Contrasting Responses of Guar Genotypes Shed Light on Multiple Component Traits of Salinity Tolerance Mechanisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Evaluations
2.2. Screening for Salinity Tolerance under Greenhouse Conditions
2.3. Ion Analysis
2.4. Selection of Candidate Genes
2.5. Expression Analyses
3. Results
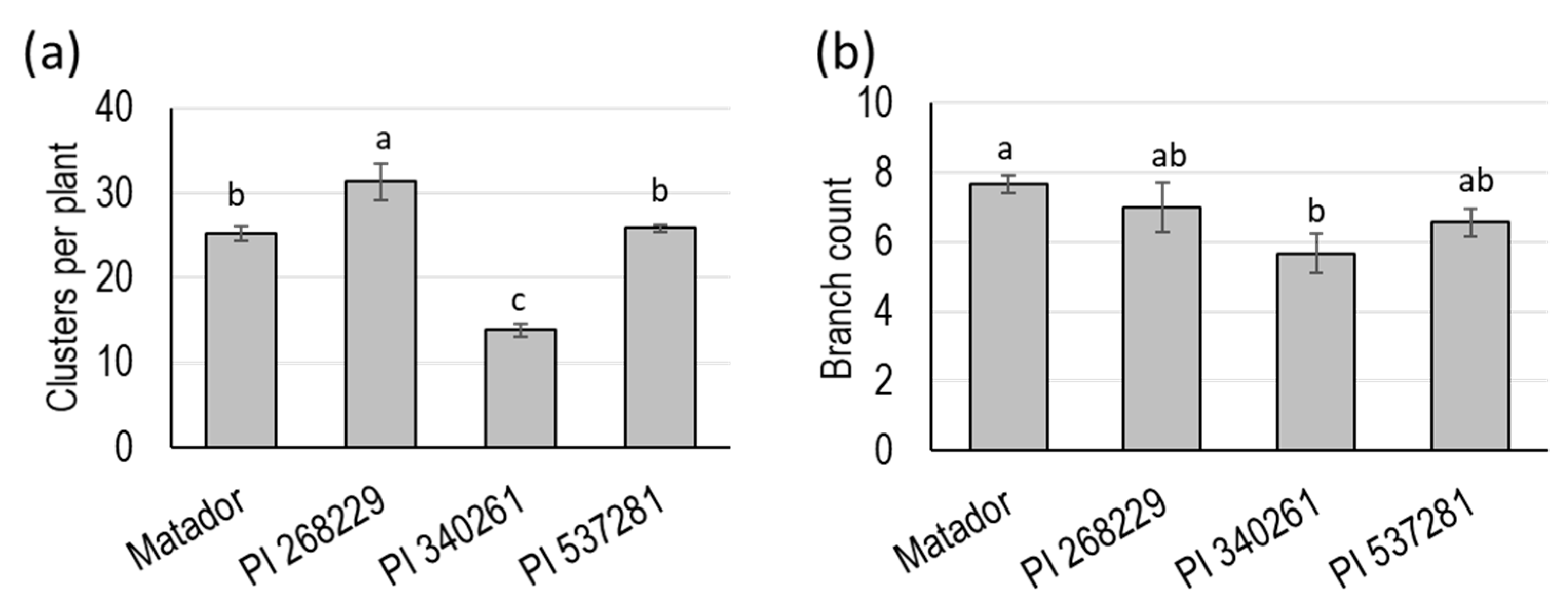
3.1. Field Evaluation of Four Guar Genotypes
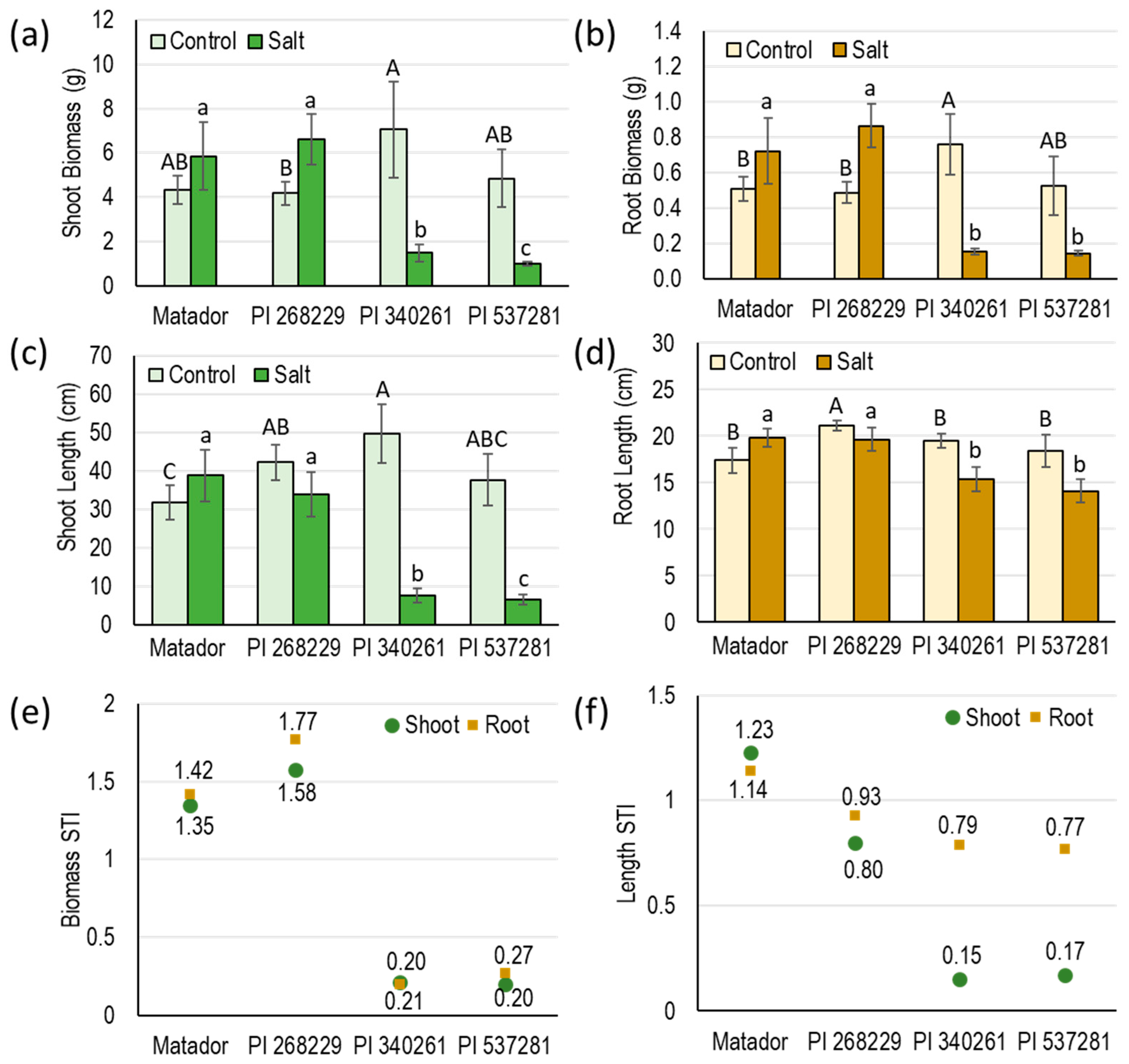
3.2. Greenhouse Evaluation of Four Guar Genotypes for Salinity Tolerance
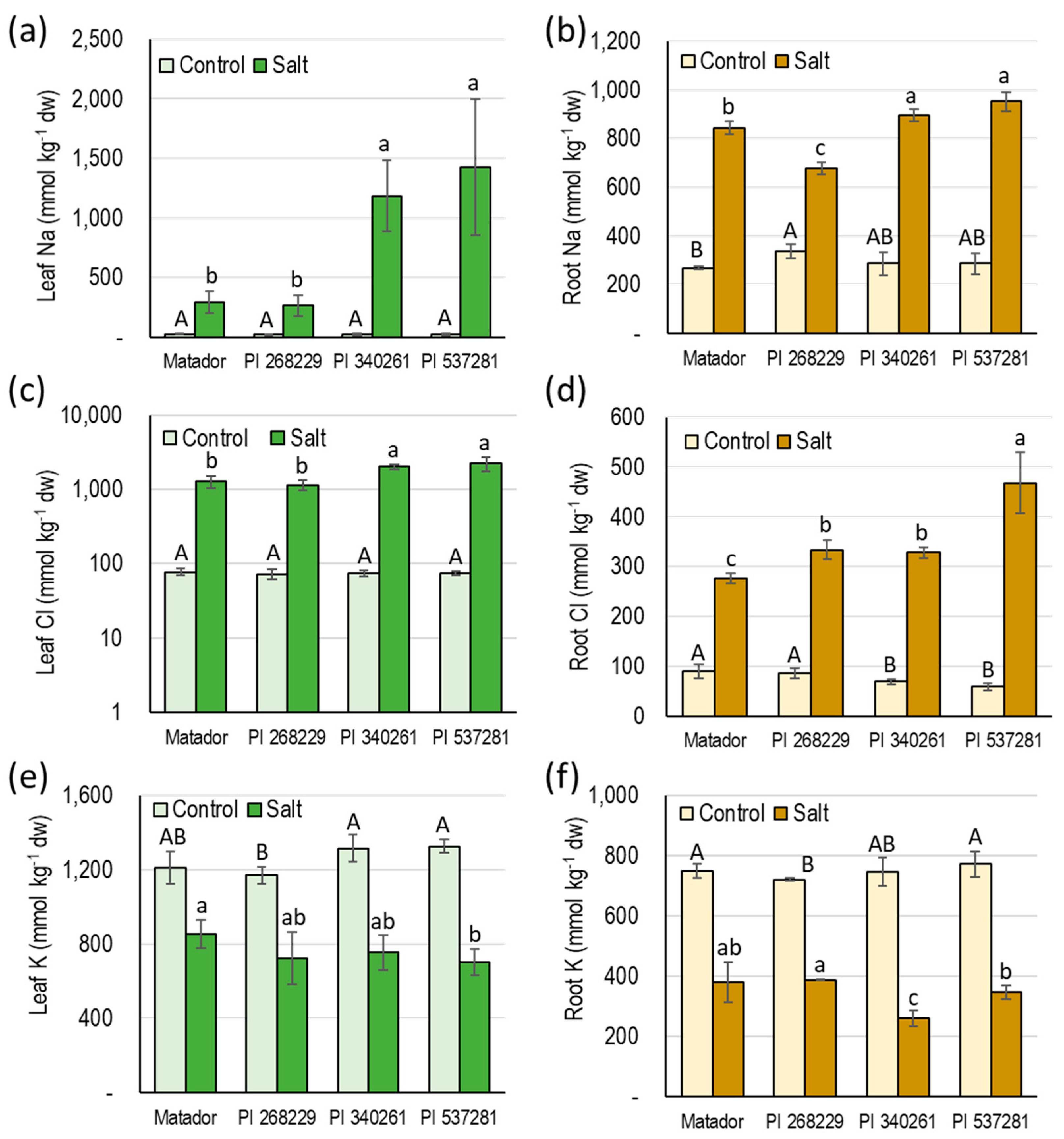
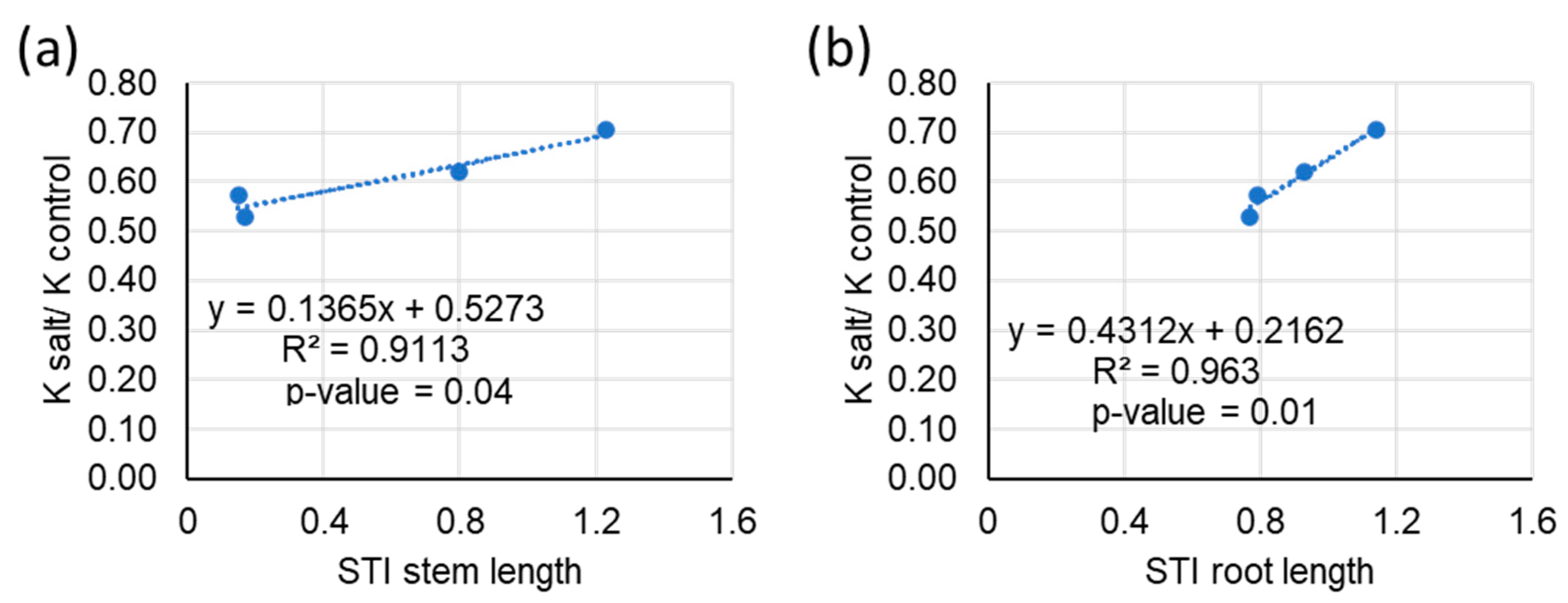
3.3. Tissue Ion Analysis
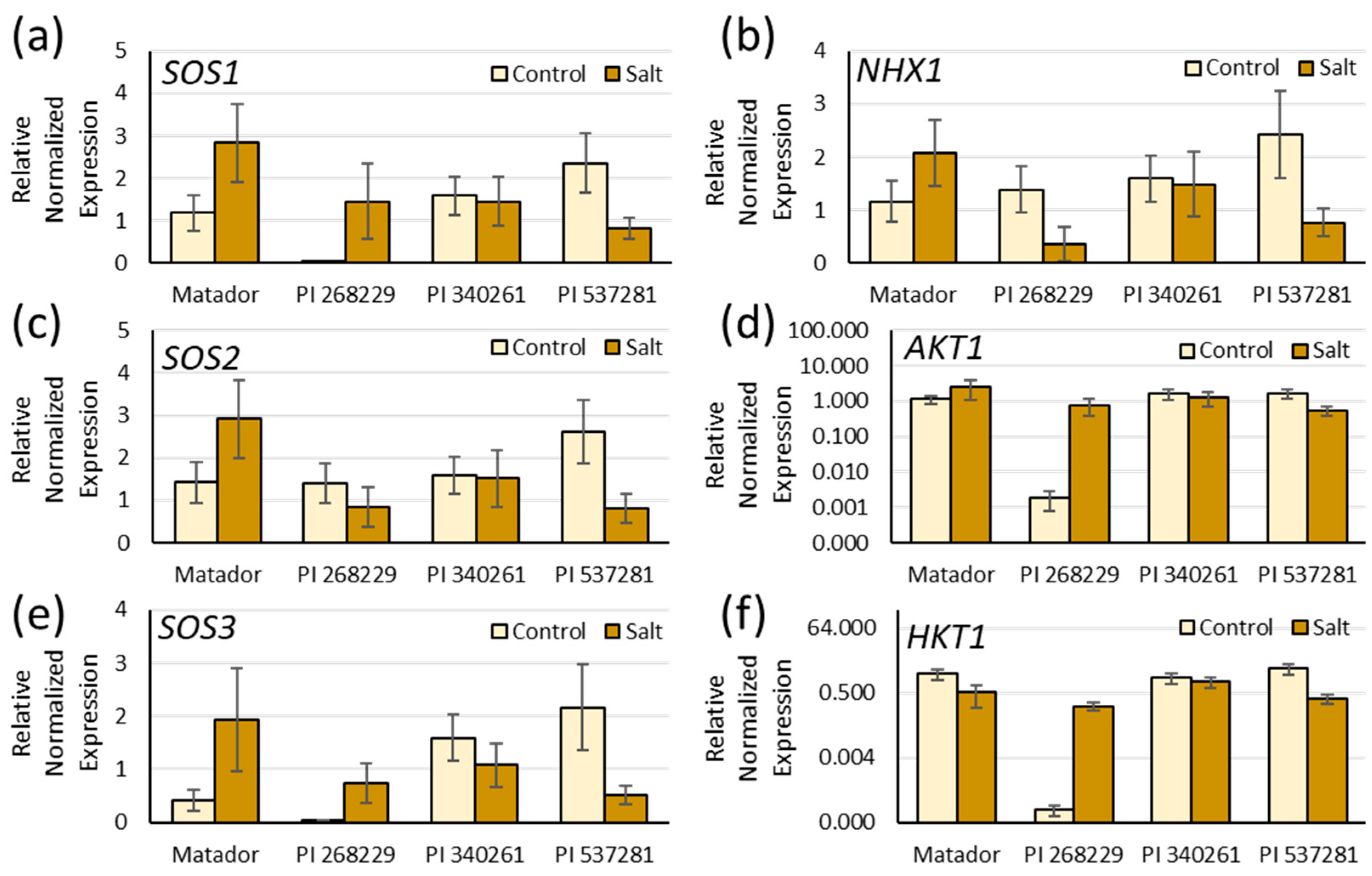
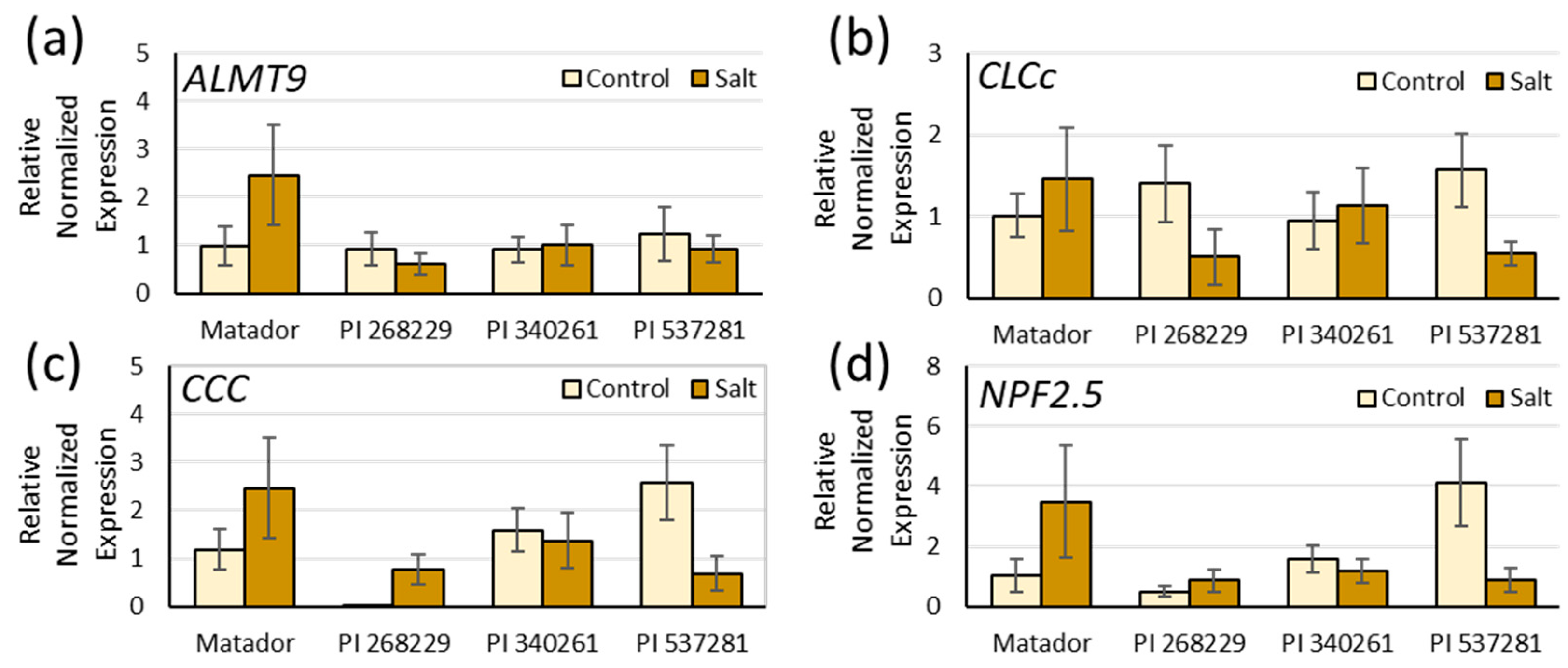
3.4. Expression Analysis under Salinity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Whistler, R.L.; Hymowitz, T. Guar: Agronomy, Production, Industrial Use, and Nutrition; Purdue University Press: West Lafayette, IN, USA, 1979. [Google Scholar]
- Morris, J.B. Morphological and reproductive characterization of guar (Cyamopsis tetragonoloba) genetic resources regenerated in Georgia, USA. Genet. Resour. Crop Evol. 2010, 57, 985–993. [Google Scholar] [CrossRef]
- Rao, S.C.; Northup, B.K. Biomass production and quality of Indian-origin forage guar in Southern Great Plains. Agron. J. 2013, 105, 945–950. [Google Scholar] [CrossRef]
