hydrogels for biomimetic cell culture scaffolds. Organs Chip 2020, 2, 100003. [Google Scholar] [CrossRef]
- Cabodi, M.; Choi, N.W.; Gleghorn, J.P.; Lee, C.S.D.; Bonassar, L.J.; Stroock, A.D. A microfluidic biomaterial. J. Am. Chem. Soc. 2005, 127, 13788–13789. [Google Scholar] [CrossRef] [PubMed]
- Omidian, H.; Park, K. Introduction to Hydrogels. In Biomedical Applications of Hydrogels Handbook; Springer: New York, NY, USA, 2010; pp. 1–16. [Google Scholar]
- Catoira, M.C.; Fusaro, L.; Di Francesco, D.; Ramella, M.; Boccafoschi, F. Overview of natural hydrogels for regenerative medicine applications. J. Mater. Sci. Mater. Med. 2019, 30, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Li, L.; Luo, C. Gel integration for microfluidic applications. Lab Chip 2016, 16, 1757–1776. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.Y.C.; Homan, K.A.; Robinson, S.S.; Kolesky, D.B.; Duarte, N.; Moisan, A.; Lewis, J.A. Renal reabsorption in 3D vascularized proximal tubule models. Proc. Natl. Acad. Sci. USA 2019, 116, 5399–5404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Yu, H.; Liu, N.; Wang, F.; Lee, G.-B.; Wang, Y.; Liu, L.; Li, W.J. Visible light induced electropolymerization of suspended hydrogel bioscaffolds in a microfluidic chip. Biomater. Sci. 2018, 6, 1371–1378. [Google Scholar] [CrossRef] [PubMed]
- Jeong, K.-H.; Park, D.; Lee, Y.-C. Polymer-based hydrogel scaffolds for skin tissue engineering applications: A mini-review. J. Polym. Res. 2017, 24, 1–10. [Google Scholar] [CrossRef]
- Tsang, V.L.; Chen, A.A.; Cho, L.M.; Jadin, K.D.; Sah, R.L.; DeLong, S.; West, J.L.; Bhatia, S.N. Fabrication of 3D hepatic tissues by additive photopatterning of cellular hydrogels. FASEB J. 2007, 21, 790–801. [Google Scholar] [CrossRef] [Green Version]
- Cuchiara, M.P.; Gould, D.J.; McHale, M.K.; Dickinson, M.E.; West, J.L. Integration of self-assembled microvascular networks with microfabricated PEG-based hydrogels. Adv. Funct. Mater. 2012, 22, 4511–4518. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Lin, B. Materials Used in Microfluidic Devices. In Encyclopedia of Microfluidics and Nanofluidics; Springer: New York, NY, USA, 2008; pp. 1065–1068. [Google Scholar]
- Bhushan, B. Introduction to Nanotechnology. In Springer Handbook of Nanotechnology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1–13. [Google Scholar]
- Maoz, B.M.; Herland, A.; Henry, O.Y.F.; Leineweber, W.D.; Yadid, M.; Doyle, J.; Mannix, R.; Kujala, V.J.; FitzGerald, E.A.; Parker, K.K.; et al. Organs-on-Chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip 2017, 17, 2294–2302. [Google Scholar] [CrossRef]
- Quirós-Solano, W.F.; Gaio, N.; Silvestri, C.; Pandraud, G.; Dekker, R.; Sarro, P.M. Metal and Polymeric Strain Gauges for Si-Based, Monolithically Fabricated Organs-on-Chips. Micromachines 2019, 10, 536. [Google Scholar] [CrossRef] [Green Version]
- Tibbe, M.P.; van der Meer, A.D.; van den Berg, A.; Stamatialis, D.; Segerink, L.I. Membranes for organs-on-chips. In Biomedical Membranes And (Bio)artificial Organs; World Scientific Publishing Co. Pte. Ltd.: Singapore, 2017; pp. 295–321. ISBN 9789813223974. [Google Scholar]
- Liu, C.; Qiao, W.; Wang, C.; Wang, H.; Zhou, Y.; Gu, S.; Xu, W.; Zhuang, Y.; Shi, J.; Yang, H. Effect of poly (lactic acid) porous membrane prepared via phase inversion induced by water droplets on 3T3 cell behavior. Int. J. Biol. Macromol. 2021, 183, 2205–2214. [Google Scholar] [CrossRef]
- Pasman, T.; Grijpma, D.; Stamatialis, D.; Poot, A. Flat and microstructured polymeric membranes in organs-on-chips. J. R. Soc. Interface 2018, 15, 20180351. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.; Lee, N.Y. A facile route for irreversible bonding of plastic-PDMS hybrid microdevices at room temperature. Lab Chip 2010, 10, 1274–1280. [Google Scholar] [CrossRef]
- Lee, K.S.; Ram, R.J. Plastic–PDMS bonding for high pressure hydrolytically stable active microfluidics. Lab Chip 2009, 9, 1618–1624. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.; Greco, G.; Cecchini, M. Polydimethylsiloxane (PDMS) irreversible bonding to untreated plastics and metals for microfluidics applications. APL Mater. 2019, 7, 081108. [Google Scholar] [CrossRef] [Green Version]
- Hesh, C.A.; Qiu, Y.; Lam, W.A. Vascularized microfluidics and the blood-endothelium interface. Micromachines 2020, 11, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J.S.; Stevens, K.R.; Yang, M.T.; Baker, B.M.; Nguyen, D.H.T.; Cohen, D.M.; Toro, E.; Chen, A.A.; Galie, P.A.; Yu, X.; et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 2012, 11, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Xia, Y.; Whitesides, G.M. Soft lithography for micro- and nanoscale patterning. Nat. Protoc. 2010, 5, 491–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sticker, D.; Rothbauer, M.; Lechner, S.; Hehenberger, M.T.; Ertl, P. Multi-layered, membrane-integrated microfluidics based on replica molding of a thiol-ene epoxy thermoset for organ-on-a-chip applications. Lab Chip 2015, 15, 4542–4554. [Google Scholar] [CrossRef]
- Liyu, D.; Nemati, S.H.; Vasdekis, A.E. Solvent-assisted prototyping of microfluidic and optofluidic microsystems in polymers. J. Polym. Sci. Part B Polym. Phys. 2016, 54, 1681–1686. [Google Scholar] [CrossRef] [Green Version]
- Sen, A.K.; Raj, A.; Banerjee, U.; Iqbal, S.R. Soft lithography, molding, and micromachining techniques for polymer micro devices. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2019; Volume 1906, pp. 13–54. [Google Scholar]
- Jung, W.; Jang, S.; Cho, S.; Jeon, H.; Jung, H. Recent Progress in Simple and Cost-Effective Top-Down Lithography for ≈10 nm Scale Nanopatterns: From Edge Lithography to Secondary Sputtering Lithography. Adv. Mater. 2020, 32, 1907101. [Google Scholar] [CrossRef]
- Khadpekar, A.J.; Khan, M.; Sose, A.; Majumder, A. Low Cost and Lithography-free Stamp fabrication for Microcontact Printing. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Cargou, S. Introduction about soft-lithography for microfluidics. Available online: https://www.elveflow.com/microfluidic-reviews/soft-lithography-microfabrication/introduction-about-soft-lithography-and-polymer-molding-for-microfluidic/ (accessed on 8 April 2021).
