Upregulation of Low-Density Lipoprotein Receptor of the Steroidogenesis Pathway in the Cumulus Cells Is Associated with the Maturation of Oocytes and Achievement of Pregnancy
Abstract
1. Introduction
2. Materials and Methods
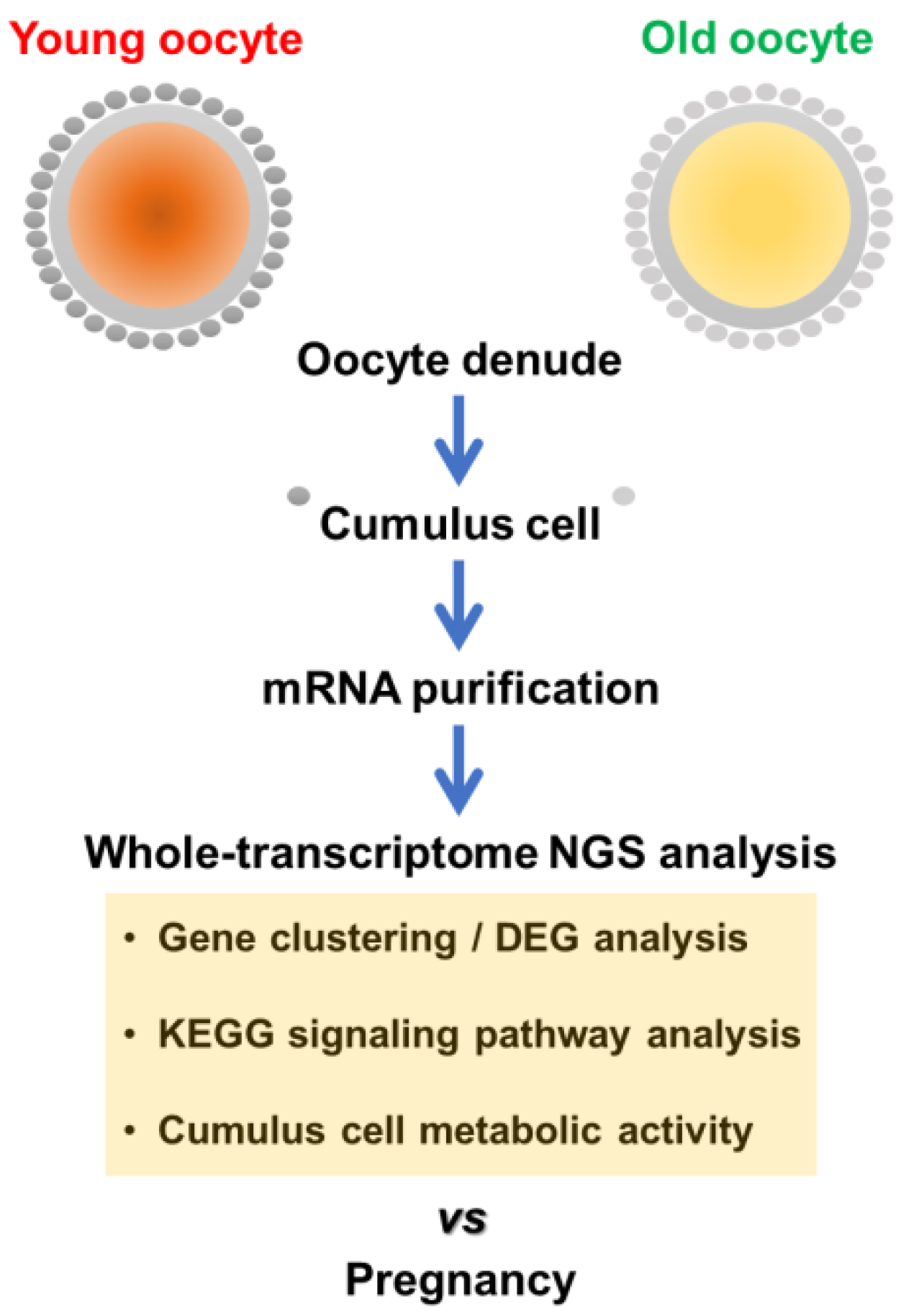
2.1. Collection of Human CCs during IVF
2.2. mRNA Extraction and mRNA-seq NGS Analysis
2.3. Bioinformatics Analysis of mRNA-seq NGS Data
2.4. Gene Ontology and Enrichment Analysis
2.5. Validation Study of NGS Data by Reverse Transcriptome-qPCR
2.6. Statistical Analyses
3. Results
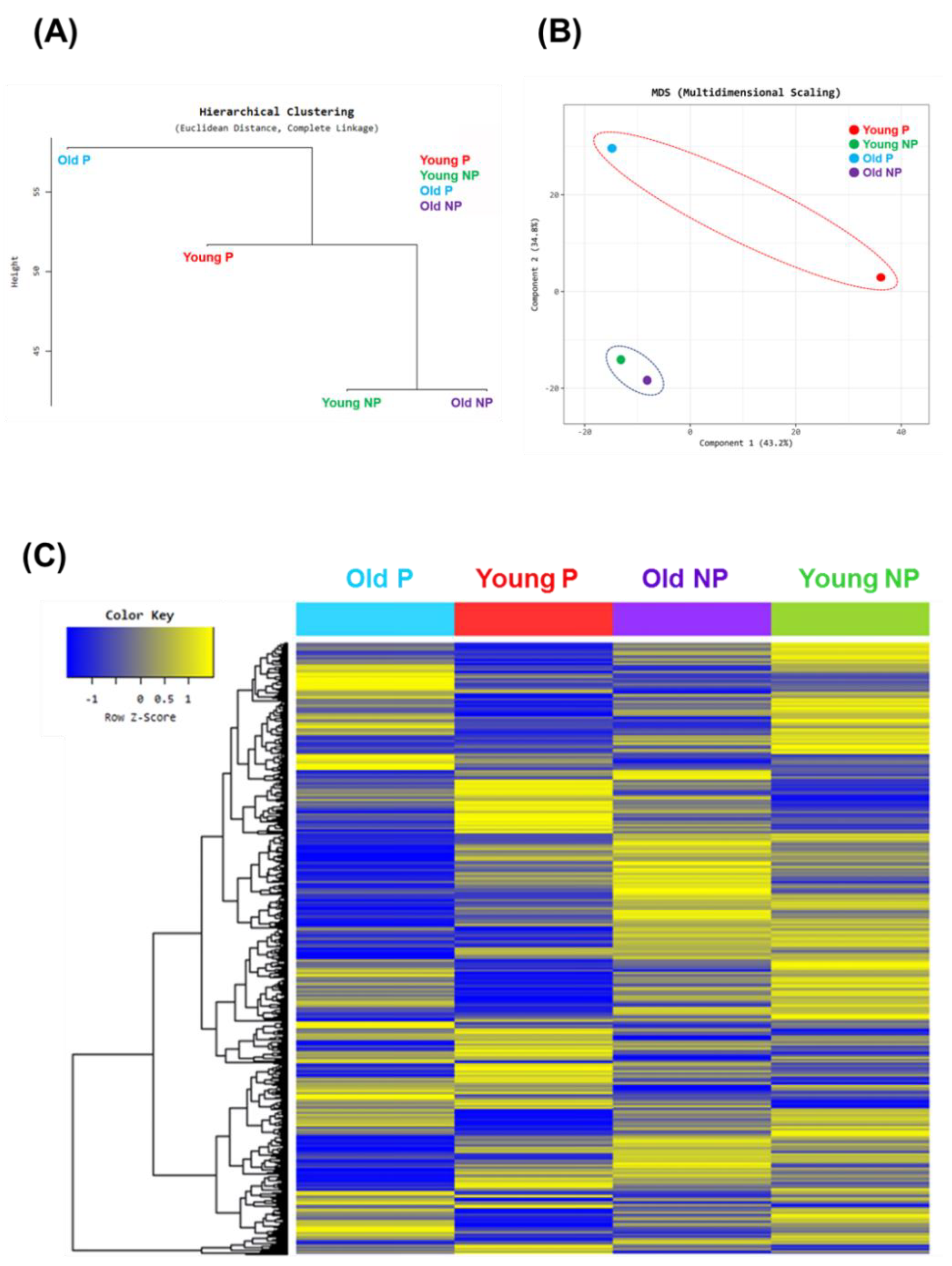
3.1. Characteristics of Clinical Samples and Oocyte Profiling Depend on the Patients
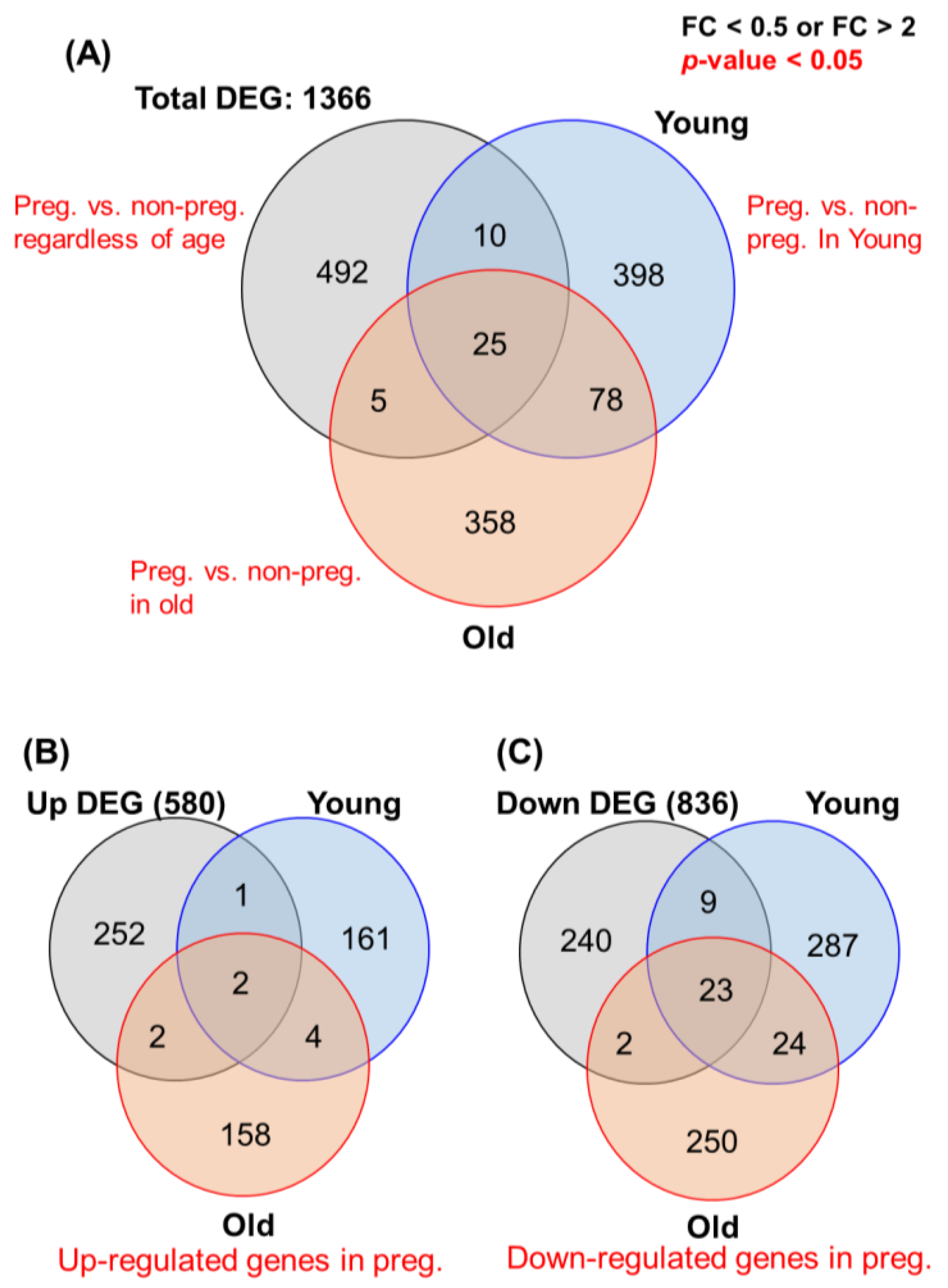
3.2. GO Analysis of Young vs Old and Pregnancy vs. Non-Pregnancy
3.3. Identification of Key Signal Map by KEGG Pathway Analysis
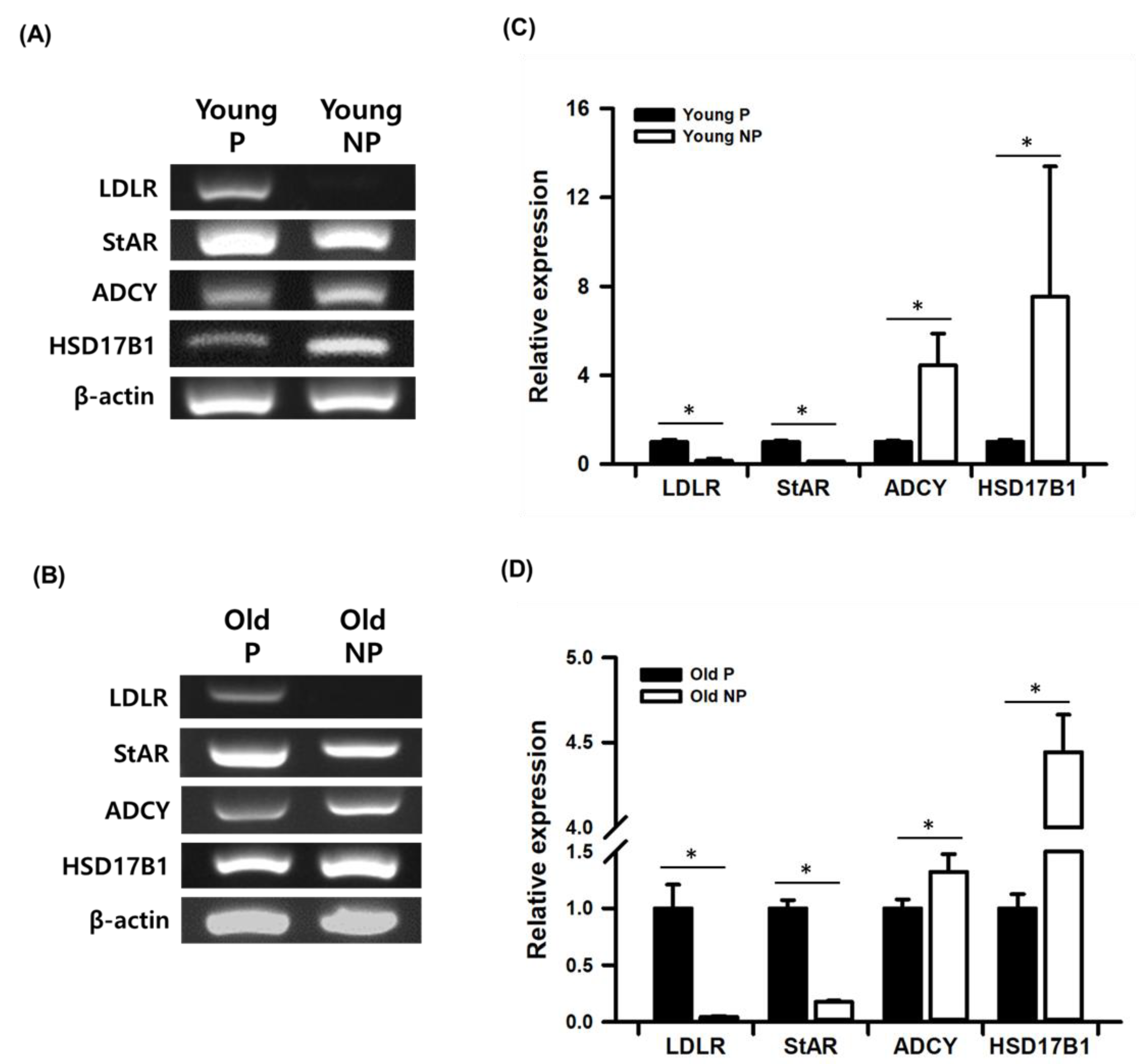
3.4. Validation Study of mRNA-seq Data by Real-Time qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Cimadomo, D.; Fabozzi, G.; Vaiarelli, A.; Ubaldi, N.; Ubaldi, F.M.; Rienzi, L. Impact of Maternal Age on Oocyte and Embryo Competence. Front. Endocrinol. 2018, 9, 327. [Google Scholar] [CrossRef] [PubMed]
- Eichenlaub-Ritter, U. Oocyte ageing and its cellular basis. Int. J. Dev. Biol. 2012, 56, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, H.; Takahashi, T.; Nagase, S. Oocyte aging underlies female reproductive aging: Biological mechanisms and therapeutic strategies. Reprod. Med. Biol. 2015, 14, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Hillier, S.G.; Smitz, J.; Eichenlaub-Ritter, U. Folliculogenesis and oogenesis: From basic science to the clinic. Mol. Hum. Reprod. 2010, 16, 617–620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yan, Z.; Qin, Q.; Nisenblat, V.; Chang, H.M.; Yu, Y.; Wang, T.; Lu, C.; Yang, M.; Yang, S.; et al. Transcriptome Landscape of Human Folliculogenesis Reveals Oocyte and Granulosa Cell Interactions. Mol. Cell. 2018, 72, 1021–1034.e1024. [Google Scholar] [CrossRef] [PubMed]
