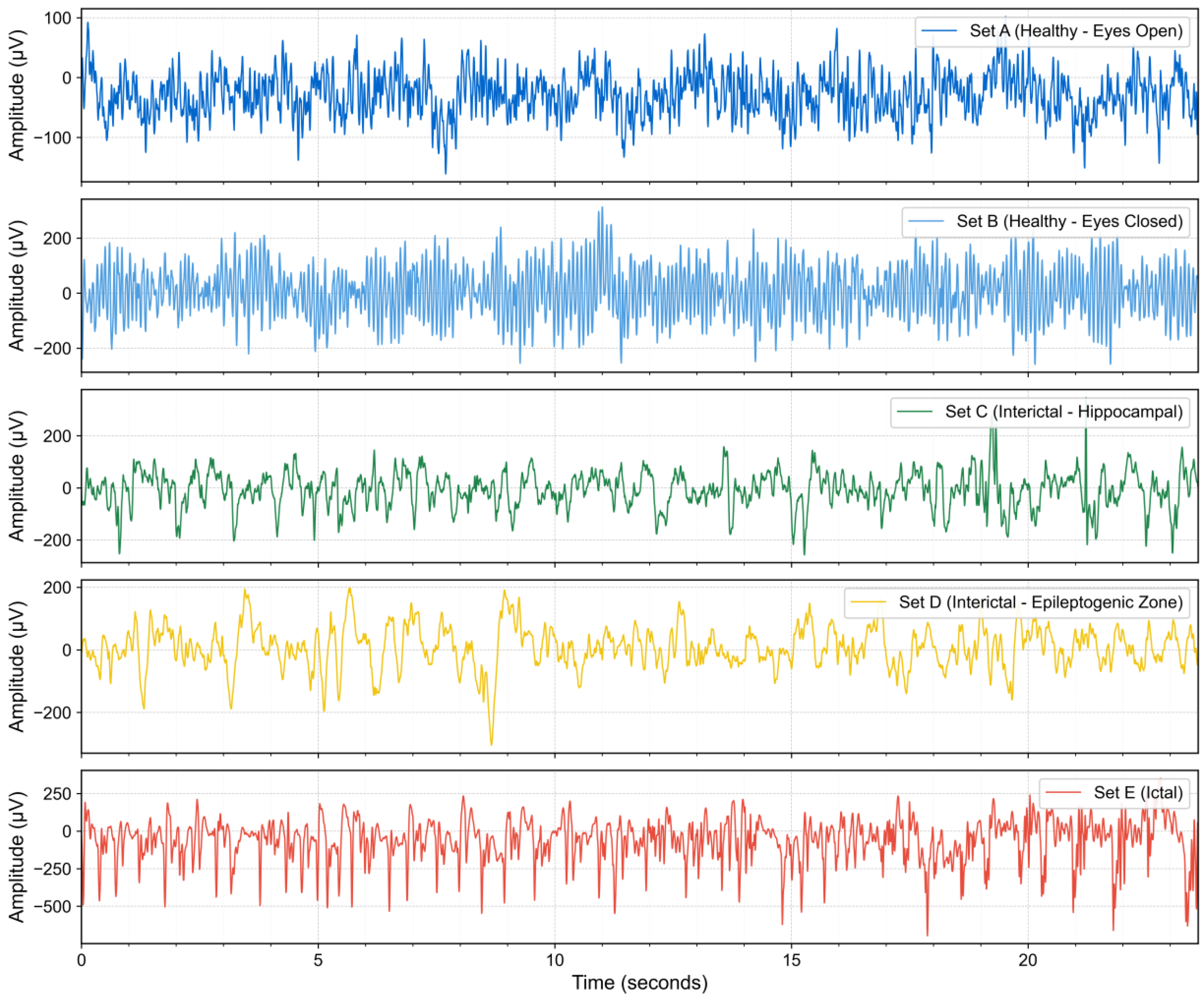
1. Introduction
Epilepsy, a common neurological disorder affecting approximately 50 million individuals globally [
1], imposes a substantial burden on patients’ quality of life and represents a significant socioeconomic challenge. The accurate diagnosis and classification of epilepsy types are paramount for the formulation of effective therapeutic strategies. Electroencephalography (EEG) serves as a cornerstone in the diagnostic toolkit for epilepsy and has been extensively adopted in clinical practice. Traditionally, neurologists rely on visual inspection of EEG recordings to identify epileptic events. Although expert-based interpretation remains the clinical gold standard, it is time-consuming, labor-intensive, and subject to inter-observer variability. The visual inspection of EEG signals by neurologists remains the clinical gold standard for epilepsy diagnosis, but it is time-consuming, labor-intensive, and prone to subjective variability.