- Alonso-Amigo, M.G. Polymer Microfabrication for Microarrays, Microreactors and Microfluidics. JALA 2016, 5, 96–101. [Google Scholar] [CrossRef]
- Paoli, R.; Samitier, J. Mimicking the kidney: A key role in organ-on-chip development. Micromachines 2016, 7, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kilic, O.; Pamies, D.; Lavell, E.; Schiapparelli, P.; Feng, Y.; Hartung, T.; Bal-Price, A.; Hogberg, H.T.; Quinones-Hinojosa, A.; Guerrero-Cazares, H.; et al. Brain-on-a-chip model enables analysis of human neuronal differentiation and chemotaxis. Lab Chip 2016, 16, 4152–4162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choe, A.; Keun Ha, S.; Choi, I.; Choi, N.; Hwan Sung, J. Microfluidic Gut-liver chip for reproducing the first pass metabolism. Biomed. Microdevices 2017. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.H.; Tjeung, R.T.; Wang, Z. Hot embossing on polymethyl methacrylate. Proc. Electron. Packag. Technol. Conf. EPTC 2006, 615–621. [Google Scholar] [CrossRef]
- Kricka, L.J.; Fortina, P.; Panaro, N.J.; Wilding, P.; Alonso-Amigo, G.; Becker, H. Fabrication of plastic microchips by hot embossing. Lab Chip 2002, 2, 1–4. [Google Scholar] [CrossRef]
- Becker, H.; Heim, U. Hot embossing as a method for the fabrication of polymer high aspect ratio structures. Sens. Actuators A Phys. 2000, 83, 130–135. [Google Scholar] [CrossRef]
- Yang, W.; Zhiwei, J. Injection moulding of polymers. In Advances in Polymer Processing; Woodhead Publishing: Sawston, UK, 2009; pp. 175–203. [Google Scholar]
- Chen, C.S.; Chen, S.C.; Liao, W.H.; Der Chien, R.; Lin, S.H. Micro injection molding of a micro-fluidic platform. Int. Commun. Heat Mass Transf. 2010, 37, 1290–1294. [Google Scholar] [CrossRef]
- Su, Y.C.; Shah, J.; Lin, L. Implementation and analysis of polymeric microstructure replication by micro injection molding. J. Micromech. Microeng. 2004, 14, 415–422. [Google Scholar] [CrossRef]
- Lee, Y.; Choi, J.W.; Yu, J.; Park, D.; Ha, J.; Son, K.; Lee, S.; Chung, M.; Kim, H.Y.; Jeon, N.L. Microfluidics within a well: An injection-molded plastic array 3D culture platform. Lab Chip 2018, 18, 2433–2440. [Google Scholar] [CrossRef]
- Nielsen, A.V.; Beauchamp, M.J.; Nordin, G.P.; Woolley, A.T. 3D Printed Microfluidics. Annu. Rev. Anal. Chem. 2020, 13, 45–65. [Google Scholar] [CrossRef]
- Aljohani, W.; Ullah, M.W.; Zhang, X.; Yang, G. Bioprinting and its applications in tissue engineering and regenerative medicine. Int. J. Biol. Macromol. 2018, 107, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Gojzewski, H.; Guo, Z.; Grzelachowska, W.; Ridwan, M.G.; Hempenius, M.A.; Grijpma, D.W.; Vancso, G.J. Layer-by-Layer Printing of Photopolymers in 3D: How Weak is the Interface? ACS Appl. Mater. Interfaces 2020, 12, 8908–8914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharjee, N.; Urrios, A.; Kang, S.; Folch, A. The upcoming 3D-printing revolution in microfluidics. Lab Chip 2016, 16, 1720–1742. [Google Scholar] [CrossRef] [Green Version]
- Ahmadian Yazdi, A.; Popma, A.; Wong, W.; Nguyen, T.; Pan, Y.; Xu, J. 3D printing: An emerging tool for novel microfluidics and lab-on-a-chip applications. Microfluid. Nanofluidics 2004, 20, 50. [Google Scholar] [CrossRef]
- Murphy, S.V.; Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef]
- Stansbury, J.W.; Idacavage, M.J. 3D printing with polymers: Challenges among expanding options and opportunities. Dent. Mater. 2016, 32, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.; Emami, K.; Wu, H.; Sun, W. Biofabrication of a three-dimensional liver micro-organ as an in vitro drug metabolism model. Biofabrication 2010, 2, 45004–45015. [Google Scholar] [CrossRef]
- Johnson, B.N.; Lancaster, K.Z.; Hogue, I.B.; Meng, F.; Kong, Y.L.; Enquist, L.W.; McAlpine, M.C. 3D printed nervous system on a chip. Lab Chip 2016, 16, 1393–1400. [Google Scholar] [CrossRef]
- Varone, A.; Nguyen, J.K.; Leng, L.; Barrile, R.; Sliz, J.; Lucchesi, C.; Wen, N.; Gravanis, A.; Hamilton, G.A.; Karalis, K.; et al. A novel organ-chip system emulates three-dimensional architecture of the human epithelia and the mechanical forces acting on it. Biomaterials 2021, 275, 120957. [Google Scholar] [CrossRef]
- Sooriyaarachchi, D.; Zhou, Y.; Maharubin, S.; Tan, G.Z. Microtube-embedded microfluidic devices for potential applications in blood brain barrier research. Procedia Manuf. 2020, 48, 294–301. [Google Scholar] [CrossRef]
- Salman, M.; Marsh, G.; Küsters, I.; Delincé, M.; Di Caprio, G.; Upadhyayula, S.; de Nola, G.; Hunt, R.; Ohashi, K.; Shimizu, F.; et al. An in-vitro BBB-on-a-chip open model of human blood-brain barrier enabling advanced optical imaging. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kratz, S.R.A.; Eilenberger, C.; Schuller, P.; Bachmann, B.; Spitz, S.; Ertl, P.; Rothbauer, M. Characterization of four functional biocompatible pressure-sensitive adhesives for rapid prototyping of cell-based lab-on-a-chip and organ-on-a-chip systems. Sci. Rep. 2019, 9, 9287. [Google Scholar] [CrossRef]
- Ewart, L.; Roth, A. Opportunities and challenges with microphysiological systems: A pharma end-user perspective. Nat. Rev. Drug Discov. 2020, 20, 327–328. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tian, T. Recent advances in an organ-on-a-chip: Biomarker analysis and applications. Anal. Methods 2018, 10, 3122–3130. [Google Scholar] [CrossRef]
- Sengul, E.; Elitas, M. Single-Cell Mechanophenotyping in Microfluidics to Evaluate Behavior of U87 Glioma Cells. Micromachines 2020, 11, 845. [Google Scholar] [CrossRef]
- Mastrangeli, M.; van den Eijnden-van Raaij, J. Organs-on-chip: The way forward. Stem Cell Rep. 2021, 16, 2037–2043. [Google Scholar] [CrossRef]
- Swanson, L.W.; Lichtman, J.W. From Cajal to Connectome and Beyond. Annu. Rev. Neurosci. 2016, 39, 197–216. [Google Scholar] [CrossRef]
- Ludwig, P.E.; Reddy, V.; Varacallo, M. Neuroanatomy, Central Nervous System (CNS); StatPearls Publishing LLC: Treasure Island, FL, USA, 2020. [Google Scholar]
- WHO. Neurological Disorders: Public Health Challenges. Arch. Neurol. 2008, 65, 154. [Google Scholar] [CrossRef]
- Gitler, A.D.; Dhillon, P.; Shorter, J. Neurodegenerative disease: Models, mechanisms, and a new hope. Dis. Model. Mech. 2017, 10, 499. [Google Scholar] [CrossRef] [Green Version]