- Assou, S.; Haouzi, D.; De Vos, J.; Hamamah, S. Human cumulus cells as biomarkers for embryo and pregnancy outcomes. Mol. Hum. Reprod. 2010, 16, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Feuerstein, P.; Cadoret, V.; Dalbies-Tran, R.; Guerif, F.; Bidault, R.; Royere, D. Gene expression in human cumulus cells: One approach to oocyte competence. Hum. Reprod. 2007, 22, 3069–3077. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wells, D. The human oocyte and cumulus cells relationship: New insights from the cumulus cell transcriptome. Mol. Hum. Reprod. 2010, 16, 715–725. [Google Scholar] [CrossRef]
- Iager, A.E.; Kocabas, A.M.; Otu, H.H.; Ruppel, P.; Langerveld, A.; Schnarr, P.; Suarez, M.; Jarrett, J.C.; Conaghan, J.; Rosa, G.J.; et al. Identification of a novel gene set in human cumulus cells predictive of an oocyte’s pregnancy potential. Fertil. Steril. 2013, 99, 745–752.e746. [Google Scholar] [CrossRef]
- Dumesic, D.A.; Meldrum, D.R.; Katz-Jaffe, M.G.; Krisher, R.L.; Schoolcraft, W.B. Oocyte environment: Follicular fluid and cumulus cells are critical for oocyte health. Fertil. Steril. 2015, 103, 303–316. [Google Scholar] [CrossRef] [PubMed]
- McReynolds, S.; Dzieciatkowska, M.; McCallie, B.R.; Mitchell, S.D.; Stevens, J.; Hansen, K.; Schoolcraft, W.B.; Katz-Jaffe, M.G. Impact of maternal aging on the molecular signature of human cumulus cells. Fertil. Steril. 2012, 98, 1574–1580.e1575. [Google Scholar] [CrossRef] [PubMed]
- Uyar, A.; Torrealday, S.; Seli, E. Cumulus and granulosa cell markers of oocyte and embryo quality. Fertil. Steril. 2013, 99, 979–997. [Google Scholar] [CrossRef] [PubMed]