To alleviate this burden, numerous automated seizure detection methods based on traditional machine learning have been proposed [
2]. These methodologies primarily integrate conventional signal processing with machine learning techniques, focusing on the two critical stages of feature extraction and classification. In the feature extraction phase, investigators typically select EEG signal features manually, guided by empirical knowledge and observational data. Features derived from time, frequency, and time–frequency domains are extensively utilized for seizure identification. For instance, Sharmila and Geethanjali [
3] pioneered the application of a combination of time-domain features, initially developed for electromyography signal analysis, such as waveform length, number of zero-crossings, and number of slope sign changes, to the detection of epileptic EEG signals. Wen and Zhang [
4] proposed a frequency-domain feature selection method that combines sample entropy with a genetic algorithm for multi-class EEG signal analysis. Kambarova et al. [
5] explored the utility of nonlinear dynamic methods, such as fractal dimension, in the analysis of electroencephalograms from healthy individuals and patients with epilepsy. AlSharabi et al. [
6] introduced a diagnostic methodology for epilepsy founded on Discrete Wavelet Transform time–frequency analysis and Shannon entropy, which entails the decomposition of EEG signals into multiple time–frequency sub-bands followed by the extraction of entropy features. Building upon entropy-based approaches, Zhang et al. [
7] proposed an automatic epileptic EEG classification approach based on differential entropy and attention mechanism. Their method decomposed EEG recordings into five sub-frequency bands and employed an improved attention model framework as the classifier for nonpatient-specific evaluation. Similarly, Akter et al. [
8] developed a multi-band entropy-based feature extraction method focusing on high-frequency components (ripple and fast ripple) from interictal iEEG. They utilized eight different entropy measures, including approximate entropy, permutation entropy, Shannon entropy, sample entropy, Tsallis entropy, phase entropy, and Reny’s entropy, combined with sparse linear discriminant analysis for feature selection in epileptic focus identification. Entropy-based methods in epilepsy detection typically employ entropy measures such as input features [
7,
8], which are then fed into conventional machine learning classifiers. In addition to the aforementioned approaches, various nonlinear features have also been extensively applied to seizure detection. Madan et al. [
9] investigated the application of the Hurst exponent, derived from the Discrete Wavelet Transform, in epilepsy detection.
The classification stage typically employs a variety of machine learning classifiers to identify epileptic seizure activity. Guo et al. [
10] utilized artificial neural networks in conjunction with waveform complexity metrics to achieve automated detection of epileptic seizures EEGs. Brinkmann et al. [
11] applied support vector machine algorithms to analyze intracranial EEG data, successfully forecasting naturally occurring seizures. Wang et al. [
12] enhanced the recognition accuracy of multi-level epileptic states by combining random forests with grid search optimization. Na et al. [
13] improved the accuracy of epilepsy diagnosis by integrating an extended K-nearest neighbors classifier with a multi-distance decision-making mechanism. Although the aforementioned methods have shown good performance in certain epilepsy classification tasks, they still face two major limitations. First, they rely on handcrafted feature extraction based on domain expertise, which may fail to capture complex nonlinear features in EEG signals [
14,
15]. Second, their generalization ability is often limited, making it difficult to adapt to new datasets or heterogeneous patient populations [
16].
To address the limitations of traditional machine learning methods, recent studies have increasingly turned to deep learning for EEG decoding. Deep learning models support end-to-end learning directly from raw or minimally preprocessed EEG signals, eliminating the need for handcrafted features and reducing the subjectivity of feature engineering [
17,
18,
19,
20,
21]. Compared to traditional methods, deep learning offers more robust and consistent diagnostic support for clinicians, as well as improved generalizability across diverse patient populations and clinical settings for patients.