- Abidi, N.; Liyanage, S.; Auld, D.; Norman, L.; Grover, K.; Augadi, S.; Singla, S.; Trostle, C. Challenges and Opportunities for Increasing Guar Production in the United States to Support Unconventional Oil and Gas Production. In Hydraulic Fracturing Impacts and Technologies; Uddameri, V., Morse, A., Tindle, K.J., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 207–225. [Google Scholar]
- Mudgil, D.; Barak, S.; Khatkar, B.S. Guar gum: Processing, properties and food applications-A Review. J. Food Sci. Technol. 2014, 51, 409–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.K. An Analysis of Guar Crop in India; Report Prepared for USDA-FAS: New Delhi, India, 2014. [Google Scholar]
- Kalyani, D.L. Performance of cluster bean genotypes under varied time of sowing. Legume Res. 2012, 35, 154–158. [Google Scholar]
- Singla, S.; Grover, K.; Angadi, S.V.; Begna, S.; Schutte, B.J.; VanLeeuwen, D. Growth and yield of guar (Cyamopsis tetragonoloba L.) genotypes under different planting dates in the semi-arid Southern High Plains. Am. J. Plant Sci. 2016, 7, 1246–1258. [Google Scholar] [CrossRef] [Green Version]
- Singla, S.; Grover, K.; Angadi, S.V.; Schutte, B.; VanLeeuwen, D. Guar stand establishment, physiology, and yield responses to planting date in southern New Mexico. Agron. J. 2016, 108, 2289–2300. [Google Scholar] [CrossRef]
- Adams, C.B.; Boote, K.J.; Shrestha, R.; MacMillan, J.; Hinson, P.O.; Trostle, C. Growth stages and developmental patterns of guar. Agron. J. 2020, 112, 4990–5001. [Google Scholar] [CrossRef]