- Wathlet, S.; Adriaenssens, T.; Segers, I.; Verheyen, G.; Janssens, R.; Coucke, W.; Devroey, P.; Smitz, J. New candidate genes to predict pregnancy outcome in single embryo transfer cycles when using cumulus cell gene expression. Fertil. Steril. 2012, 98, 432–439. [Google Scholar] [CrossRef]
- Gilchrist, R.B.; Lane, M.; Thompson, J.G. Oocyte-secreted factors: Regulators of cumulus cell function and oocyte quality. Hum. Reprod. Update. 2008, 14, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Hennet, M.L.; Combelles, C.M. The antral follicle: A microenvironment for oocyte differentiation. Int. J. Dev. Biol. 2012, 56, 819–831. [Google Scholar] [CrossRef]
- Assidi, M.; Montag, M.; Van der Ven, K.; Sirard, M.A. Biomarkers of human oocyte developmental competence expressed in cumulus cells before ICSI: A preliminary study. J. Assist. Reprod. Genet. 2011, 28, 173–188. [Google Scholar] [CrossRef][Green Version]
- Gebhardt, K.M.; Feil, D.K.; Dunning, K.R.; Lane, M.; Russell, D.L. Human cumulus cell gene expression as a biomarker of pregnancy outcome after single embryo transfer. Fertil. Steril. 2011, 96, 47–52.e42. [Google Scholar] [CrossRef]
- Canosa, S.; Bergandi, L.; Macri, C.; Charrier, L.; Paschero, C.; Carosso, A.; Di Segni, N.; Silvagno, F.; Gennarelli, G.; Benedetto, C.; et al. Morphokinetic analysis of cleavage stage embryos and assessment of specific gene expression in cumulus cells independently predict human embryo development to expanded blastocyst: A preliminary study. J. Assist. Reprod. Genet. 2020, 37, 1409–1420. [Google Scholar] [CrossRef]
- Kordus, R.J.; LaVoie, H.A. Granulosa cell biomarkers to predict pregnancy in ART: Pieces to solve the puzzle. Reproduction 2017, 153, R69–R83. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, J.; Wen, H.; Feng, Y.; Zhang, X.; Xiang, H.; Cao, Y.; Tong, X.; Ji, Y.; Xue, Z. Analyzing the Transcriptome Profile of Human Cumulus Cells Related to Embryo Quality via RNA Sequencing. Biomed. Res. Int. 2018, 2018, 9846274. [Google Scholar] [CrossRef]