Currently, Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs) represent the most frequently employed deep learning models in epilepsy detection. CNNs have demonstrated significant advantages in this domain, primarily through their capacity to automatically extract high-level features from EEG signals, thereby circumventing the limitations of manual feature engineering inherent in traditional machine learning and enabling end-to-end automated diagnostic workflows. By leveraging multiple convolutional and pooling layers, CNNs can effectively learn intricate patterns and temporal dependencies within EEG signals. However, this methodology is confronted with challenges, including substantial requirements for training data, high computational complexity, and a propensity for overfitting when data are scarce. Moreover, the inherent constraints imposed by CNN kernel sizes often impede the capture of long-range dependencies crucial in EEG time-series analysis. In contrast to CNNs, which are predominantly oriented towards spatial feature extraction, RNNs, as deep learning architectures expressly tailored for time-series data, exhibit distinct advantages in epilepsy detection. They are particularly well suited for processing physiological data such as EEG signals, which are characterized by temporal dependencies and variable lengths. Long Short-Term Memory (LSTM) networks, a specialized RNN variant, effectively address the short-term memory and vanishing gradient problems of conventional RNNs through sophisticated gating mechanisms, enabling the capture of long-term temporal dependencies in EEG signals. Gated Recurrent Units (GRUs), another RNN variant, offer reduced model complexity by consolidating gating structures while maintaining robust sequence modeling capabilities. The inherent architecture of RNNs is naturally suited for the detection and prediction of time-series events like epileptic seizures, as they can learn complex temporal patterns within EEG signals. Nevertheless, RNN-based approaches also possess limitations, including protracted training durations, sensitivity to sequence length, high computational demands, and potential susceptibility to gradient-related issues when processing long sequences [
22]. Beyond CNNs and RNNs, recent studies have explored alternative deep learning paradigms tailored to the unique dynamics of brain signals. Li et al. [
23] proposed a graph-generative network to dynamically model evolving brain connectivity patterns. Ghosh et al. [
24] introduced a deep oscillatory neural network that uses Hopf oscillators to capture frequency-specific oscillatory dynamics. Similarly, Hadad et al. [
25] employed a biologically inspired spiking neural network to decode cognitive states from EEG signals using event-driven spike processing.
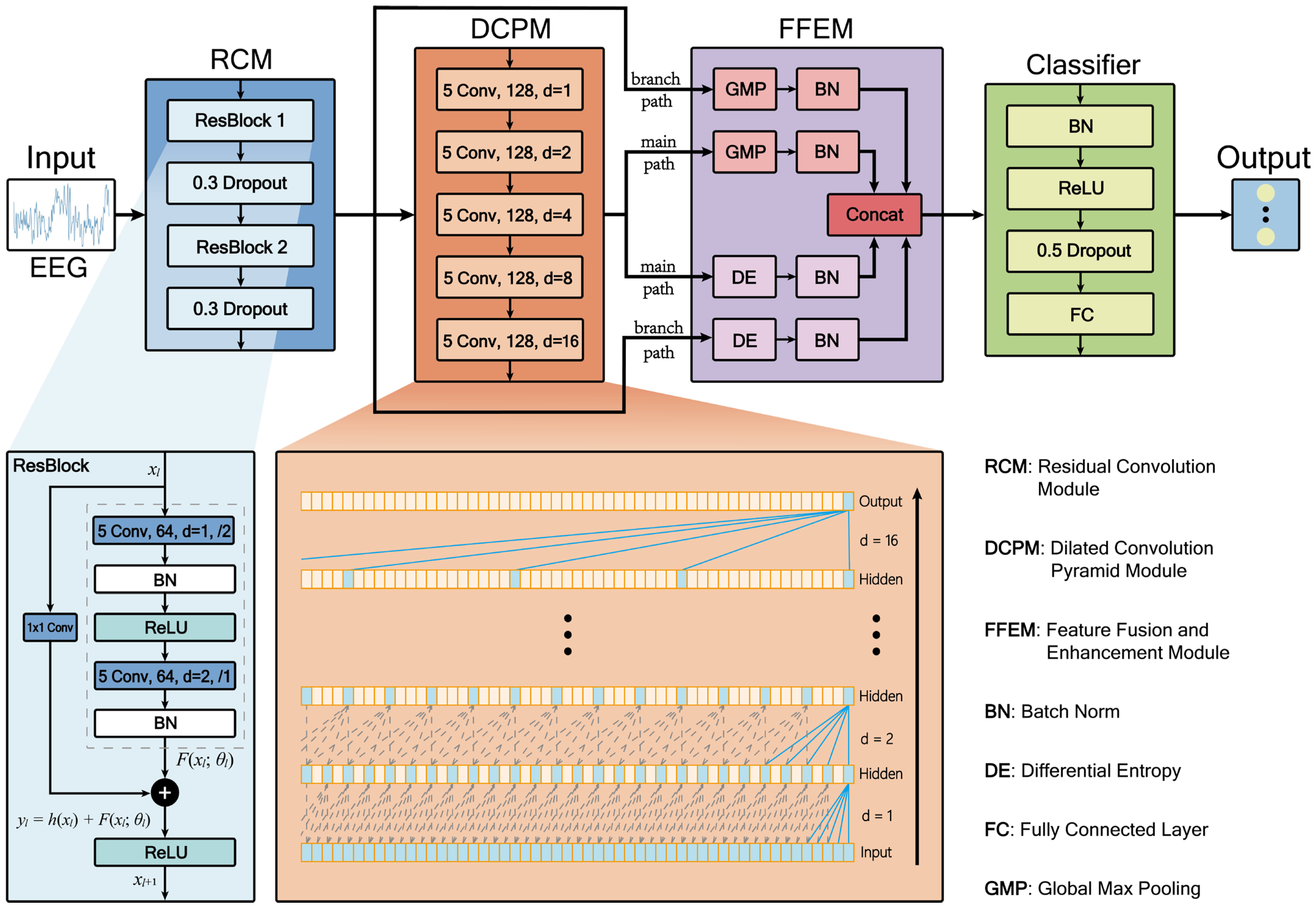
Despite significant advancements in deep learning methodologies, several key challenges persist in automated epileptic EEG classification. Existing models often struggle to simultaneously capture local detail features and long-range temporal dependencies in EEG signals effectively, while traditional entropy-based features are predominantly used as static inputs, failing to dynamically characterize the statistical complexity of deep network activations. Additionally, multi-scale feature fusion processes frequently encounter issues such as scale mismatch and redundant information interference. To address these challenges, we propose RDPNet, a novel multi-scale Residual Dilated Pyramid Network, with the following key contributions:
- (1)
