Application of Recurrence Plot Analysis to Examine Dynamics of Biological Molecules on the Example of Aggregation of Seed Mucilage Components
Abstract
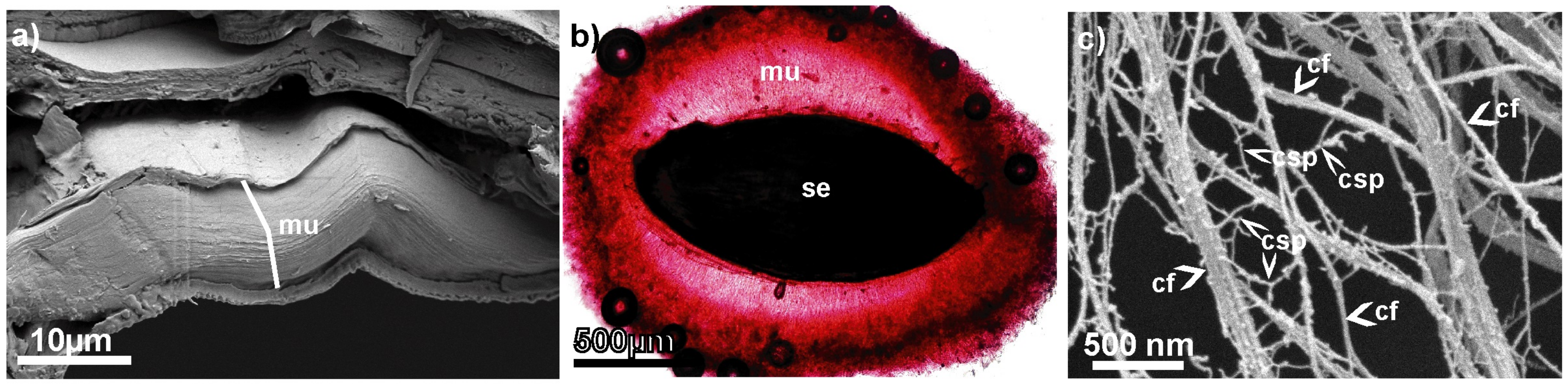
1. Introduction
2. Materials and Methods
2.1. Physical Model with Reference to Quantum Mechanics
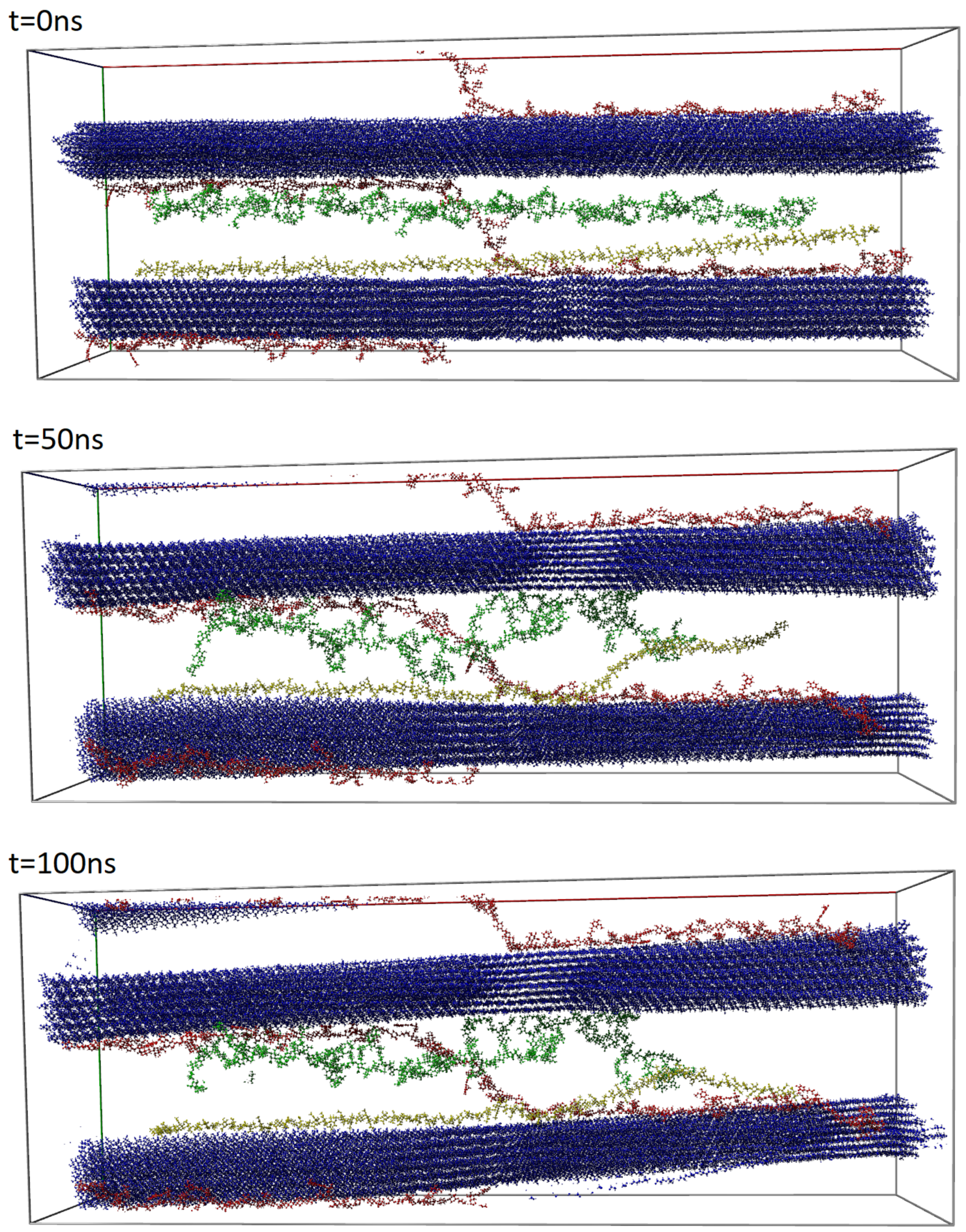
2.2. Molecular Dynamics Simulation
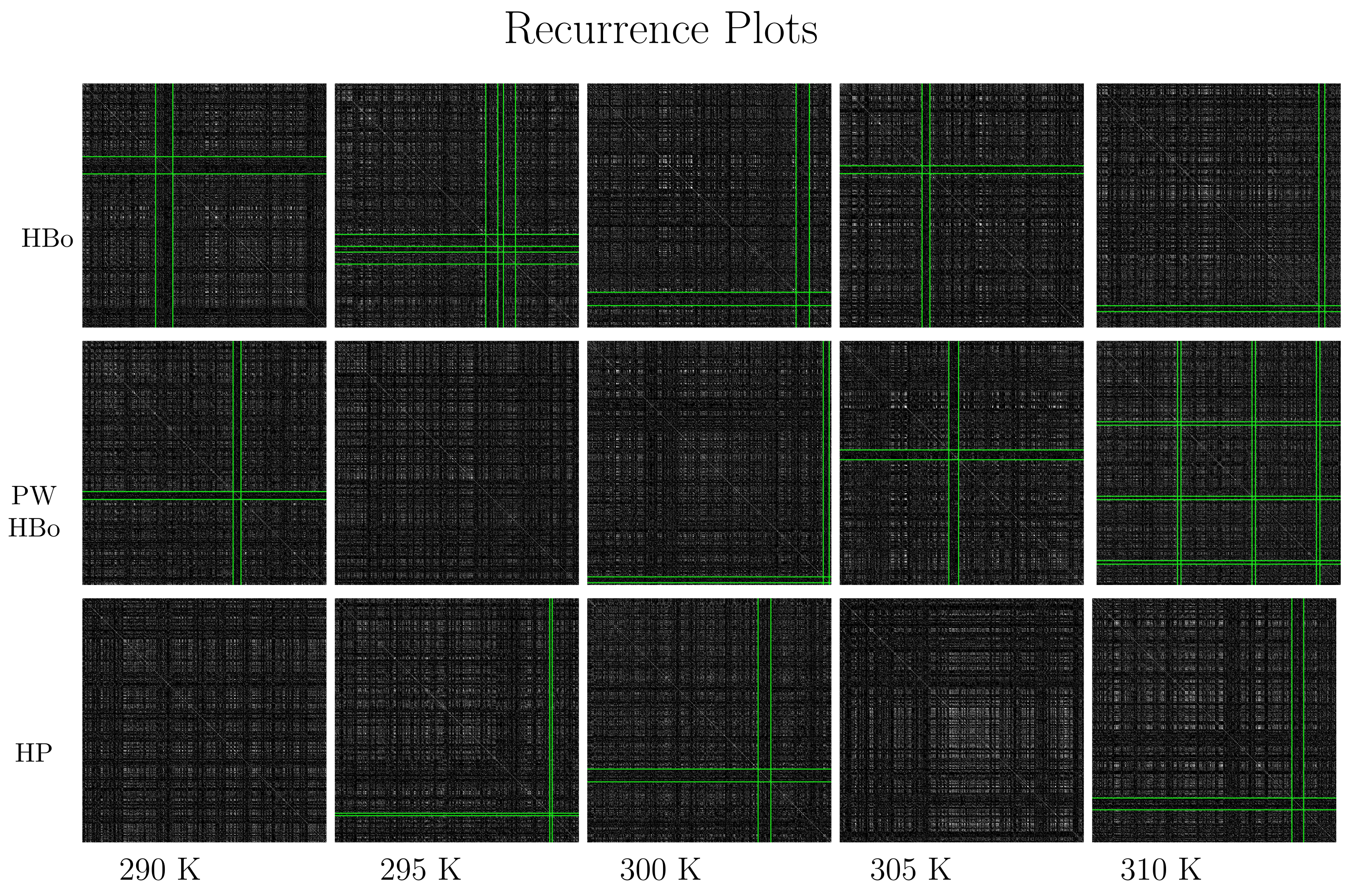
2.3. Recurrence Plot Method
2.3.1. Input
2.3.2. Proceeding
- Data compression. In the first step, we perform a compression of time series into the following sub-series:i.e., by putting at the beginning, spacing by , and keeping at the end. Such series (of length ) are numbered by i, where () corresponds to the initial part of , while corresponds to the rear part of , etc.Henceforth, the procedure is parameterized as follows:
- parameter —delay time;
- parameter d—embedding dimension of the system.