- Scarica, C. Improving oocyte and embryo competence assessment through granulosa cells transcriptome analysis. Curr. Trends Clin. Embriol. 2016, 3, 12–18. [Google Scholar] [CrossRef]
- Wathlet, S.; Adriaenssens, T.; Segers, I.; Verheyen, G.; Van de Velde, H.; Coucke, W.; Ron El, R.; Devroey, P.; Smitz, J. Cumulus cell gene expression predicts better cleavage-stage embryo or blastocyst development and pregnancy for ICSI patients. Hum. Reprod. 2011, 26, 1035–1051. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, D.; Zhang, Z.; Wei, Z.; Cao, Y. Molecular signature in human cumulus cells related to embryonic developmental potential. Reprod. Sci. 2015, 22, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Molinari, E.; Bar, H.; Pyle, A.M.; Patrizio, P. Transcriptome analysis of human cumulus cells reveals hypoxia as the main determinant of follicular senescence. Mol. Hum. Reprod. 2016, 22, 866–876. [Google Scholar] [CrossRef]
- Gardner, D.K.; Schoolcraft, W.B. A randomized trial of blastocyst culture and transfer in in-vitro fertilization: Reply. Hum. Reprod. 1999, 14, 1663A. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Sherman, B.T.; Huang da, W.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012, 28, 1805–1806. [Google Scholar] [CrossRef]
- Bronson, R.A.; Fusi, F.M. Integrins and human reproduction. Mol. Hum. Reprod. 1996, 2, 153–168. [Google Scholar] [CrossRef]
- Drake, J.M.; Friis, R.R.; Dharmarajan, A.M. The role of sFRP4, a secreted frizzled-related protein, in ovulation. Apoptosis 2003, 8, 389–397. [Google Scholar] [CrossRef]