We designed a novel network architecture for seizure detection by integrating residual convolutional module and dilated convolutional pyramids to jointly capture local features and global temporal dependencies in EEG signals.
- (2)
We introduced an entropy-guided dual-pathway fusion strategy that combined global max pooling with dynamically computed differential entropy features, enhancing the discriminability and statistical robustness of multi-scale representations.
- (3)
We conducted comprehensive experiments on the University of Bonn and Temple University Hospital EEG Seizure Corpus (TUSZ) benchmark datasets, and we demonstrated that RDPNet consistently outperformed several baseline methods in terms of classification accuracy and generalization across diverse clinical scenarios.
The remainder of this paper is organized as follows:
Section 2 presents related research and the current state of epilepsy detection technology.
Section 3 details the research methodology and experimental results, encompassing an introduction to the dataset, data preprocessing procedures, the architectural design of the proposed RDPNet along with its constituent components, ten-fold cross-validation results, ablation studies, parameter sensitivity analysis, feature visualization, and comprehensive comparative evaluation with baseline methods.
Section 4 discusses the limitations of the study and future research directions.
Section 5 summarizes the main contributions and findings of the entire paper. Through these sections, we will comprehensively demonstrate the efficacy of the proposed model and its prospective applications in the field of epilepsy detection.
2. Related Work
In the domain of neural network-based epilepsy classification, deep learning methodologies have demonstrated substantial developmental potential. Initial research endeavors predominantly focused on the application of foundational CNNs. Acharya et al. [
26] pioneered the application of deep convolutional neural networks to seizure detection in EEG signals, proposing an end-to-end automated analysis framework capable of autonomously learning and identifying features requisite for classification directly from raw EEG data. Truong et al. [
27] introduced a CNN-based method for seizure prediction, which extracts critical feature information through time-frequency domain transformation of EEG signals; this approach can automatically generate optimized features for each patient to optimally classify preictal and interictal segments. The method exhibits robust universality and generalization capabilities, consequently lowering the implementation threshold and expertise prerequisites for epilepsy prediction technologies. Pachori and Gandhi [
28] proposed a methodology integrating Fourier-Bessel Series Expansion (FBSE) with CNN classification for epileptic seizure detection. Their approach decomposes EEG signals into five rhythmic components using FBSE, applies Euclidean distance metrics to generate image representations, and subsequently classifies these images through a CNN architecture. This work demonstrates the potential of combining traditional signal processing techniques with deep learning frameworks for seizure detection. Zhao et al. [
29] designed a one-dimensional CNN employing larger convolutional kernels for seizure detection, constructing an end-to-end architecture comprising three convolutional blocks and three fully connected layers. This model integrates batch normalization and dropout layers within conventional convolutional blocks to augment model learning capacity and prevent overfitting.