- Singh, J.; Guzman, I.; Begna, S.; Trostle, C.; Angadi, S. Germination and early growth response of guar cultivars to low temperatures. Ind. Crop. Prod. 2021, 159, 113082. [Google Scholar] [CrossRef]
- Alshameri, A.; Al-Qurainy, F.; Gaafar, A.R.; Khan, S.; Nadeem, M.; Alansi, S. Identification of heat-responsive genes in guar [Cyamopsis tetragonoloba (L.) Taub]. Int. J. Genom. 2020, 2020, 3126592. [Google Scholar] [CrossRef]
- Al-Qurainy, F.; Alshameri, A.; Gaafar, A.R.; Khan, S.; Nadeem, M.; Alameri, A.A.; Tarroum, M.; Ashraf, M. Comprehensive stress-based de novo transcriptome assembly and annotation of guar (Cyamopsis tetragonoloba (L.) Taub.): An important industrial and forage crop. Int. J. Genom. 2019, 2019, 7295859. [Google Scholar] [CrossRef] [Green Version]
- Jamil, A.; Riaz, S.; Ashraf, M.; Foolad, M.R. Gene expression profiling of plants under salt stress. Critic. Rev. Plant Sci. 2011, 30, 435–458. [Google Scholar] [CrossRef]
- Munns, R.; Husain, S.; Rivelli, A.R.; James, R.A.; Condon, A.G.T.; Lindsay, M.P.; Lagudah, E.S.; Schachtman, D.P.; Hare, R.A. Avenues for increasing salt tolerance of crops, and the role of physiologically based selection traits. In Progress in Plant Nutrition: Plenary Lectures of the XIV International Plant Nutrition Colloquium: Food Security and Sustainability of Agro-Ecosystems through Basic and Applied Research; Horst, W.J., Bürkert, A., Claassen, N., Flessa, H., Frommer, W.B., Goldbach, H., Merbach, W., Olfs, H.W., Römheld, V., Sattelmacher, B., et al., Eds.; Springer: Dordrecht, The Netherlands, 2002; pp. 93–105. [Google Scholar]