In our approach, the particular value of each of these parameters was computed by algorithmic methods discussed in [71], namely time delay was computed by the first non-significant auto-correlation introduced therein, and dimensional embedding was computed by the Cao method [72]. If we compare more time series, majority voting among data inputs is performed to determine one value of each parameter for a fair comparison. Generally, the recurrence plot method is dedicated to data originating from a chaotic physical system. Hence, subsequent elements of the data series are supposed to carry similar information. Besides, after a long enough evolution, a chaotic system is supposed to return to the neighborhood of the starting point, and the corresponding data record is supposed to reflect repeating information.Referring to the toy example of the artificial input vector in Equation (1) and assuming and (and ), we have in the following sub-series: - Sub-series transformation. From sub-series (), one creates a matrix () with zero and ones entries as follows:The idea beyond this representation relies on the comparison of subsequently compressed series ( and ) [73] with the norm . Ideally, such a distance should be smaller than the threshold value () determining the accuracy of compression in the previous step. In practice, the particular value of reveals the local dynamics of data. In detail, maps as in Equation (4) display interesting patterns that can be analyzed later on by sophisticated methods (see [74]). The fraction of zeros in from Equation (4) is the recurrence rate in the literature, and there is no single broadly accepted method concerning its determination.
- Recurrence plot creation. A recurrence plot is created by turning the zeros and ones in Equation (4) into white and black pixels (dots). The recurrence plot naturally visualizes data in a time window with a length of d and spacing of . Such recurrence plots can be analyzed either manually (the state-of-the-art approach) or in an automatic manner (a new approach if advanced tools of image processing are applied). Here, we are left with the open question as to what particular information is tied to the recurrence plot and how it depends on parameters (and the recurrence rate in particular).
- Entropy. To assess information tied to the recurrence plots, we first use the most straightforward approach, namely the entropy approach as proposed in [27] and by us in [71]. Following this, we compute the Shannon entropy from the distribution of specific features of the recurrence plot. Following [27], we estimate topological entropy, which measures the total complexity of the orbit structure of the chaotic system, as the Shannon entropy of the distribution of parallel-to-diagonal lines. In detail, a histogram is made for the lengths of zero sequences along the diagonal direction. Normalizing the histogram and thinking of it as a probability distribution (), a Shannon entropy can be derived through the following equation:where is the probability of a sequence of length j and the summation is over all sequence lengths. Then, we analyze such entropy for the entire range of the recurrence rate (from 0 to 1) to assess whether the information tied to the recurrence plot varies with the recurrence rate and select its most suitable value.
- For , we have , , and ; and
- For , we have , , and .
- Recurrence plot analysis and automatic pattern recognition. If we refer in more detail to the recurrence plot layout, we can conclude that in regions close to its diagonal (i close to in Equation (4)) sub-series from close time instants are analyzed, while in the regions away from the diagonal (i is far from ), series from distinct time instants are analyzed. Furthermore, if we observe the dark cross hitting the diagonal at certain subsequent sub-series (i.e., ), the dynamics are tied to such that should differ from the dynamics of the rest of the data series. Such is interesting from the point of view of pattern recognition and data analysis. To search for automatically, we introduce the following cross-detection procedure:
- (a)
- Compute the column-wise weight of the recurrence plot in Equation (4) (), and create weight vector ;
- (b)
- Smooth to by applying the simple moving average (SMA) of size k, where k is the parameter;
- (c)
- Normalize to by subtracting the mean and dividing by the standard deviation;
- (d)
- Determine the k parameter for SMA to maximize the highest value of the normalized vector, namely
- (e)
- Save a k value equal to the width of dark crosses;
- (f)
- Determine cross centers as local maxima of the smoothed that are greater than the threshold standard score (number of standard deviations).
Converting into a standard score enables us to work with a vector of unitless, standardized values. This facilitates comparison across different datasets and enhances the method’s versatility. With standard scores established, it becomes feasible to suggest a default threshold value for cross-detection, such as or ( denotes standard deviation). An alternative solution would require the inspection of each dataset individually and would be more challenging to automate. Please, keep in mind that this method can be easily generalized for white cross-detection.