Although foundational CNN models have demonstrated promising results in epilepsy detection, they continue to face limitations when handling more complex EEG signals, particularly in capturing multi-scale and long-range temporal patterns. To address these challenges, researchers have explored advanced CNN variants, notably Residual Networks (ResNets) and dilated convolutions, which offer improved representational capacity and feature extraction capabilities. ResNets have been introduced to mitigate the degradation and vanishing gradient problems associated with deep networks. For instance, Gao et al. [
30] employed a ResNet152 architecture for epileptic EEG classification. In their approach, EEG signals were first transformed into power spectral density energy diagrams, and the residual blocks of ResNet152 were leveraged to extract deep features. In parallel, to effectively expand the receptive field for capturing long-range dependencies in time-series data without incurring additional computational costs, dilated convolutions have been introduced into the field of seizure prediction. Hussein et al. [
31] proposed a novel “semi-dilated convolution” module, which mapped EEG signals to two-dimensional wavelet scalograms and applied dilated convolutions to handle the resulting nonsquare inputs, thereby improving seizure prediction accuracy. Building on this idea, Gao et al. [
32] further developed a spatiotemporal multi-scale convolutional network that utilizes dilated convolutions with different dilation rates in parallel across both temporal and spatial dimensions. This design enables the aggregation of multi-scale information from local to global levels, significantly enhancing patient-specific seizure prediction performance.
Although CNNs and their variants are effective in extracting spatial features, they are limited in modeling the temporal dependencies inherent in EEG signals. To address this, RNNs and their advanced variants, such as LSTM and GRU networks, have been widely adopted due to their ability to capture sequential patterns and mitigate gradient-related issues in long time-series data. Tsiouris et al. [
33] employed LSTM networks for long-term seizure prediction using continuous EEG recordings spanning several hours. Their approach incorporated features from both time and frequency domains, as well as interchannel correlations and graph-theoretic measures, across varying prediction windows from 15 min to 2 h. Zhang et al. [
34] proposed a seizure detection method based on a Bidirectional GRU (BiGRU) network, which captures both forward and backward temporal dependencies to improve classification performance. Similarly, Najafi et al. [
35] developed an RNN-LSTM-based model to distinguish between focal and generalized epilepsy, leveraging time–frequency features with a focus on theta-band activity.
As research has progressed, investigators have recognized the limitations of singular network architectures, leading to the development of more robust hybrid frameworks that amalgamate the spatial feature extraction capabilities of CNNs with the temporal modeling strengths of RNNs. Roy et al. [
36] introduced ChronoNet, a specialized RNN-based architecture for abnormal EEG detection. ChronoNet integrates stacked one-dimensional convolutional layers with deep GRU layers. Xu et al. [
37] developed a one-dimensional CNN-LSTM model for epileptic seizure recognition, wherein a CNN effectively extracts features from normalized EEG sequence data, which are subsequently processed by LSTM layers to further extract temporal characteristics. Zhao et al. [
38] introduced the ResBiLSTM hybrid deep learning method; this model initially employs a one-dimensional ResNet to adeptly extract local spatial features from EEG signals, following which the acquired features are input into a bidirectional Long Short-Term Memory (BiLSTM) network layer to model temporal dependencies, achieving end-to-end seizure detection through the integration of three residual blocks and the BiLSTM network. Moreover, Sun et al. [
39] proposed a causal spatiotemporal model that integrates transfer entropy-based causal graphs with GAT and BiLSTM to enhance epileptic seizure detection by capturing both interchannel causal relationships and spatiotemporal dynamics. Despite these advancements, the nonlinear structure and multi-scale temporal characteristics of EEG signals continue to pose challenges for accurate seizure detection. Drawing on the strengths of residual connections, dilated convolutions, and entropy-based measures, we designed a hybrid architecture to improve automated epileptic EEG classification.
4. Discussion
This section presents a comprehensive discussion of the experimental findings of RDPNet, focusing on its architectural components, entropy-based feature modeling, parameter sensitivity, temporal window selection, computational efficiency, and interpretability through feature visualization.
Ablation studies confirm the importance of each module. Removing the RCM resulted in noticeable performance degradation, indicating that residual connections help alleviate gradient vanishing and facilitate the learning of deeper representations. This is particularly important for modeling the complex nonlinear characteristics of epileptic EEG signals. The removal of the DCPM led to the most significant decline in performance, emphasizing the critical role of multi-scale temporal modeling. By progressively expanding the receptive field, dilated convolutions allow the model to capture temporal dynamics without increasing the number of parameters, which is essential for recognizing seizure patterns. Furthermore, excluding the differential entropy features from the FFEM reduced the classification performance, suggesting that quantifying statistical uncertainty contributes additional discriminative information. This highlights the value of integrating information-theoretic descriptors to enhance the model’s ability to distinguish between different epileptic states.