- Francois, L.E.; Donovan, T.J.; Maas, E.V. Salinity effects on emergence, vegetative growth, and seed yield of guar. Agron. J. 1990, 82, 587–592. [Google Scholar] [CrossRef]
- Ashraf, M.Y.; Akhtar, K.; Sarwar, G.; Ashraf, M. Role of the rooting system in salt tolerance potential of different guar accessions. Agron. Sustain. Dev. 2005, 25, 243–249. [Google Scholar] [CrossRef]
- Suthar, J.D.; Rajpar, I.; Ganjegunte, G.K.; Shah, Z.-U.-H.; Niu, G.; Grover, K. Germination, growth, and ion uptake of 15 guar accessions under elevated salinity. Agrosyst. Geosci. Environ. 2019, 2, 190020. [Google Scholar] [CrossRef] [Green Version]
- Teolis, I.; Liu, W.; Peffley, E.B. Salinity effects on seed germination and plant growth of guar. Crop Sci. 2009, 49, 637–642. [Google Scholar] [CrossRef]
- Khator, K.; Mahawar, L.; Shekhawat, G.S. NaCl induced oxidative stress in legume crops of Indian Thar Desert: An insight in the cytoprotective role of HO1, NO and antioxidants. Physiol. Mol. Biol. Plants 2020, 26, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Robina, S.; Sheela, A. Salt stress effects on germination, early seedling growth and ion uptake of Cyamopsis seedlings. Curr. Agric. 2006, 30, 31–37. [Google Scholar]
- Suthar, J.D.; Rajpar, I.; Ganjegunte, G.K.; Shah, Z.-u.-h. Evaluation of guar (Cyamopsis tetragonoloba L.) genotypes performance under different irrigation water salinity levels: Growth parameters and seed yield. Ind. Crop. Prod. 2018, 123, 247–253. [Google Scholar] [CrossRef]