- Winner, B.; Kohl, Z.; Gage, F.H. Neurodegenerative disease and adult neurogenesis. Eur. J. Neurosci. 2011, 33, 1139–1151. [Google Scholar] [CrossRef]
- Slanzi, A.; Iannoto, G.; Rossi, B.; Zenaro, E.; Constantin, G. In vitro Models of Neurodegenerative Diseases. Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Nichols, K.; Koppes, R.; Koppes, A. Recent advancements in microphysiological systems for neural development and disease. Curr. Opin. Biomed. Eng. 2020, 14, 42–51. [Google Scholar] [CrossRef]
- Shepherd, G.M.; Marenco, L.; Hines, M.L.; Migliore, M.; McDougal, R.A.; Carnevale, N.T.; Newton, A.J.H.; Surles-Zeigler, M.; Ascoli, G.A. Neuron Names: A Gene- and Property-Based Name Format, With Special Reference to Cortical Neurons. Front. Neuroanat. 2019, 13, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndyabawe, K.; Kisaalita, W.S. Engineering microsystems to recapitulate brain physiology on a chip. Drug Discov. Today 2019, 24, 1725–1730. [Google Scholar] [CrossRef]
- Dauth, S.; Maoz, B.M.; Sheehy, S.P.; Hemphill, M.A.; Murty, T.; Macedonia, M.K.; Greer, A.M.; Budnik, B.; Parker, K.K. Neurons derived from different brain regions are inherently different in vitro: A novel multiregional brain-on-a-chip. J. Neurophysiol. 2017, 117, 1320–1341. [Google Scholar] [CrossRef] [PubMed]
- Stafstrom, C.E.; Carmant, L. Seizures and Epilepsy: An Overview for Neuroscientists. Cold Spring Harb. Perspect. Med. 2015, 5, 1–19. [Google Scholar] [CrossRef]
- Manford, M. Recent advances in epilepsy. J. Neurol. 2017, 264, 1811. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Sternberg, A.R.; Ghiasvand, S.; Berdichevsky, Y. Epilepsy-on-a-chip system for antiepileptic drug discovery. IEEE Trans. Biomed. Eng. 2019, 66, 1231–1241. [Google Scholar] [CrossRef]
- Park, J.; Lee, K.; Jeong, S.; Hyun, K.; Justin, C.; Lee, S. Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of Alzheimer’s disease. Lab Chip 2014. [Google Scholar] [CrossRef]
- Anderson, W.A.; Bosak, A.; Hogberg, H.T.; Hartung, T.; Moore, M.J. Advances in 3D neuronal microphysiological systems: Towards a functional nervous system on a chip. Vitr. Cell. Dev. Biol. Anim. 2021. [Google Scholar] [CrossRef]
- Lee, S.; Chung, M.; Lee, S.R.; Jeon, N.L. 3D brain angiogenesis model to reconstitute functional human blood–brain barrier in vitro. Biotechnol. Bioeng. 2020, 117, 748–762. [Google Scholar] [CrossRef]
- Daneman, R.; Prat, A. The Blood–Brain Barrier. Cold Spring Harb. Perspect. Biol. 2015, 7, a020412. [Google Scholar] [CrossRef] [Green Version]
- Banks, W.A. The blood brain barrier. Neuroimmune Pharmacol. 2008, 21–38. [Google Scholar] [CrossRef]
- Sahtoe, D.D.; Coscia, A.; Mustafaoglu, N.; Miller, L.M.; Olal, D.; Vulovic, I.; Yu, T.-Y.; Goreshnik, I.; Lin, Y.-R.; Clark, L.; et al. Transferrin receptor targeting by de novo sheet extension. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Morad, G.; Carman, C.V.; Hagedorn, E.J.; Perlin, J.R.; Zon, L.I.; Mustafaoglu, N.; Park, T.-E.; Ingber, D.E.; Daisy, C.C.; Moses, M.A. Tumor-Derived Extracellular Vesicles Breach the Intact Blood–Brain Barrier via Transcytosis. ACS Nano 2019, 13, 13853–13865. [Google Scholar] [CrossRef] [PubMed]
- Wevers, N.R.; Kasi, D.G.; Gray, T.; Wilschut, K.J.; Smith, B.; van Vught, R.; Shimizu, F.; Sano, Y.; Kanda, T.; Marsh, G.; et al. A perfused human blood–brain barrier on-a-chip for high-throughput assessment of barrier function and antibody transport. Fluids Barriers CNS 2018, 15, 23. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Lin, B.; Shao, W.; Zhu, Z.; Ji, T.; Yang, C. In Vitro and in Vivo Studies on the Transport of PEGylated Silica Nanoparticles across the Blood–Brain Barrier. ACS Appl. Mater. Interfaces 2014, 6, 2131–2136. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, E.A.; Noorani, B.; Alqahtani, F.; Bhalerao, A.; Raut, S.; Sivandzade, F.; Cucullo, L. Understanding the brain uptake and permeability of small molecules through the BBB: A technical overview. J. Cereb. Blood Flow Metab. 2021, 0271678X20985946. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Yoon, J.Y. In situ sensors for blood-brain barrier (BBB) on a chip. Sens. Actuators Rep. 2021, 3, 100031. [Google Scholar] [CrossRef]
- Jain, A. COVID-19 and lung pathology. Indian J. Pathol. Microbiol. 2020, 63, 171–172. [Google Scholar] [CrossRef]
- Wyss Institute at Harvard Lung-on-a-Chip. Available online: https://wyss.harvard.edu/media-post/lung-on-a-chip/ (accessed on 23 January 2021).
- Huang, D.; Liu, T.; Liao, J.; Maharjan, S.; Xie, X.; Pérez, M.; Anaya, I.; Wang, S.; Mayer, A.T.; Kang, Z.; et al. Reversed-engineered human alveolar lung-on-a-chip model. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Kim, J.; Lee, C.; Kim, I.; Ro, J.; Kim, J.; Min, Y.; Park, J.; Sunkara, V.; Park, Y.-S.; Michael, I.; et al. Three-Dimensional Human Liver-Chip Emulating Premetastatic Niche Formation by Breast Cancer-Derived Extracellular Vesicles. ACS Nano 2020, 14, 14971–14988. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, A.; Duche, D.; Ouedraogo, G.; Nahmias, Y. Challenges and Opportunities in the Design of Liver-on-Chip Microdevices. Annu. Rev. Biomed. Eng. 2019, 21, 219–239. [Google Scholar] [CrossRef] [PubMed]
- Pindera, M.Z.; Ding, H.; Athavale, M.M.; Chen, Z. Accuracy of 1D microvascular flow models in the limit of low Reynolds numbers. Microvasc. Res. 2009, 77, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Cong, Y.; Han, X.; Wei, W.; Lu, Y.; Liu, T.; Zhao, W.; Lin, B.; Luo, Y.; Zhang, X. A liver-on-a-chip for hepatoprotective activity assessment. Biomicrofluidics 2020, 14, 064107. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, F. Kidney-on-a-Chip; Elsevier Inc.: Amsterdam, The Netherlands, 2019; Volume 2, ISBN 9780128172025. [Google Scholar]
- Tang, X.; Lieske, J.C. Acute and chronic kidney injury in nephrolithiasis. Curr. Opin. Nephrol. Hypertens. 2014, 23, 385–390. [Google Scholar] [CrossRef] [Green Version]
- Britannica The Editors of Encyclopaedia. “Nephron”. Available online: https://www.britannica.com/science/nephron (accessed on 29 January 2021).