Parameter sensitivity analysis of convolution kernel sizes validates the rationality of the model design, indicating that appropriate kernel size selection is crucial for capturing the feature scales of EEG signals. Experimental results show that the optimal combination of residual block and dilated convolution kernels achieves an effective balance between local feature extraction and long-range dependency modeling. This finding emphasizes the importance of considering signal-specific characteristics in deep learning architecture design, providing valuable guidance for parameter selection in physiological signal processing.
Table 7 reveals the critical impact of temporal window length selection on model performance. As window length increased from 1 s to 3 s, the five-class accuracy improved from 88.87% to a peak of 95.1%. When window length further extended to 4 s, accuracy decreased to 94.95%. The FLOPs increased linearly with window length, rising from 25.86 million operations for the 1-s window to 101.74 million operations for the 4-s window. The single-sample inference time increased slightly with window length, from 1.806 ms to 1.879 ms. The results indicate that the 3-s temporal window achieves the optimal balance between accuracy and computational overhead.
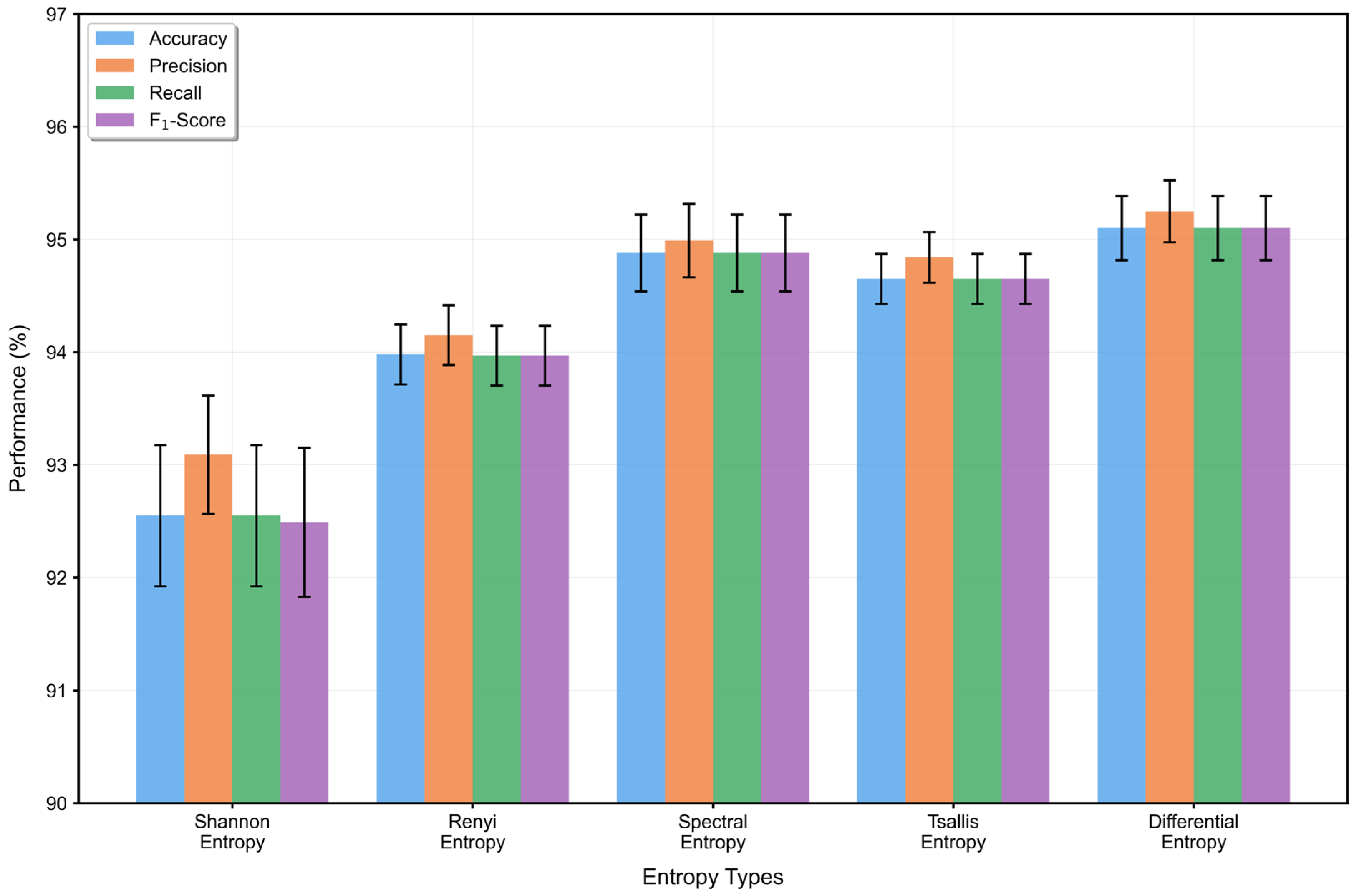
From an information-theoretic perspective, entropy serves as a fundamental measure of uncertainty and holds unique value in feature representation. While existing studies typically use entropy as handcrafted prior knowledge derived from raw EEG signals, our approach computes differential entropy on dual-pathway features processed through residual networks and dilated convolutional pyramids. Unlike traditional entropy metrics that rely on discretization, differential entropy operates directly on continuous-valued deep feature maps, allowing for a more accurate characterization of the statistical complexity and uncertainty embedded in learned representations.