- Rasheed, M.J.Z.; Ahmad, K.; Ashraf, M.; Qurainy, F.A.; Khan, S.; Athar, H.U.R. Screening of diverse local germplasm of guar (Cyamposis tetragonoloba (l) taub) for salt tolerance: A possible approach to utilize salt-affected soils. Pak. J. Bot. 2015, 47, 1721–1726. [Google Scholar]
- EPA 600/4-79-020. Methods For Chemical Analysis of Water And Wastes; Method 325.2; Environmental Protection Agency: Cincinnati, OH, USA, 1983. [Google Scholar]
- Tanwar, U.K.; Pruthi, V.; Randhawa, G.S. RNA-Seq of guar (Cyamopsis tetragonoloba, L. Taub.) leaves: De novo transcriptome assembly, functional annotation and development of genomic resources. Front. Plant Sci. 2017, 8, 91. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, P.S.; Kaur, N.; Randhawa, G.S. Identification of reference genes for qRT-PCR gene expression studies during seed development and under abiotic stresses in Cyamopsis tetragonoloba. Crop Sci. 2019, 59, 252–265. [Google Scholar] [CrossRef]
- Sandhu, D.; Kaundal, A. Dynamics of salt tolerance: Molecular perspectives. In Biotechnologies of Crop Improvement, Volume 3: Genomic Approaches; Gosal, S.S., Wani, S.H., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 25–40. [Google Scholar]