2.3.3. Output
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FF | Force field |
| HBo | Bydrogen bond |
| PW HBo | Polysaccharides–water hydrogen bond |
| HP | Hydrophobic–polar |
| MD | Molecular dynamics |
| AIMD | Ab initio molecular dynamics |
| PDB | Protein Data Bank |
| HG | Homogalacturonan |
| RG1 | Rhamnogalacturonan I |
References
- Phan, J.L.; Burton, R.A. New Insights into the Composition and Structure of Seed Mucilage. In Annual Plant Reviews Online; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2018; pp. 63–104. [Google Scholar] [CrossRef]
- Kreitschitz, A. Biological Properties of Fruit and Seed Slime Envelope: How to Live, Fly, and Not Die. In Functional Surfaces in Biology: Little Structures with Big Effects Volume 1; Gorb, S.N., Ed.; Springer: Dordrecht, The Netherlands, 2009; pp. 11–30. [Google Scholar] [CrossRef]
- Ralet, M.C.; Crépeau, M.J.; Vigouroux, J.; Tran, J.; Berger, A.; Sallé, C.; Granier, F.; Botran, L.; North, H.M. Xylans Provide the Structural Driving Force for Mucilage Adhesion to the Arabidopsis Seed Coat. Plant Physiol. 2016, 171, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Kučka, M.; Ražná, K.; Harenčár, V.; Kolarovičová, T. Plant Seed Mucilage—Great Potential for Sticky Matter. Nutraceuticals 2022, 2, 253–269. [Google Scholar] [CrossRef]
- Western, T.L. The sticky tale of seed coat mucilages: Production, genetics, and role in seed germination and dispersal. Seed Sci. Res. 2012, 22, 1–25. [Google Scholar] [CrossRef]
- Yokoyama, R.; Shinohara, N.; Asaoka, R.; Narukawa, H.; Nishitani, K. The Biosynthesis and Function of Polysaccharide Components of the Plant Cell Wall. In Plant Cell Wall Patterning and Cell Shape; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014; Chapter 1; pp. 1–34. [Google Scholar] [CrossRef]
- Houston, K.; Tucker, M.R.; Chowdhury, J.; Shirley, N.; Little, A. The plant cell wall: A complex and dynamic structure as revealed by the responses of genes under stress conditions. Front. Plant Sci. 2016, 7, 984. [Google Scholar] [CrossRef] [PubMed]
- Kreitschitz, A.; Gorb, S.N. The micro- and nanoscale spatial architecture of the seed mucilage—Comparative study of selected plant species. PLoS ONE 2018, 13, e0200522. [Google Scholar] [CrossRef] [PubMed]
- Haughn, G.W.; Western, T.L. Arabidopsis seed coat mucilage is a specialized cell wall that can be used as a model for genetic analysis of plant cell wall structure and function. Front. Plant Sci. 2012, 3, 64. [Google Scholar] [CrossRef]
- Kreitschitz, A.; Gorb, S.N. How does the cell wall ‘stick’ in the mucilage? A detailed microstructural analysis of the seed coat mucilaginous cell wall. Flora 2017, 229, 9–22. [Google Scholar] [CrossRef]
- Kruszewska, N.; Weber, P.; Gadomski, A.; Domino, K. A Method of Mechanical Control of Structure-property Relationship in Grains-containing Material Systems. Acta Phys. Pol. B 2013, 44, 1049. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, J.; Wang, X.; Durachko, D.; Zhang, S.; Cosgrove, D. Molecular insights into the complex mechanics of plant epidermal cell walls. Science 2021, 372, 706–711. [Google Scholar] [CrossRef]
- Zhao, Z.; Crespi, V.H.; Kubicki, J.D.; Cosgrove, D.J.; Zhong, L. Molecular dynamics simulation study of xyloglucan adsorption on cellulose surfaces: Effects of surface hydrophobicity and side-chain variation. Cellulose 2014, 21, 1025–1039. [Google Scholar] [CrossRef]
- Huang, R.; Becker, A.A.; Jones, I.A. A finite strain fibre-reinforced viscoelasto-viscoplastic model of plant cell wall growth. J. Eng. Math. 2015, 95, 121–154. [Google Scholar] [CrossRef]
- Khodayari, A.; Thielemans, W.; Hirn, U.; Van Vuure, A.W.; Seveno, D. Cellulose-hemicellulose interactions—A nanoscale view. Carbohydr. Polym. 2021, 270, 118364. [Google Scholar] [CrossRef]