Compared with existing methods, RDPNet offers pronounced technical advantages through its multi-scale hybrid architecture. Unlike CNN-LSTM pipelines that rely on sequential processing, RDPNet leverages residual and dilated convolutions to effectively model both short- and long-range temporal dependencies in EEG signals. The dual-pathway fusion mechanism further aggregates local and long-range cues, while FFEM introduces a differential entropy computation method for deep feature maps that dynamically computes differential entropy to quantify the statistical variability of deep feature maps, improving the recognition capability for subtle epileptic state differences. Due to the published baselines report only task-level ten-fold means and omit per-fold statistics, paired tests such as the Wilcoxon signed-rank could not be applied; we therefore adopted a conservative one-sided criterion (α = 0.05): a baseline is considered to perform worse when its mean accuracy falls below the lower bound of RDPNet’s t-based 95% confidence interval. Under this conservative evaluation criterion, RDPNet demonstrates superior performance compared to baseline methods on both binary and multi-class epileptic-EEG tasks; notably, on the severely imbalanced seven-class TUSZ benchmark, every competing method fell outside RDPNet’s CI (Acc: 95.14–96.32%; F1,w: 95.13–96.31%), indicating the model’s robustness and clinical relevance.
To address the concern regarding computational efficiency, we conducted a quantitative comparison between RDPNet and ReBiLSTM, one of the best-performing and publicly available baseline models, since most other baselines do not provide open-source implementations. RDPNet contains approximately 569 k parameters with a computational complexity of 75.27 M FLOPs, and it achieves an average inference time of 1.843 ms per sample. In contrast, ReBiLSTM has 315 k parameters, 45.99 M FLOPs, and an inference time of 1.231 ms.
Although RDPNet incurs a slightly higher computational cost, with an additional 0.61 milliseconds in inference time per sample, this overhead is considered acceptable and worthwhile in the context of critical clinical applications such as real-time epileptic seizure detection. Experimental results show that RDPNet achieved a 3.83% improvement in accuracy over ReBiLSTM on the five-class task of the Bonn dataset and a 0.7% gain on the seven-class classification task for different seizure types using the clinically sourced TUSZ dataset. These findings demonstrate that RDPNet offers a favorable balance between accuracy and efficiency, making it a strong candidate for deployment in medical scenarios that demand both high diagnostic precision and real-time responsiveness.
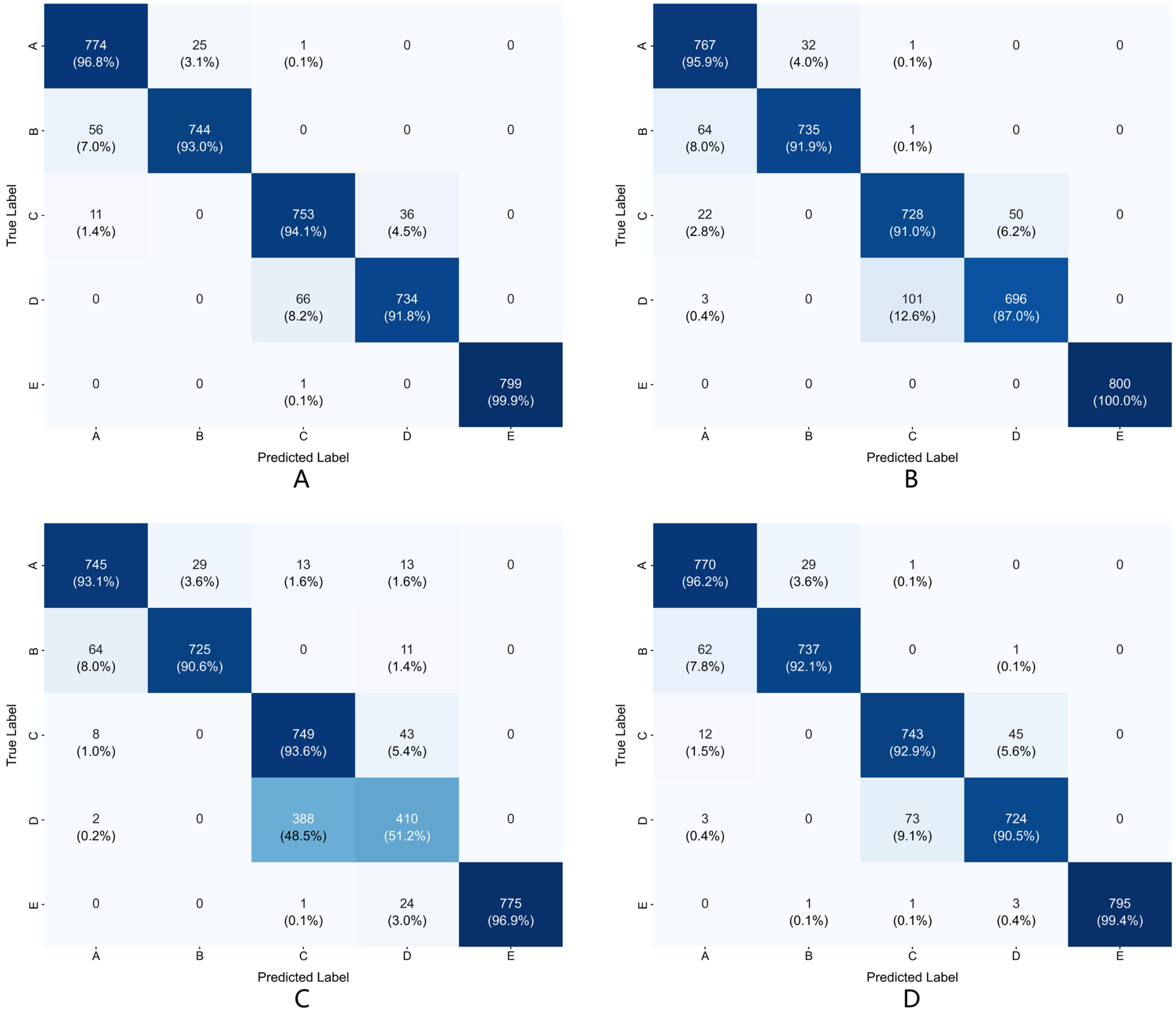