- Sandhu, D.; Cornacchione, M.V.; Ferreira, J.F.; Suarez, D.L. Variable salinity responses of 12 alfalfa genotypes and comparative expression analyses of salt-response genes. Sci. Rep. 2017, 7, 42958. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Termaat, A. Whole plant responses to salinity. Aust. J. Plant Physiol. 1986, 13, 143–160. [Google Scholar] [CrossRef]
- Al-Niemi, T.S.; Campbell, W.F.; Rumbaugh, M.D. Response of alfalfa cultivars to salinity during germination and post-germination growth. Crop Sci. 1992, 32, 976–980. [Google Scholar] [CrossRef]
- Wang, M.; Zheng, Q.; Shen, Q.; Guo, S. The critical role of potassium in plant stress response. Int. J. Mol. Sci. 2013, 14, 7370–7390. [Google Scholar] [CrossRef] [Green Version]
- Sandhu, D.; Kaundal, A.; Acharya, B.R.; Forest, T.; Pudussery, M.V.; Liu, X.; Ferreira, J.F.S.; Suarez, D.L. Linking diverse salinity responses of 14 almond rootstocks with physiological, biochemical, and genetic determinants. Sci. Rep. 2020, 10, 21087. [Google Scholar] [CrossRef] [PubMed]
- Rubio, F.; Gassmann, W.; Schroeder, J.I. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 1995, 270, 1660–1663. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Hu, J.; Zhou, X.-R.; Yuan, H.-J.; Kumar, T.; Luan, S.; Wang, S.-M. ZxAKT1 is essential for K+ uptake and K+/Na+ homeostasis in the succulent xerophyte Zygophyllum xanthoxylum. Plant J. 2017, 90, 48–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baetz, U.; Eisenach, C.; Tohge, T.; Martinoia, E.; De Angeli, A. Vacuolar chloride fluxes impact ion content and distribution during early salinity stress. Plant Physiol. 