- Heinonen, E.; Henriksson, G.; Lindström, M.E.; Vilaplana, F.; Wohlert, J. Xylan adsorption on cellulose: Preferred alignment and local surface immobilizing effect. Carbohydr. Polym. 2022, 285, 119221. [Google Scholar] [CrossRef] [PubMed]
- Del Mundo, J.T.; Rongpipi, S.; Yang, H.; Ye, D.; Kiemle, S.N.; Moffitt, S.L.; Troxel, C.L.; Toney, M.F.; Zhu, C.; Kubicki, J.D.; et al. Grazing-incidence diffraction reveals cellulose and pectin organization in hydrated plant primary cell wall. Sci. Rep. 2023, 13, 5421. [Google Scholar] [CrossRef] [PubMed]
- Gastegger, M.; Marquetand, P. Molecular Dynamics with Neural Network Potentials. In Machine Learning Meets Quantum Physics; Springer: Cham, Switzerland, 2020; pp. 233–252. [Google Scholar] [CrossRef]
- MacKerell, A.D.J.; Bashford, D.; Bellott, M.; Dunbrack, R.L.J.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Popelier, P.L.A. Non-covalent interactions from a Quantum Chemical Topology perspective. J. Mol. Model. 2022, 28, 276. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Schade, R.; Kenter, T.; Elgabarty, H.; Lass, M.; Schütt, O.; Lazzaro, A.; Pabst, H.; Mohr, S.; Hutter, J.; Kühne, T.D.; et al. Towards electronic structure-based ab-initio molecular dynamics simulations with hundreds of millions of atoms. Parallel Comput. 2022, 111, 102920. [Google Scholar] [CrossRef]
- Gao, Y.; Mei, Y.; Zhang, J.Z.H. Treatment of Hydrogen Bonds in Protein Simulations. In Advanced Materials for Renewable Hydrogen Production, Storage and Utilization; Liu, J., Ed.; IntechOpen: Rijeka, Croatia, 2015; Chapter 5. [Google Scholar] [CrossRef]
- Iftimie, R.; Minary, P.; Tuckerman, M.E. Ab initio molecular dynamics: Concepts, recent developments, and future trends. Proc. Natl. Acad. Sci. USA 2005, 102, 6654–6659. [Google Scholar] [CrossRef]
- Thiel, M.; Romano, M.C.; Kurths, J. How much information is contained in a recurrence plot? Phys. Lett. A 2004, 330, 343–349. [Google Scholar] [CrossRef]
- Marwan, N.; Romano, M.C.; Thiel, M.; Kurths, J. Recurrence plots for the analysis of complex systems. Phys. Rep. 2007, 438, 237–329. [Google Scholar] [CrossRef]
- Marwan, N.; Donges, J.F.; Zou, Y.; Donner, R.V.; Kurths, J. Complex network approach for recurrence analysis of time series. Phys. Lett. A 2009, 373, 4246–4254. [Google Scholar] [CrossRef]
- Rawald, T.; Sips, M.; Marwan, N. PyRQA—Conducting recurrence quantification analysis on very long time series efficiently. Comput. Geosci. 2017, 104, 101–108. [Google Scholar] [CrossRef]
- Lehn, J.M. Toward Self-Organization and Complex Matter. Science 2002, 295, 2400–2403. [Google Scholar] [CrossRef] [PubMed]
- Wołek, K.; Cieplak, M. Self-assembly of model proteins into virus capsids. J. Phys. Condens. Matter 2017, 29, 474003. [Google Scholar] [CrossRef]
- Gadomski, A.; Rubí, J.M.; Łuczka, J.; Ausloos, M. On temperature- and space-dimension dependent matter agglomerations in a mature growing stage. Chem. Phys. 2005, 310, 153–161. [Google Scholar] [CrossRef]
- Herrmann, K.; Gerngross, O.; Abitz, W. Zur röntgenographischen Strukturerforschung des Gelatinemicells. Z. Phys. Chem. 1930, 10B, 371–394. [Google Scholar] [CrossRef]
- Yan, L.; Zhu, Q. Direct observation of the fringed micelles structure of cellulose molecules solvated in dimethylacetamide/LiCl system. Polym. Int. 2002, 51, 738–739. [Google Scholar] [CrossRef]
- Zhang, M.C.; Guo, B.H.; Xu, J. A Review on Polymer Crystallization Theories. Crystals 2017, 7, 4. [Google Scholar] [CrossRef]
- Kruszewska, N.; Gadomski, A. Revealing sol–gel type main effects by exploring a molecular cluster behavior in model in-plane amphiphilic aggregations. Phys. A Stat. Mech. Its Appl. 2010, 389, 3053–3068. [Google Scholar] [CrossRef]