Feature visualization analysis provides intuitive evidence for the model’s effectiveness. Visualization results show that the model’s feature representation capability presents progressive improvement across different processing stages, particularly achieving effective separation for difficult-to-distinguish categories. This separation capability has important significance for clinical applications, as accurately distinguishing different epileptic states is the foundation for diagnosis and treatment decisions.
However, this study also has some limitations that need to be addressed in future work. First, the model’s generalization capability needs further validation on larger-scale and more diverse datasets. While benchmark datasets provide standardized platforms for model evaluation, signal characteristics in real clinical environments may be more complex and variable, including different acquisition devices, electrode configurations, and patient population characteristics. Second, the multi-module architecture incurs relatively high computational overhead, which may become a limiting factor in real-time monitoring applications. Third, while the cross-validation strategy employed in this study ensures statistical reliability, the lack of strict subject-independent validation may limit the conclusiveness of the model’s generalization performance assessment on unseen patients. Fourth, the model’s interpretability mechanisms need further enhancement to improve its acceptability and credibility in clinical practice. Medical applications have high requirements for the transparency of model decision-making processes, particularly requiring intuitive mechanisms for clinical reasoning explanations of individual predictions.
Future research can be extended in several directions. First, this can include developing stronger generalization techniques, such as domain adaptation and transfer learning methods, to improve the model’s applicability across different datasets and clinical environments. Cross-dataset validation and multi-center clinical trials will be important steps for validating the model’s practical utility. Second, the work can delve into exploring lightweight architectural designs to reduce computational complexity, making the model more suitable for deployment in resource-constrained environments, particularly portable monitoring devices. Third, conducting rigorous subject-independent evaluations using patient-level data splitting strategies to better assess the model’s generalization performance in real-world deployment scenarios will be necessary. Fourth, we can explore integrating interpretability mechanisms, such as Grad-CAM and other attention visualization and feature importance analysis methods, to enhance the transparency of the model’s decision-making process and promote its application in clinical practice. Meanwhile, combining domain knowledge and clinical experience to develop explanation frameworks that better align with medical diagnostic logic will help improve the model’s acceptance in clinical environments.