- Ashammakhi, N.; Wesseling-Perry, K.; Hasan, A.; Elkhammas, E.; Zhang, Y.S. Kidney-on-a-chip: Untapped opportunities. Kidney Int. 2018, 94, 1073–1086. [Google Scholar] [CrossRef] [PubMed]
- Sakolish, C.M.; Philip, B.; Mahler, G.J. A human proximal tubule-on-a-chip to study renal disease and toxicity. Biomicrofluidics 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.J.; Suh, K.Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 2010, 10, 36–42. [Google Scholar] [CrossRef]
- Kim, S.; Takayama, S. Organ-on-a-chip and the kidney. Kidney Res. Clin. Pract. 2015, 34, 165–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashammakhi, N.; Nasiri, R.; de Barros, N.R.; Tebon, P.; Thakor, J.; Goudie, M.; Shamloo, A.; Martin, M.G.; Khademhosseni, A. Gut-on-a-chip: Current progress and future opportunities. Biomaterials 2020, 255, 120196. [Google Scholar] [CrossRef]
- Guinane, C.M.; Cotter, P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Therap. Adv. Gastroenterol. 2013, 6, 295–308. [Google Scholar] [CrossRef] [Green Version]
- Shanks, N.; Greek, R.; Greek, J. Are animal models predictive for humans? Philos. Ethics Humanit. Med. 2009, 4, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Kasendra, M.; Luc, R.; Yin, J.; Manatakis, D.V.; Kulkarni, G.; Lucchesi, C.; Sliz, J.; Apostolou, A.; Sunuwar, L.; Obrigewitch, J.; et al. Duodenum Intestine-Chip for preclinical drug assessment in a human relevant model. eLife 2020, 9, e50135. [Google Scholar] [CrossRef] [PubMed]
- Gijzen, L.; Marescotti, D.; Raineri, E.; Nicolas, A.; Lanz, H.L.; Guerrera, D.; van Vught, R.; Joore, J.; Vulto, P.; Peitsch, M.C.; et al. An Intestine-on-a-Chip Model of Plug-and-Play Modularity to Study Inflammatory Processes. SLAS Technol. Transl. Life Sci. Innov. 2020, 25, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Sontheimer-Phelps, A.; Chou, D.B.; Tovaglieri, A.; Ferrante, T.C.; Duckworth, T.; Fadel, C.; Frismantas, V.; Sutherland, A.D.; Jalili-Firoozinezhad, S.; Kasendra, M.; et al. Human Colon-on-a-Chip Enables Continuous In Vitro Analysis of Colon Mucus Layer Accumulation and Physiology. CMGH 2020, 9, 507–526. [Google Scholar] [CrossRef] [Green Version]
- Gazzaniga, F.S.; Camacho, D.M.; Wu, M.; Silva Palazzo, M.F.; Dinis, A.L.M.; Grafton, F.N.; Cartwright, M.J.; Super, M.; Kasper, D.L.; Ingber, D.E. Harnessing Colon Chip Technology to Identify Commensal Bacteria That Promote Host Tolerance to Infection. Front. Cell. Infect. Microbiol. 2021, 11, 105. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Wen, H.; Yu, Y.; Li, M.; Fu, X.; Huang, S. Gut-on-chip: Recreating human intestine in vitro. J. Tissue Eng. 2020, 11. [Google Scholar] [CrossRef]
- WHO. Cardiovascular Diseases. Available online: https://www.who.int/health-topics/cardiovascular-diseases#tab=tab_1 (accessed on 31 January 2021).
- Ribas, J.; Sadeghi, H.; Manbachi, A.; Leijten, J.; Brinegar, K.; Zhang, Y.S.; Ferreira, L.; Khademhosseini, A. Cardiovascular Organ-on-a-Chip Platforms for Drug Discovery and Development. Appl. Vitr. Toxicol. 2016, 2, 82–96. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Vidan, E.; Bergman, G.W. Cardiac Motion of Coronary Arteries: Variability in the Rest Period and Implications for Coronary MR Angiography1. Radiology 1999, 213, 751–758. [Google Scholar] [CrossRef]
- Polini, A.; Prodanov, L.; Bhise, N.S.; Manoharan, V.; Dokmeci, M.R.; Khademhosseini, A. Organs-on-a-chip: A new tool for drug discovery. Expert Opin. Drug Discov. 2014, 9, 335–352. [Google Scholar] [CrossRef]
- Abulaiti, M.; Yalikun, Y.; Murata, K.; Sato, A.; Sami, M.M.; Sasaki, Y.; Fujiwara, Y.; Minatoya, K.; Shiba, Y.; Tanaka, Y.; et al. Establishment of a heart-on-a-chip microdevice based on human iPS cells for the evaluation of human heart tissue function. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Feinberg, A.W.; Feigel, A.; Shevkoplyas, S.S.; Sheehy, S.; Whitesides, G.M.; Parker, K.K. Muscular thin films for building actuators and powering devices. Science 2007, 317, 1366–1370. [Google Scholar] [CrossRef] [Green Version]
- Florencio-Silva, R.; Sasso, G.R.D.S.; Sasso-Cerri, E.; Simões, M.J.; Cerri, P.S. Biology of Bone Tissue: Structure, Function, and Factors That Influence Bone Cells. Biomed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Porwit, A.; McCullough, J.J.; Erber, W.N. Blood and Bone Marrow Pathology; ClinicalKey 2012; Churchill Livingstone: London, UK, 2011; ISBN 9780702031472. [Google Scholar]
- Lindberg, M.R.; Lamps, L.W. Bone Marrow; Elsevier: Amsterdam, The Netherlands, 2018; ISBN 978-0-323-54803-8. [Google Scholar]
- Chou, D.B.; Frismantas, V.; Milton, Y.; David, R.; Pop-Damkov, P.; Ferguson, D.; MacDonald, A.; Vargel Bölükbaşı, Ö.; Joyce, C.E.; Moreira Teixeira, L.S.; et al. On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology. Nat. Biomed. Eng. 2020, 4, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Bahmaee, H.; Owen, R.; Boyle, L.; Perrault, C.M.; Garcia-Granada, A.A.; Reilly, G.C.; Claeyssens, F. Design and Evaluation of an Osteogenesis-on-a-Chip Microfluidic Device Incorporating 3D Cell Culture. Front. Bioeng. Biotechnol. 2020, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Marturano-Kruik, A.; Nava, M.M.; Yeager, K.; Chramiec, A.; Hao, L.; Robinson, S.; Guo, E.; Raimondi, M.T.; Vunjak-Novakovic, G. Human bone perivascular niche-on-a-chip for studying metastatic colonization. Proc. Natl. Acad. Sci. USA 2018, 115, 1256–1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sriram, G.; Alberti, M.; Dancik, Y.; Wu, B.; Wu, R.; Feng, Z.; Ramasamy, S.; Bigliardi, P.L.; Bigliardi-Qi, M.; Wang, Z. Full-thickness human skin-on-chip with enhanced epidermal morphogenesis and barrier function. Mater. Today 2018, 21, 326–340. [Google Scholar] [CrossRef]
- Wufuer, M.; Lee, G.H.; Hur, W.; Jeon, B.; Kim, B.J.; Choi, T.H.; Lee, S.H. Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sutterby, E.; Thurgood, P.; Baratchi, S.; Khoshmanesh, K.; Pirogova, E. Microfluidic Skin-on-a-Chip Models: Toward Biomimetic Artificial Skin. Small 2020, 16. [Google Scholar] [CrossRef]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-chips: Into the next decade. Nat. Rev. Drug Discov. 2020, 20, 345–361. [Google Scholar] [CrossRef]
- Zhu, J.; Zhu, J. Application of Organ-on-Chip in Drug Discovery. J. Biosci. Med. 2020, 8, 119–134. [Google Scholar] [CrossRef] [Green Version]
- Meet Chip: Female Reproductive System | National Center for Advancing Translational Sciences. Available online: https://ncats.nih.gov/tissuechip/chip/female (accessed on 29 September 2021).