- Hernandez Gifford, J.A. The role of WNT signaling in adult ovarian folliculogenesis. Reproduction 2015, 150, R137–R148. [Google Scholar] [CrossRef]
- Maman, E.; Yung, Y.; Cohen, B.; Konopnicki, S.; Dal Canto, M.; Fadini, R.; Kanety, H.; Kedem, A.; Dor, J.; Hourvitz, A. Expression and regulation of sFRP family members in human granulosa cells. Mol. Hum. Reprod. 2011, 17, 399–404. [Google Scholar] [CrossRef]
- Rusovici, R.; LaVoie, H.A. Expression and distribution of AP-1 transcription factors in the porcine ovary. Biol. Reprod. 2003, 69, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, S.; Lacey, M.; Whitehead, S.A. Interactions between interleukin-1 beta, nitric oxide and prostaglandin E2 in the rat ovary: Effects on steroidogenesis. Eur. J. Endocrinol. 1997, 137, 293–300. [Google Scholar] [CrossRef][Green Version]
- Duffy, D.M.; McGinnis, L.K.; Vandevoort, C.A.; Christenson, L.K. Mammalian oocytes are targets for prostaglandin E2 (PGE2) action. Reprod. Biol. Endocrinol. 2010, 8, 131. [Google Scholar] [CrossRef]
- Goverde, H.J. The enhancement by prostaglandin E2 of cumulus cell outgrowth in vitro. Prostaglandins 1993, 45, 241–247. [Google Scholar] [CrossRef]
- Takahashi, M.; Aizawa, Y. Effect of prostaglandin E2 and F2 alpha on steroid biosynthesis in rat ovary (author’s transl). Nihon Yakurigaku Zasshi 1979, 75, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Anahory, T.; Dechaud, H.; Bennes, R.; Marin, P.; Lamb, N.J.; Laoudj, D. Identification of new proteins in follicular fluid of mature human follicles. Electrophoresis 2002, 23, 1197–1202. [Google Scholar] [CrossRef]
- Iwai, T.; Fujii, S.; Nanbu, Y.; Nonogaki, H.; Konishi, I.; Mori, T.; Masutani, H.; Yodoi, J. Expression of adult T-cell leukaemia-derived factor, a human thioredoxin homologue, in the human ovary throughout the menstrual cycle. Virchows Arch. A 1992, 420, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Kugu, K.; Dharmarajan, A.M.; Preutthipan, S.; Wallach, E.E. Role of calcium/calmodulin-dependent protein kinase II in gonadotrophin-induced ovulation in in vitro perfused rabbit ovaries. J. Reprod. Fertil. 