2016, 172, 1167–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Qiu, J.; Jayakannan, M.; Xu, B.; Li, Y.; Mayo, G.M.; Tester, M.; Gilliham, M.; Roy, S.J. AtNPF2.5 modulates chloride (Cl−) efflux from roots of Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 2013. [Google Scholar] [CrossRef] [Green Version]
- Colmenero-Flores, J.M.; Martinez, G.; Gamba, G.; Vazquez, N.; Iglesias, D.J.; Brumos, J.; Talon, M. Identification and functional characterization of cation-chloride cotransporters in plants. Plant J. 2007, 50, 278–292. [Google Scholar] [CrossRef] [PubMed]
- Jossier, M.; Kroniewicz, L.; Dalmas, F.; Le Thiec, D.; Ephritikhine, G.; Thomine, S.; Barbier-Brygoo, H.; Vavasseur, A.; Filleur, S.; Leonhardt, N. The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance. Plant J. 2010, 64, 563–576. [Google Scholar] [CrossRef] [PubMed]
| Treatment | ECiw (dS m−1) | Ion Concentration (mmolc L−1) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Cl− | SO42− | NO3− | PO43− | Na+ | K+ | Ca2+ | Mg2+ | ||
| Control | 1.46 | 1.41 | 1.44 | 5.39 | 1.5 | 1.88 | 6.59 | 3.35 | 2.1 |
| Saline | 13.65 | 128.35 | 27.32 | 5.39 | 1.5 | 106.88 | 6.59 | 29.6 | 23 |
| S. No. | Trait | p-Value |
|---|---|---|
| 1 | Single plant above-ground dry biomass (g) | 0.419 |
| 2 | Clusters per plant | 0.001 * |
| 3 | Pods per plant | 0.176 |
| 4 | Pods per cluster | 0.210 |
| 5 | Total pod weight per plant (g) | 0.256 |