- Wright, C.B.; Becker, S.M.; Low, L.A.; Tagle, D.A.; Sieving, P.A. Improved Ocular Tissue Models and Eye-On-A-Chip Technologies Will Facilitate Ophthalmic Drug Development. J. Ocul. Pharmacol. Ther. 2020, 36, 25. [Google Scholar] [CrossRef] [Green Version]
- Peng, Z.; Zhou, L.; Wong, J.K.W.; Chan, Y.K. Eye-on-a-chip (EOC) models and their role in the future of ophthalmic drug discovery. Expert Rev. Ophthalmol. 2020, 15, 259–261. [Google Scholar] [CrossRef]
- Mancini, V.; Pensabene, V. Organs-on-chip models of the female reproductive system. Bioengineering 2019, 6, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blundell, C.; Tess, E.R.; Schanzer, A.S.R.; Coutifaris, C.; Su, E.J.; Parry, S.; Huh, D. A microphysiological model of the human placental barrier. Lab Chip 2016, 16, 3065–3073. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-X.; Liang, G.-T.; Yan, W.; Zhang, Q.; Wang, W.; Zhou, X.-M.; Liu, D.-Y. Artificial Uterus on a Microfluidic Chip. Chin. J. Anal. Chem. 2013, 41, 467–472. [Google Scholar] [CrossRef]
- Nagashima, J.B.; El Assal, R.; Songsasen, N.; Demirci, U. Evaluation of an ovary-on-a-chip in large mammalian models: Species specificity and influence of follicle isolation status. J. Tissue Eng. Regen. Med. 2018, 12, e1926–e1935. [Google Scholar] [CrossRef] [PubMed]
- Kyoto University. Eye Blinking on-a-Chip: A New Approach Could Lead to “Cornea-on-a-Chip” Devices That More Accurately Test the Effects of Drugs on the Human Eye; ScienceDaily: Kyoto, Japan, 2020. [Google Scholar]
- Polini, A.; del Mercato, L.L.; Barra, A.; Zhang, Y.S.; Calabi, F.; Gigli, G. Towards the development of human immune-system-on-a-chip platforms. Drug Discov. Today 2019, 24, 517–525. [Google Scholar] [CrossRef]