1995, 103, 273–278. [Google Scholar] [CrossRef]
- Soto, E.; Silavin, S.L.; Tureck, R.W.; Strauss, J.F., 3rd. Stimulation of progesterone synthesis in luteinized human granulosa cells by human chorionic gonadotropin and 8-bromo-adenosine 3’,5’-monophosphate: The effect of low density lipoprotein. J. Clin. Endocrinol. Metab. 1984, 58, 831–837. [Google Scholar] [CrossRef]
- Argov, N.; Sklan, D. Expression of mRNA of lipoprotein receptor related protein 8, low density lipoprotein receptor, and very low density lipoprotein receptor in bovine ovarian cells during follicular development and corpus luteum formation and regression. Mol. Reprod. Dev. 2004, 68, 169–175. [Google Scholar] [CrossRef]
- Morita, Y.; Wada-Hiraike, O.; Yano, T.; Shirane, A.; Hirano, M.; Hiraike, H.; Koyama, S.; Oishi, H.; Yoshino, O.; Miyamoto, Y.; et al. Resveratrol promotes expression of SIRT1 and StAR in rat ovarian granulosa cells: An implicative role of SIRT1 in the ovary. Reprod. Biol. Endocrinol. 2012, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Sandhoff, T.W.; McLean, M.P. Hormonal regulation of steroidogenic acute regulatory (StAR) protein messenger ribonucleic acid expression in the rat ovary. Endocrine 1996, 4, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Christenson, L.K.; Strauss, J.F., 3rd. Steroidogenic acute regulatory protein: An update on its regulation and mechanism of action. Arch. Med. Res. 2001, 32, 576–586. [Google Scholar] [CrossRef]