| 6 | Pod width (mm) | 0.751 |
| 7 | Pod length (cm) | 0.157 |
| 8 | Pod thickness (mm) | 0.456 |
| 9 | Seeds per pod | 0.369 |
| 10 | Seeds per plant | 0.179 |
| 11 | Seed weight per plant (g) | 0.352 |
| 12 | 100-seed Weight (g) | 0.146 |
| 13 | Single plant harvest index | 0.664 |
| 14 | Dry above-ground biomass kg/ha | 0.447 |
| 15 | Seed yield kg/ha | 0.123 |
| 16 | Harvest index (kg/ha) | 0.471 |
| 17 | Plant height 65 days after planting (cm) | 0.094 |
| 18 | Plant width 65 days after planting (cm) | 0.767 |
| 19 | Branch count 65 days after planting | 0.049 * |
| 20 | Plant count per meter 50 days after planting | 0.993 |
| 21 | Plant height (cm) | 0.249 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sandhu, D.; Pallete, A.; Pudussery, M.V.; Grover, K.K. Contrasting Responses of Guar Genotypes Shed Light on Multiple Component Traits of Salinity Tolerance Mechanisms. Agronomy 2021, 11, 1068. https://doi.org/10.3390/agronomy11061068
Sandhu D, Pallete A, Pudussery MV, Grover KK. Contrasting Responses of Guar Genotypes Shed Light on Multiple Component Traits of Salinity Tolerance Mechanisms. Agronomy. 2021; 11(6):1068. https://doi.org/10.3390/agronomy11061068
Chicago/Turabian StyleSandhu, Devinder, Andrew Pallete, Manju V. Pudussery, and Kulbhushan K. Grover. 2021. "Contrasting Responses of Guar Genotypes Shed Light on Multiple Component Traits of Salinity Tolerance Mechanisms" Agronomy 11, no. 6: 1068. https://doi.org/10.3390/agronomy11061068