- Herland, A.; Maoz, B.M.; Das, D.; Somayaji, M.R.; Prantil-Baun, R.; Novak, R.; Cronce, M.; Huffstater, T.; Jeanty, S.S.F.; Ingram, M.; et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat. Biomed. Eng. 2020, 4, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Shanti, A.; Hallfors, N.; Petroianu, G.A.; Planelles, L.; Stefanini, C. Lymph Nodes-On-Chip: Promising Immune Platforms for Pharmacological and Toxicological Applications. Front. Pharmacol. 2021, 2132. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Kankala, R.K.; Wang, S.-B.; Chen, A.-Z. Multi-Organs-on-Chips: Towards Long-Term Biomedical Investigations. Molecules 2019, 24, 675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elufioye, T.O.; Badal, S. Chapter 1—Background to Pharmacognosy. In Pharmacognosy; Badal, S., Delgoda, R.B.T.-P., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 3–13. ISBN 978-0-12-802104-0. [Google Scholar]
- Palaninathan, V.; Kumar, V.; Maekawa, T.; Liepmann, D.; Paulmurugan, R.; Eswara, J.R.; Ajayan, P.M.; Augustine, S.; Malhotra, B.D.; Viswanathan, S.; et al. Multi-organ on a chip for personalized precision medicine. MRS Commun. 2018, 8, 652–667. [Google Scholar] [CrossRef]
- Liu, D.; Jiao, S.; Wei, J.; Zhang, X.; Pei, Y.; Pei, Z.; Li, J.; Du, Y. Investigation of absorption, metabolism and toxicity of ginsenosides compound K based on human organ chips. Int. J. Pharm. 2020, 587, 119669. [Google Scholar] [CrossRef]
- Novak, R.; Ingram, M.; Marquez, S.; Das, D.; Delahanty, A.; Herland, A.; Maoz, B.M.; Jeanty, S.S.F.; Somayaji, M.R.; Burt, M.; et al. Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat. Biomed. Eng. 2020, 4, 407–420. [Google Scholar] [CrossRef]