- Lastro, M.; Collins, S.; Currie, W.B. Adenylyl cyclases in oocyte maturation: A characterization of AC isoforms in bovine cumulus cells. Mol. Reprod. Dev. 2006, 73, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Mehlmann, L.M. Stops and starts in mammalian oocytes: Recent advances in understanding the regulation of meiotic arrest and oocyte maturation. Reproduction 2005, 130, 791–799. [Google Scholar] [CrossRef]
- Jones, M.R.; Mathur, R.; Cui, J.; Guo, X.; Azziz, R.; Goodarzi, M.O. Independent confirmation of association between metabolic phenotypes of polycystic ovary syndrome and variation in the type 6 17beta-hydroxysteroid dehydrogenase gene. J. Clin. Endocrinol. Metab. 2009, 94, 5034–5038. [Google Scholar] [CrossRef]
- Qin, K.; Ehrmann, D.A.; Cox, N.; Refetoff, S.; Rosenfield, R.L. Identification of a functional polymorphism of the human type 5 17beta-hydroxysteroid dehydrogenase gene associated with polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 2006, 91, 270–276. [Google Scholar] [CrossRef]


| Young Preg (n = 8) | Young Non-Preg (n = 8) | Old Preg (n = 8) | Old Non-Preg (n = 8) | |
|---|---|---|---|---|
| Age (years) | 31.16 ± 1.16 | 33.8 ± 1.78 | 39.83 ± 0.75 | 40.83 ± 1.83 |
| Serum AMH (ng/mL) | 4.49 ± 2.79 | 2.80 ± 1.27 | 1.95 ± 1.01 | 1.99 ± 0.97 |
| BMI (kg/m2) | 23.30 ± 3.41 | 21.49 ± 2.56 | 24.20 ± 3.15 | 22.33 ± 2.56 |
| Basal FSH (mIU/mL) | 7.96 ± 2.99 | 8.36 ± 2.20 | 8.23 ± 2.16 | 7.89 ± 1.47 |
| No. of previous IVF cycles | 2.6 ± 1.67 | 2.0 ± 1.22 | 1.0 ± 0.81 | 2.16 ± 1.60 |
| Total Gonadotropin dose | 3030 ± 497 | 2308 ± 820 | 2525 ± 318 | 2995 ± 359 |
| No. of antral follicle count | 11.25 ± 5.31 | 8.88 ± 7.64 | 8.80 ± 3.63 | 9.33 ± 4.96 |
| No. of retrieved oocytes | 13.16 ± 6.43 | 10.4 ± 2.30 | 8.66 ± 5.88 | 10.16 ± 4.99 |
| Maturation rate (%) | 77.01 ± 15.31 * | 64.77 ± 7.08 | 87.12 ± 16.11 * | 81.22 ± 17.78 |
| No. of MII oocytes | 9.5 ± 3.93 | 6.8 ± 2.04 | 5.0 ± 3.37 | 6.66 ± 4.84 |