- Hong, Y.l.; Koga, T.; Miyoshi, T. Chain Trajectory and Crystallization Mechanism of a Semicrystalline Polymer in Melt- and Solution-Grown Crystals As Studied Using 13C–13C Double-Quantum NMR. Macromolecules 2015, 48, 3282–3293. [Google Scholar] [CrossRef]
- Gadomski, A. (Nano)Granules-Involving Aggregation at a Passage to the Nanoscale as Viewed in Terms of a Diffusive Heisenberg Relation. Entropy 2024, 26, 76. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Yakubov, G.E.; Gilbert, E.P.; Sewell, K.; van de Meene, A.M.L.; Stokes, J.R. Multi-scale assembly of hydrogels formed by highly branched arabinoxylans from Plantago ovata seed mucilage studied by USANS/SANS and rheology. Carbohydr. Polym. 2019, 207, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Haken, H.; Wolf, H.C. Fundamentals of the Quantum Theory of Chemical Bonding. In The Physics of Atoms and Quanta: Introduction to Experiments and Theory; Springer: Berlin/Heidelberg, Germany, 1996; pp. 399–420. [Google Scholar] [CrossRef]
- Li, X.Z.; Walker, B.; Michaelides, A. Quantum nature of the hydrogen bond. Proc. Natl. Acad. Sci. USA 2011, 108, 6369–6373. [Google Scholar] [CrossRef]
- Wohlert, M.; Benselfelt, T.; Wågberg, L.; Furó, I.; Berglund, L.A.; Wohlert, J. Cellulose and the role of hydrogen bonds: Not in charge of everything. Cellulose 2022, 29, 1–23. [Google Scholar] [CrossRef]
- Lindman, B.; Karlström, G.; Stigsson, L. On the mechanism of dissolution of cellulose. J. Mol. Liq. 2010, 156, 76–81. [Google Scholar] [CrossRef]
- Gómez, S.; Rojas-Valencia, N.; Gómez, S.A.; Cappelli, C.; Merino, G.; Restrepo, A. A molecular twist on hydrophobicity. Chem. Sci. 2021, 12, 9233–9245. [Google Scholar] [CrossRef] [PubMed]
- Kreitschitz, A.; Haase, E.; Gorb, S.N. The role of mucilage envelope in the endozoochory of selected plant taxa. Sci. Nat. 2020, 108, 2. [Google Scholar] [CrossRef]
- Gawkowska, D.; Cybulska, J.; Zdunek, A. Structure-Related Gelling of Pectins and Linking with Other Natural Compounds: A Review. Polymers 2018, 10, 762. [Google Scholar] [CrossRef]
- Said, N.S.; Olawuyi, I.F.; Lee, W.Y. Pectin Hydrogels: Gel-Forming Behaviors, Mechanisms, and Food Applications. Gels 2023, 9, 732. [Google Scholar] [CrossRef] [PubMed]
- Facas, G.G.; Maliekkal, V.; Zhu, C.; Neurock, M.; Dauenhauer, P.J. Cooperative Activation of Cellulose with Natural Calcium. JACS Au 2021, 1, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Ratcliffe, I.; Williams, P.A.; Luo, S.; Chen, J.; Liu, C. The influence of pH and monovalent ions on the gelation of pectin from the fruit seeds of the creeping fig plant. Food Hydrocoll. 2021, 111, 106219. [Google Scholar] [CrossRef]
- Sanna, N.; Chillemi, G.; Grandi, A.; Castelli, S.; Desideri, A.; Barone, V. New Hints on the pH-Driven Tautomeric Equilibria of the Topotecan Anticancer Drug in Aqueous Solutions from an Integrated Spectroscopic and Quantum-Mechanical Approach. J. Am. Chem. Soc. 2005, 127, 15429–15436. [Google Scholar] [CrossRef] [PubMed]
- Wybranowski, T.; Cyrankiewicz, M.; Ziomkowska, B.; Kruszewski, S. The HSA affinity of warfarin and flurbiprofen determined by fluorescence anisotropy measurements of camptothecin. Biosystems 2008, 94, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Gadomski, A.; Zielińska-Raczyńska, S. Information and Statistical Measures in Classical vs. Quantum Condensed-Matter and Related Systems. Entropy 2020, 22, 645. [Google Scholar] [CrossRef] [PubMed]
- Gadomski, A.; Kruszewska, N. Matter-Aggregating Low-Dimensional Nanostructures at the Edge of the Classical vs. Quantum Realm. Entropy 2023, 25, 1. [Google Scholar] [CrossRef]
- Nelson, E. Derivation of the Schrödinger Equation from Newtonian Mechanics. Phys. Rev. 1966, 150, 1079–1085. [Google Scholar] [CrossRef]