- Schimek, K.; Frentzel, S.; Luettich, K.; Bovard, D.; Rütschle, I.; Boden, L.; Rambo, F.; Erfurth, H.; Dehne, E.M.; Winter, A.; et al. Human multi-organ chip co-culture of bronchial lung culture and liver spheroids for substance exposure studies. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Mastrangeli, M.; Millet, S.; ORCHID partners; van den Eijnden-van Raaij, J. Organ-on-Chip In Development:Towards a roadmap for Organs-on-Chip. Preprints 2019. [Google Scholar] [CrossRef]
- Esch, M.B.; Smith, A.S.T.; Prot, J.M.; Oleaga, C.; Hickman, J.J.; Shuler, M.L. How multi-organ microdevices can help foster drug development. Adv. Drug Deliv. Rev. 2014, 69–70, 158–169. [Google Scholar] [CrossRef] [Green Version]
- Jivani, R.R.; Lakhtaria, G.J.; Patadiya, D.D.; Patel, L.D.; Jivani, N.P.; Jhala, B.P. Biomedical microelectromechanical systems (BioMEMS): Revolution in drug delivery and analytical techniques. Saudi Pharm. J. SPJ 2016, 24, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Fischer, A.C.; Forsberg, F.; Lapisa, M.; Bleiker, S.J.; Stemme, G.; Roxhed, N.; Niklaus, F. Integrating MEMS and ICs. Microsyst. Nanoeng. 2015, 1, 15005. [Google Scholar] [CrossRef] [Green Version]
- Sip, C.G.; Folch, A. Stable chemical bonding of porous membranes and poly(dimethylsiloxane) devices for long-term cell culture. Biomicrofluidics 2014, 8, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
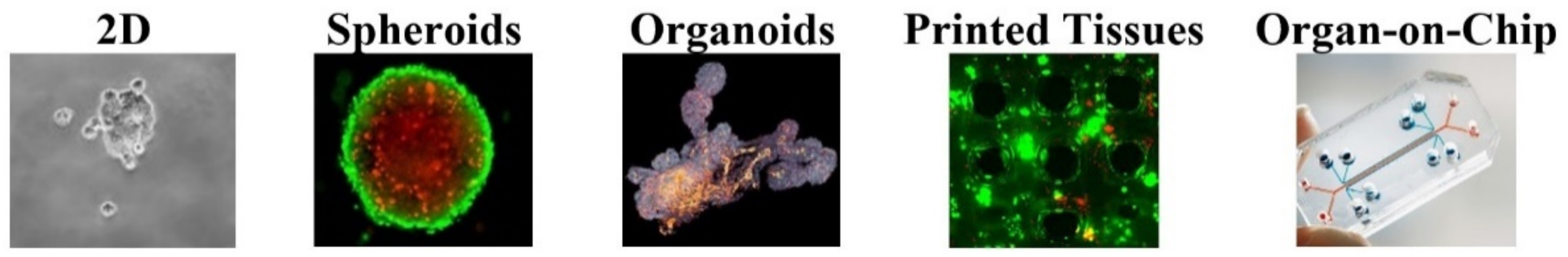
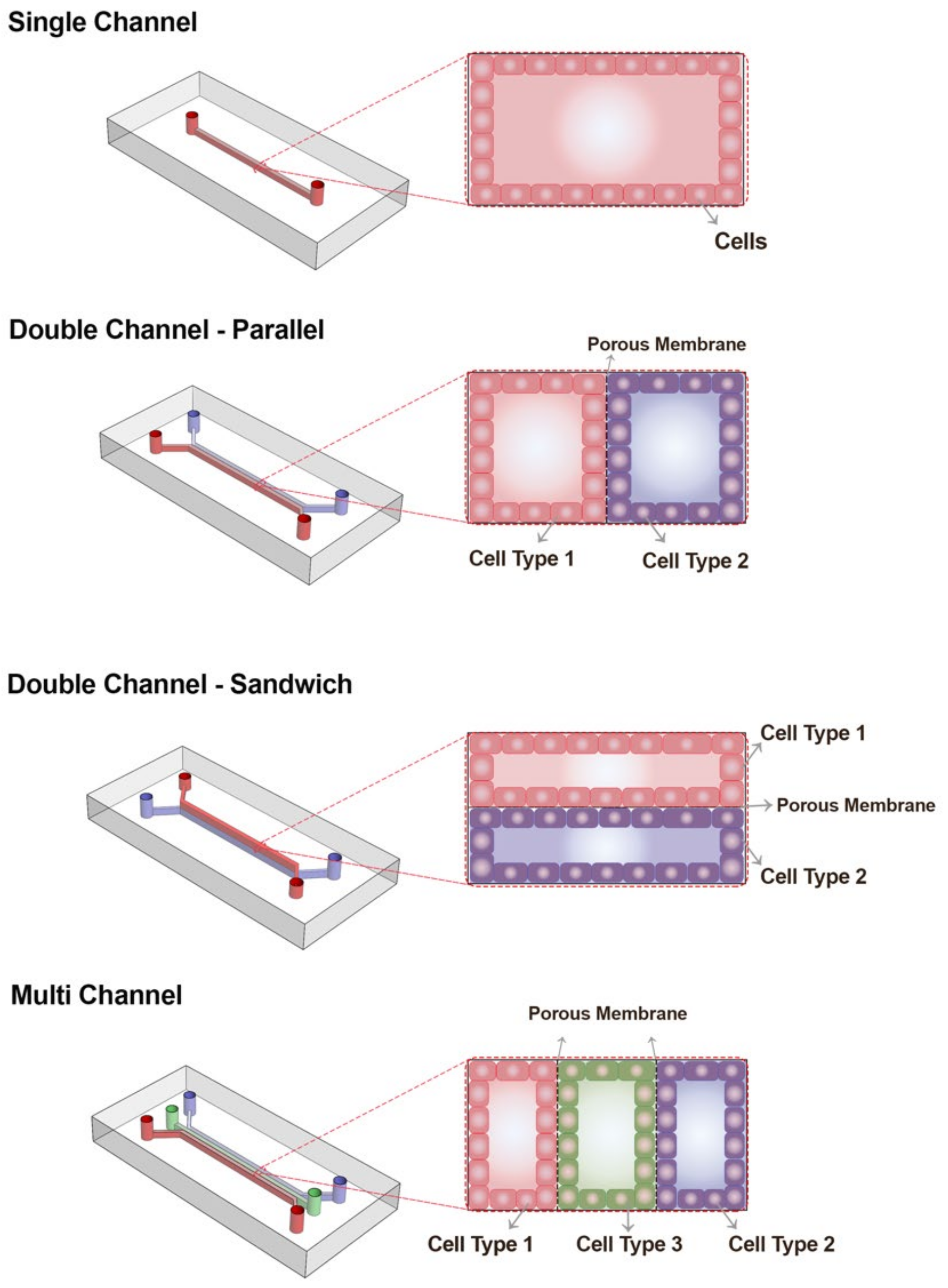
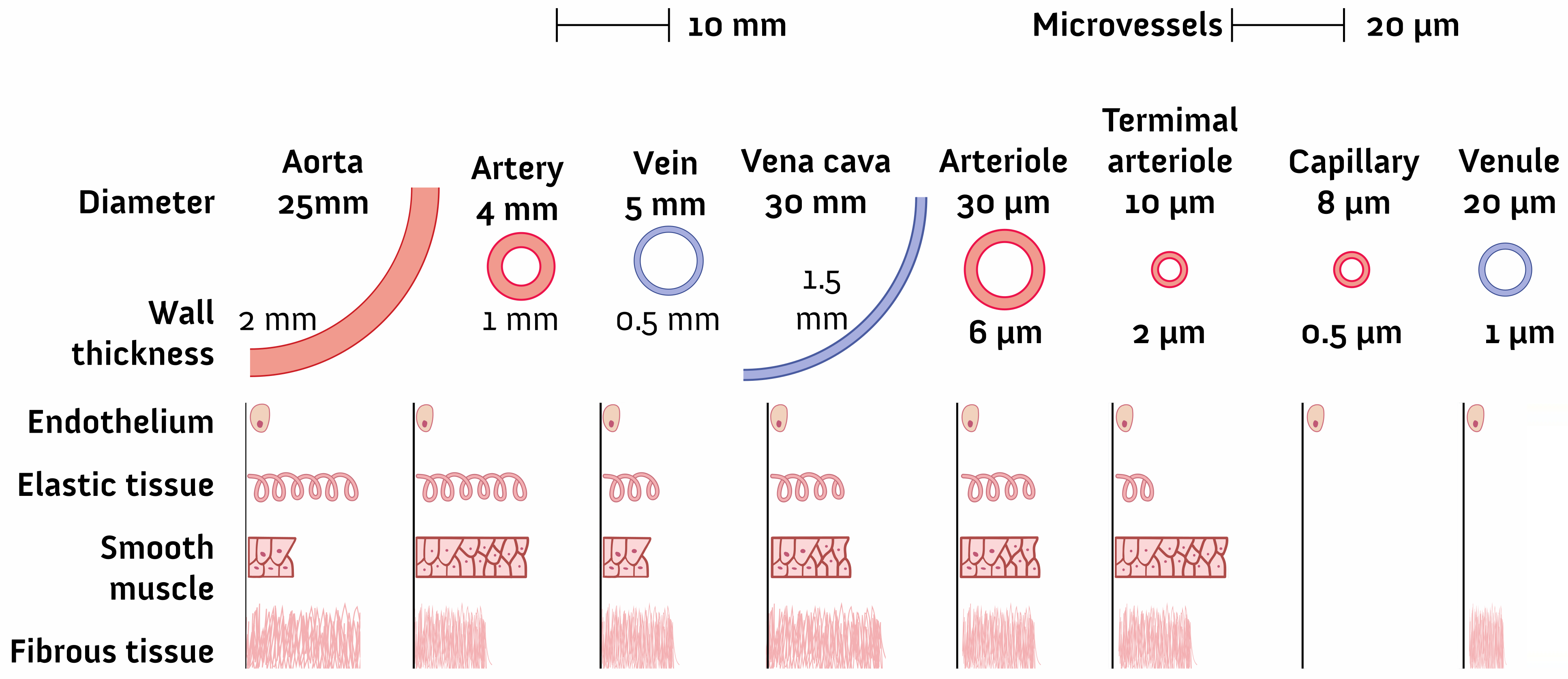
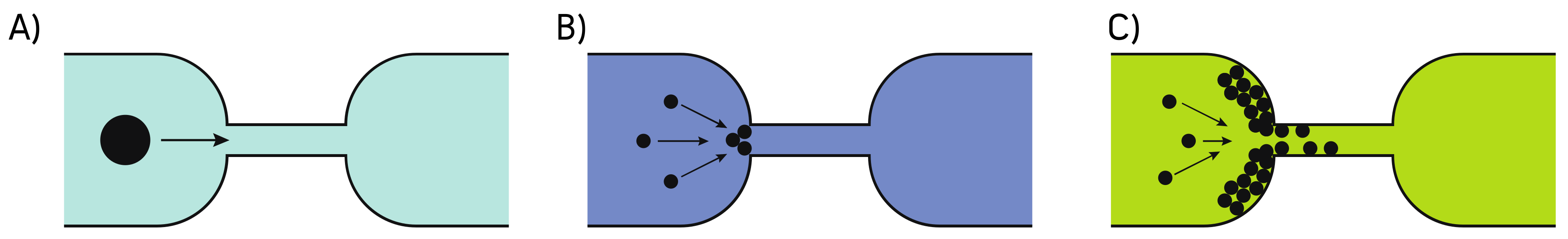

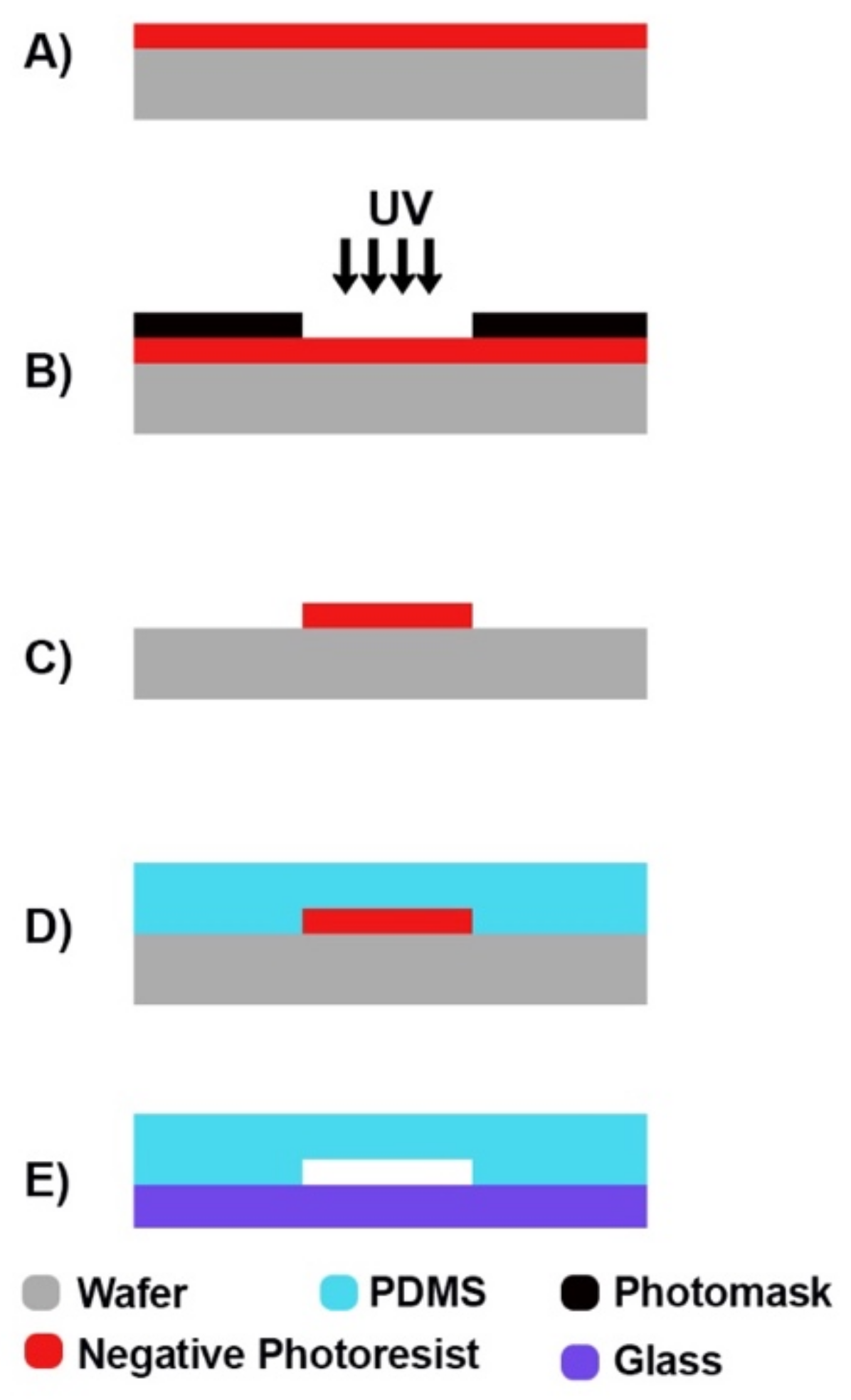
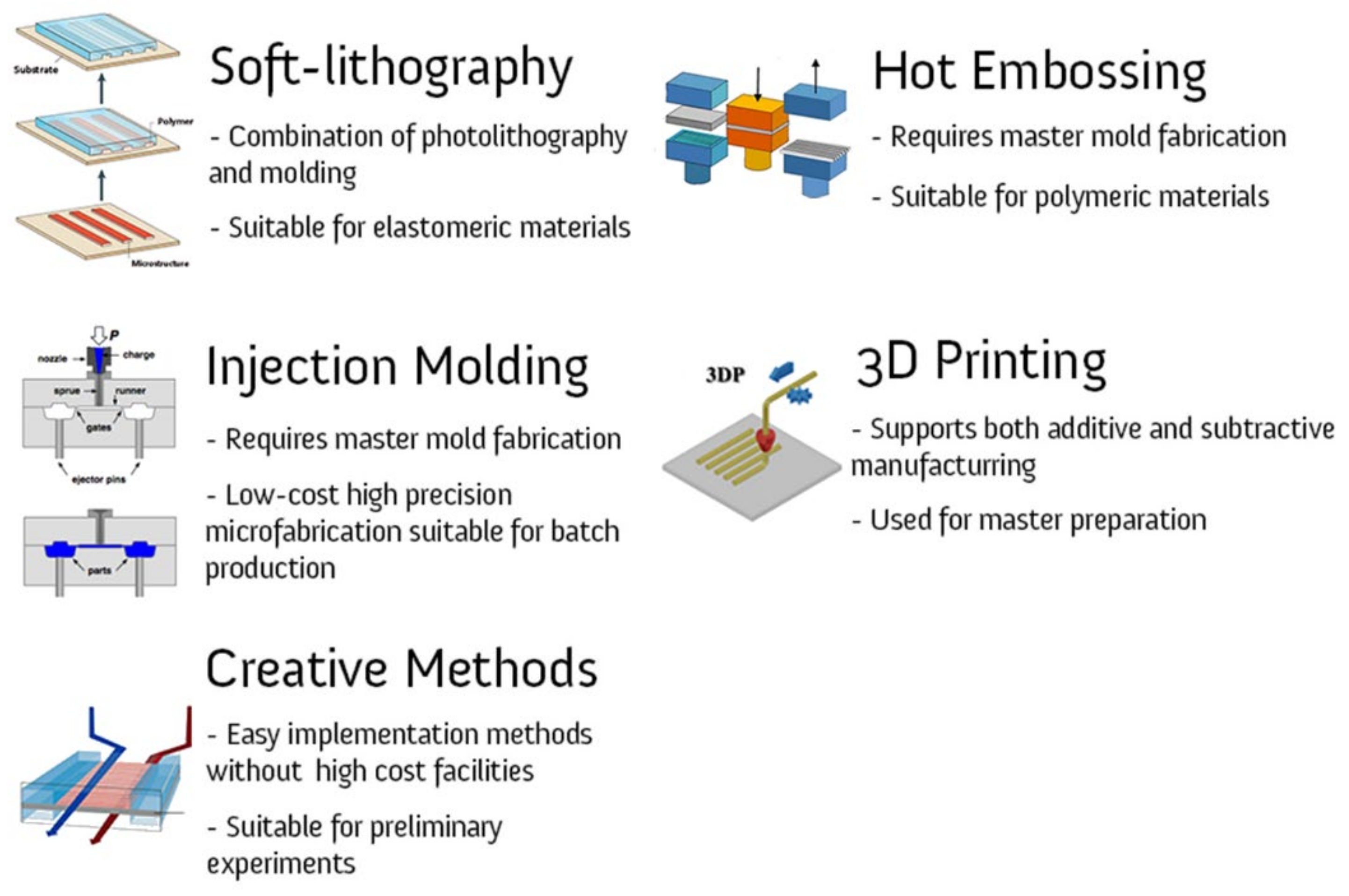