| Fertilization rate (%) | 91.05 ± 8.61 | 68.85 ± 32.23 | 83.33 ± 23.57 | 75.82 ± 17.93 |
| No. of transferred embryo | 1.16 ± 0.40 | 1.63 ± 0.92 | 2.33 ± 0.81 | 2.00 ± 0.00 |
| Stage of transferred embryo | ||||
| BL (%)/CL (%) | 100 */0 | 62.5/37.5 | 83.33 */16.67 | 66.66/33.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.J.; Kim, Y.S.; Kim, Y.J.; Lee, H.R.; Choi, K.H.; Park, E.A.; Kang, K.Y.; Yoon, T.K.; Hwang, S.; Ko, J.J.; et al. Upregulation of Low-Density Lipoprotein Receptor of the Steroidogenesis Pathway in the Cumulus Cells Is Associated with the Maturation of Oocytes and Achievement of Pregnancy. Cells 2021, 10, 2389. https://doi.org/10.3390/cells10092389
Kim MJ, Kim YS, Kim YJ, Lee HR, Choi KH, Park EA, Kang KY, Yoon TK, Hwang S, Ko JJ, et al. Upregulation of Low-Density Lipoprotein Receptor of the Steroidogenesis Pathway in the Cumulus Cells Is Associated with the Maturation of Oocytes and Achievement of Pregnancy. Cells. 2021; 10(9):2389. https://doi.org/10.3390/cells10092389
Chicago/Turabian StyleKim, Myung Joo, Young Sang Kim, Yu Jin Kim, Hye Ran Lee, Kyoung Hee Choi, Eun A Park, Ki Ye Kang, Tae Ki Yoon, Sohyun Hwang, Jung Jae Ko, and et al. 2021. "Upregulation of Low-Density Lipoprotein Receptor of the Steroidogenesis Pathway in the Cumulus Cells Is Associated with the Maturation of Oocytes and Achievement of Pregnancy" Cells 10, no. 9: 2389. https://doi.org/10.3390/cells10092389
APA StyleKim, M. J., Kim, Y. S., Kim, Y. J., Lee, H. R., Choi, K. H., Park, E. A., Kang, K. Y., Yoon, T. K., Hwang, S., Ko, J. J., Kim, Y. S., & Lee, J. H. (2021). Upregulation of Low-Density Lipoprotein Receptor of the Steroidogenesis Pathway in the Cumulus Cells Is Associated with the Maturation of Oocytes and Achievement of Pregnancy. Cells, 10(9), 2389. https://doi.org/10.3390/cells10092389