- Gomes, T.C.F.; Skaf, M.S. Cellulose-Builder: A toolkit for building crystalline structures of cellulose. J. Comput. Chem. 2012, 33, 1338–1346. [Google Scholar] [CrossRef]
- Ding, S.Y.; Himmel, M.E. The Maize Primary Cell Wall Microfibril: A New Model Derived from Direct Visualization. J. Agric. Food Chem. 2006, 54, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.M.; Mota, T.R.; Salatta, F.V.; Marchiosi, R.; Gomez, L.D.; McQueen-Mason, S.J.; Ferrarese-Filho, O.; dos Santos, W.D. Designing xylan for improved sustainable biofuel production. Plant Biotechnol. J. 2019, 17, 2225–2227. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks, C.L., III; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef]
- Harholt, J.; Suttangkakul, A.; Vibe Scheller, H. Biosynthesis of Pectin. Plant Physiol. 2010, 153, 384–395. [Google Scholar] [CrossRef]
- Ochoa-Villarreal, M.; Aispuro-Hernández, E.; Vargas-Arispuro, I.; Martínez-Téllez, M.A. Plant Cell Wall Polymers: Function, Structure and Biological Activity of Their Derivatives. In Polymerization; IntechOpen: London, UK, 2012. [Google Scholar] [CrossRef]
- Nepogodiev, S.A.; Field, R.A.; Damager, I. Approaches to Chemical Synthesis of Pectic Oligosaccharides. In Annual Plant Reviews; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2010; pp. 65–92. [Google Scholar] [CrossRef]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; González-Outeiriño, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef] [PubMed]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.E.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins Struct. Funct. Bioinform. 2006, 65, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Essmann, U.; Perera, L.E.; Berkowitz, M.L.; Darden, T.A.; Lee, H.C.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Siódmiak, J.; Bełdowski, P.; Augé, W.; Ledziński, D.; Śmigiel, S.; Gadomski, A. Molecular dynamic analysis of hyaluronic acid and phospholipid interaction in tribological surgical adjuvant design for osteoarthritis. Molecules 2017, 22, 1436. [Google Scholar] [CrossRef]
- Kruszewska, N.; Bełdowski, P.; Domino, K.; Lambert, K.D. Investigating conformation changes and network formation of mucin in joints functioning in human locomotion. In Multiscale (Loco)motion—Toward Its Active-Matter Addressing Physical Principles; Gadomski, A., Ed.; UTP Publishing Department: Bydgoszcz, Poland, 2019; pp. 121–138. [Google Scholar]
- Krieger, E.; Vriend, G. YASARA View—Molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef] [PubMed]
- Weber, P.; Bełdowski, P.; Gadomski, A.; Domino, K.; Sionkowski, P.; Ledziński, D. Statistical method for analysis of interactions between chosen protein and chondroitin sulfate in an aqueous environment. arXiv 2022, arXiv:2202.07461. [Google Scholar]
- Cao, L. Practical method for determining the minimum embedding dimension of a scalar time series. Phys. D Nonlinear Phenom. 1997, 110, 43–50. [Google Scholar] [CrossRef]
- Eckmann, J.P.; Kamphorst, S.O.; Ruelle, D. Recurrence plots of dynamical systems. World Sci. Ser. Nonlinear Sci. Ser. A 1995, 16, 441–446. [Google Scholar]
- Goswami, B. A brief introduction to nonlinear time series analysis and recurrence plots. Vibration 2019, 2, 332–368. [Google Scholar] [CrossRef]
- Hanley, S.J.; Revol, J.F.; Godbout, L.; Gray, D.G. Atomic force microscopy and transmission electron microscopy of cellulose from Micrasterias denticulata; evidence for a chiral helical microfibril twist. Cellulose 1997, 4, 209–220. [Google Scholar] [CrossRef]
- Ye, D.; Rongpipi, S.; Kiemle, S.N.; Barnes, W.J.; Chaves, A.M.; Zhu, C.; Norman, V.A.; Liebman-Peláez, A.; Hexemer, A.; Toney, M.F.; et al. Preferred crystallographic orientation of cellulose in plant primary cell walls. Nat. Commun. 2020, 11, 4720. [Google Scholar] [CrossRef]