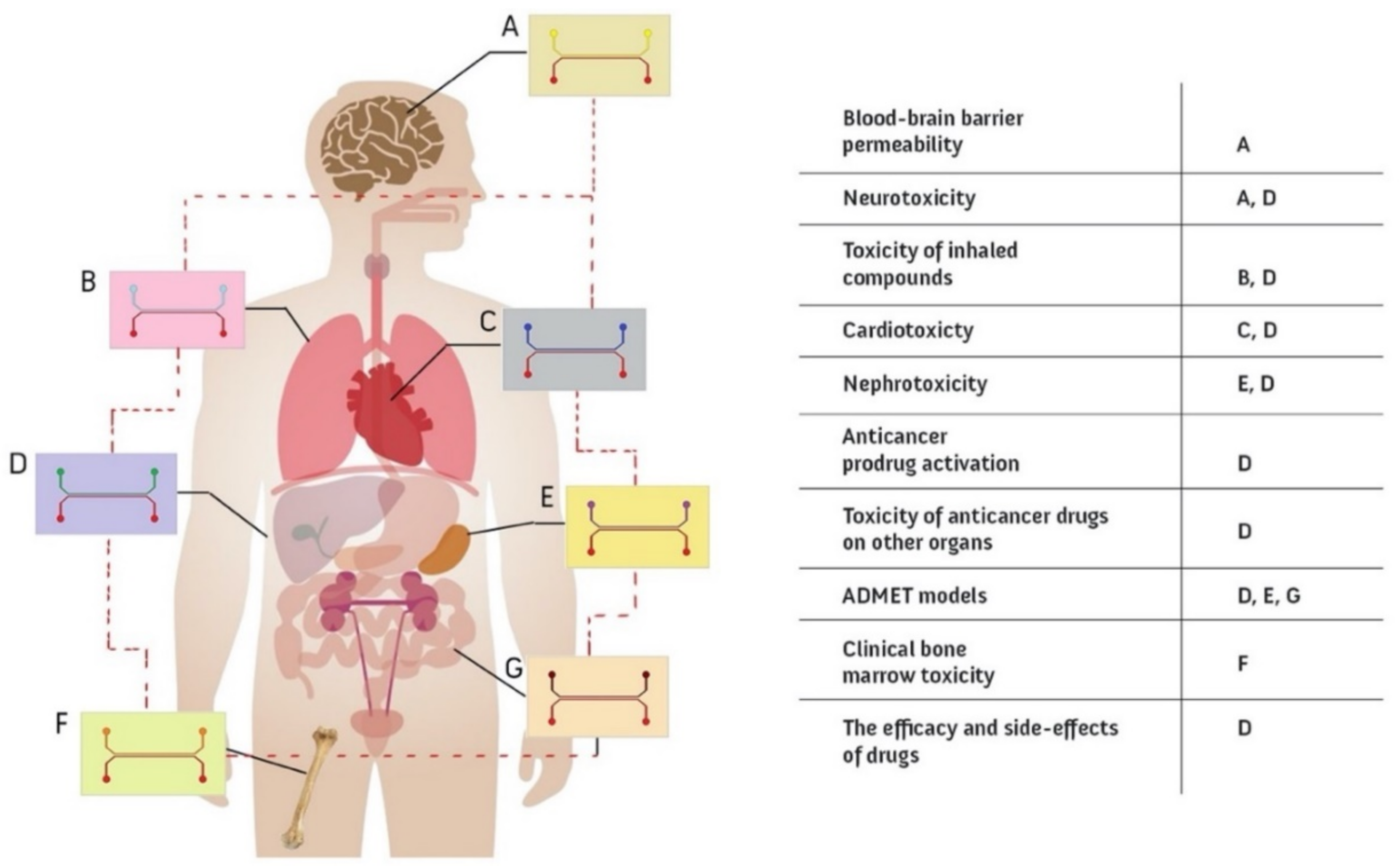
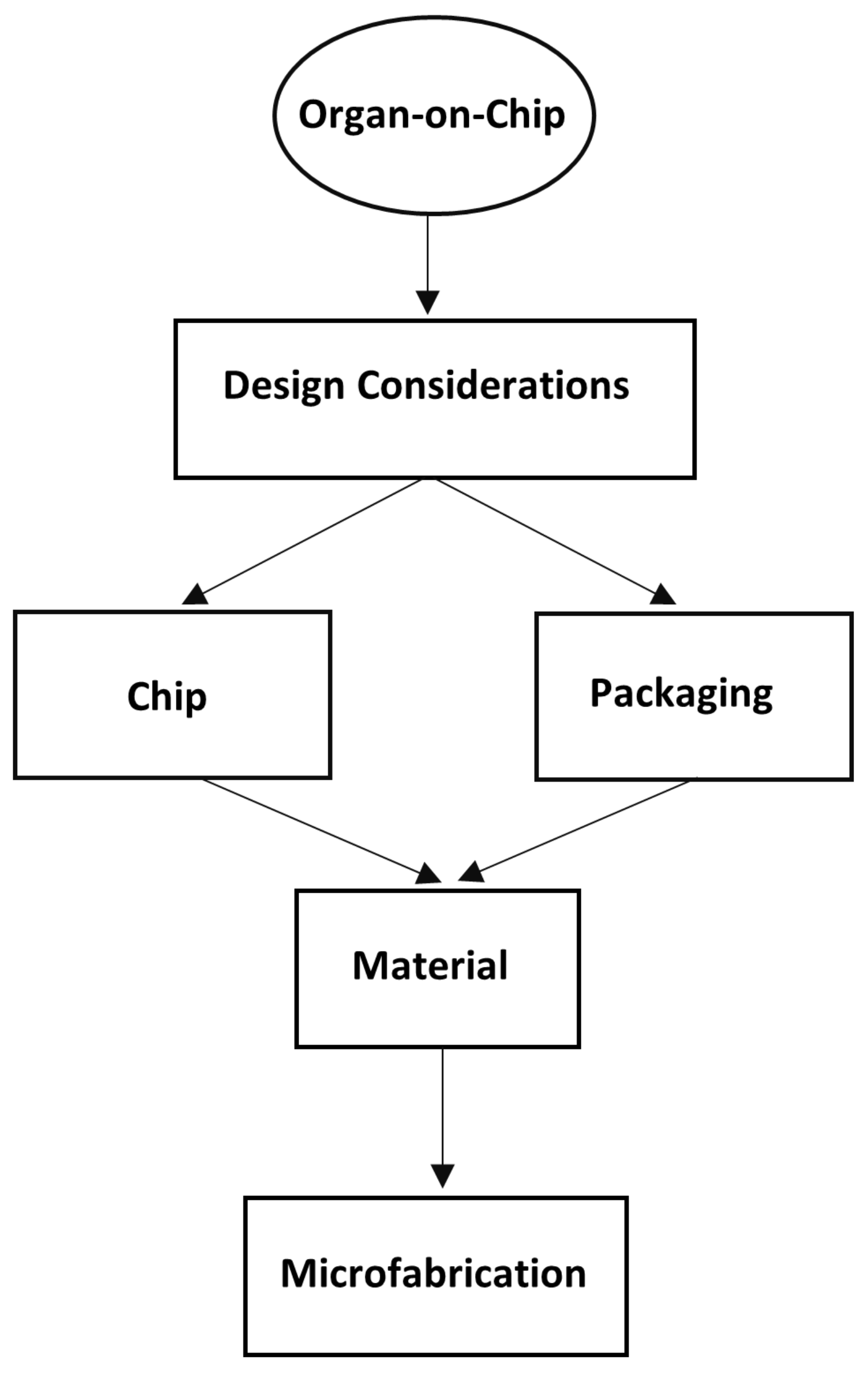
| Studied Organ | Flow Rate | Refs. |
|---|---|---|
| Blood-Brain Barrier | 100 µL/h | [49] |
| 10 µL/min | [78] | |
| 16 µL/min | [79] | |
| 2.5 mL/h | [80] | |
| Lung | 30 µL/h | [81] |
| 60 µL/h | [82,83] | |
| Gut | 30 µL/h | [84,85] |
| 60 µL/h | [44] | |
| Heart | 40 µL/h | [86] |
| Ref. | Type | Available Flow Rate Range (mL/min) |
|---|---|---|
| [89] | Peristaltic micropump | 1.66 × 10−4–3600 |
| [90] | Syringe pumps | 1 × 10−6–0.127 |
| [91] | Electrokinetic pump | 1.8 × 10−3–0.01 |
| [92] | Capillary pump | 5.05 × 10−4–210 |
| Method | Cost | Facility requirement | Precision | Capability for surface treatment | |
|---|---|---|---|---|---|
| 3D printing | Additive, Removal, and Patterning | ||||
| Soft lithography | One step patterning and removal and molding | ||||
| Hot Embossing | One step patterning and removal and molding | ||||
| Injection molding | One step patterning and removal and molding | ||||
| Miscellaneous | --- |
 Positive;
Positive;  Moderate;
Moderate;  Negative.
Negative.Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tajeddin, A.; Mustafaoglu, N. Design and Fabrication of Organ-on-Chips: Promises and Challenges. Micromachines 2021, 12, 1443. https://doi.org/10.3390/mi12121443
Tajeddin A, Mustafaoglu N. Design and Fabrication of Organ-on-Chips: Promises and Challenges. Micromachines. 2021; 12(12):1443. https://doi.org/10.3390/mi12121443
Chicago/Turabian StyleTajeddin, Alireza, and Nur Mustafaoglu. 2021. "Design and Fabrication of Organ-on-Chips: Promises and Challenges" Micromachines 12, no. 12: 1443. https://doi.org/10.3390/mi12121443