- Hadden, J.A.; French, A.D.; Woods, R.J. Unraveling cellulose microfibrils: A twisted tale. Biopolymers 2013, 99, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Altaner, C.M.; Jarvis, M.C. Modelling polymer interactions of the `molecular Velcro’ type in wood under mechanical stress. J. Theor. Biol. 2008, 253, 434–445. [Google Scholar] [CrossRef]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Voiniciuc, C.; Schmidt, M.H.W.; Berger, A.; Yang, B.; Ebert, B.; Scheller, H.V.; North, H.M.; Usadel, B.; Günl, M. MUCILAGE-RELATED10 produces galactoglucomannan that maintains pectin and cellulose architecture in Arabidopsis seed mucilage. Plant Physiol. 2015, 169, 403–420. [Google Scholar] [CrossRef] [PubMed]
- Durell, S.R.; Ben-Naim, A. Temperature Dependence of Hydrophobic and Hydrophilic Forces and Interactions. J. Phys. Chem. B 2021, 125, 13137–13146. [Google Scholar] [CrossRef]
- Zbilut, J.P.; Zaldivar-Comenges, J.M.; Strozzi, F. Recurrence quantification based Liapunov exponents for monitoring divergence in experimental data. Phys. Lett. A 2002, 297, 173–181. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, J.Q.; Liu, Y.; Hu, B.; Yang, Y.; Zhao, L.; Lu, Q. Effect of temperature on the interactions between cellulose and lignin via molecular dynamics simulations. Cellulose 2022, 29, 6565–6578. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, E.; Zhu, Y. Image segmentation evaluation: A survey of methods. Artif. Intell. Rev. 2020, 53, 5637–5674. [Google Scholar] [CrossRef]
- Zou, Z.; Chen, K.; Shi, Z.; Guo, Y.; Ye, J. Object detection in 20 years: A survey. Proc. IEEE 2023, 111, 257–276. [Google Scholar] [CrossRef]
- Zhao, Z.Q.; Zheng, P.; Xu, S.t.; Wu, X. Object detection with deep learning: A review. IEEE Trans. Neural Netw. Learn. Syst. 2019, 30, 3212–3232. [Google Scholar] [CrossRef]
- Kirichenko, L.; Zinchenko, P.; Radivilova, T. Classification of time realizations using machine learning recognition of recurrence plots. In International Scientific Conference “Intellectual Systems of Decision Making and Problem of Computational Intelligence”; Springer: Cham, Switzerland, 2020; pp. 687–696. [Google Scholar]
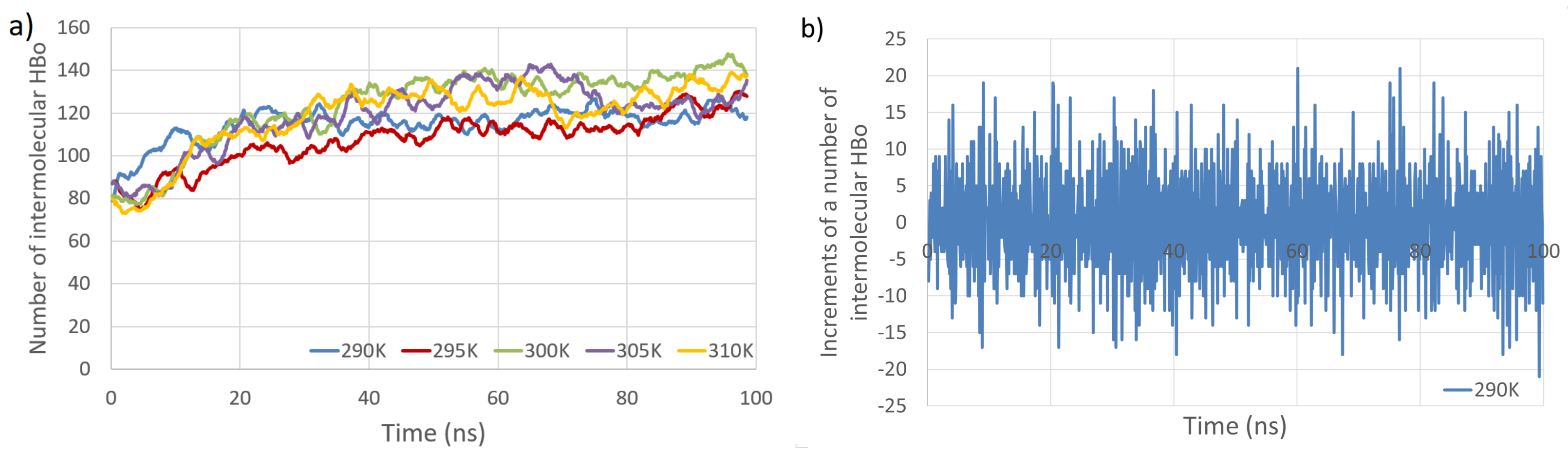
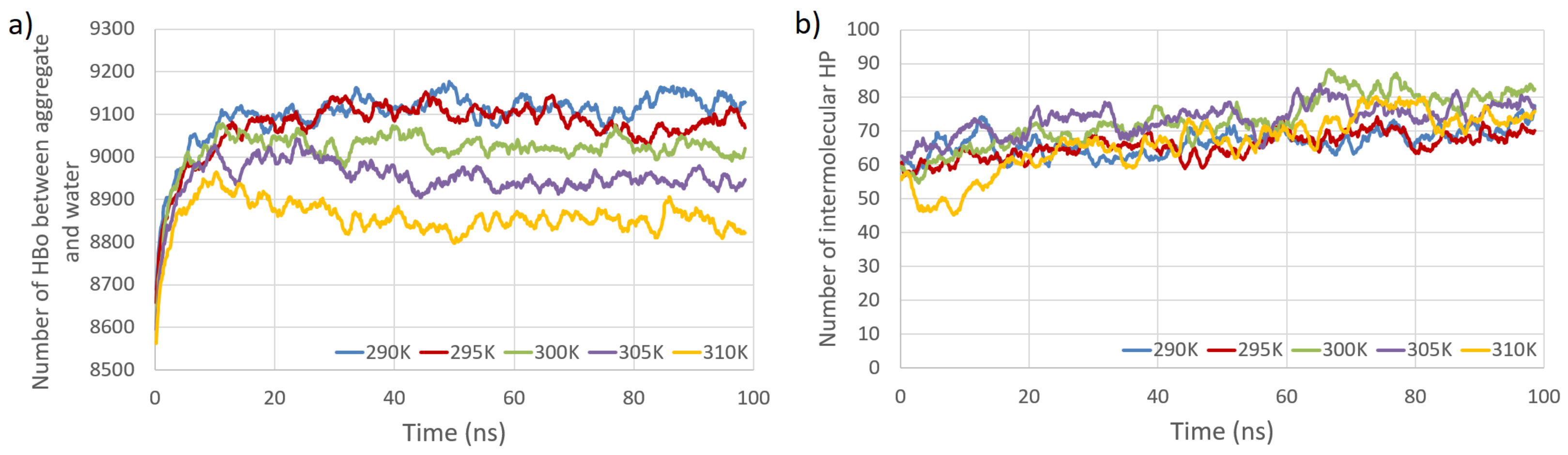

| Interaction Type | Entropy | n.o. Bonds | Entropy | n.o. Bonds | ||
|---|---|---|---|---|---|---|
| Mean | Mean | |||||
| Intermolecular HBo | 114.8 | 9.9 | 118.7 | 17.4 | ||
| PW HBo | 9098.4 | 84.0 | 8855.3 | 69.4 | ||
| Intermolecular HP | 66.7 | 6.8 | 66.7 | 10.2 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sionkowski, P.; Kruszewska, N.; Kreitschitz, A.; Gorb, S.N.; Domino, K. Application of Recurrence Plot Analysis to Examine Dynamics of Biological Molecules on the Example of Aggregation of Seed Mucilage Components. Entropy 2024, 26, 380. https://doi.org/10.3390/e26050380
Sionkowski P, Kruszewska N, Kreitschitz A, Gorb SN, Domino K. Application of Recurrence Plot Analysis to Examine Dynamics of Biological Molecules on the Example of Aggregation of Seed Mucilage Components. Entropy. 2024; 26(5):380. https://doi.org/10.3390/e26050380
Chicago/Turabian StyleSionkowski, Piotr, Natalia Kruszewska, Agnieszka Kreitschitz, Stanislav N. Gorb, and Krzysztof Domino. 2024. "Application of Recurrence Plot Analysis to Examine Dynamics of Biological Molecules on the Example of Aggregation of Seed Mucilage Components" Entropy 26, no. 5: 380. https://doi.org/10.3390/e26050380
APA StyleSionkowski, P., Kruszewska, N., Kreitschitz, A., Gorb, S. N., & Domino, K. (2024). Application of Recurrence Plot Analysis to Examine Dynamics of Biological Molecules on the Example of Aggregation of Seed Mucilage Components. Entropy, 26(5), 380. https://doi.org/10.3390/e26050380









