ABCDP: Approximate Bayesian Computation with Differential Privacy
Abstract
:1. Introduction
- We provide a novel ABC framework, ABCDP, which combines the sparse vector technique (SVT) [17] with the rejection ABC paradigm. The resulting ABCDP framework can improve the trade-off between the privacy and accuracy of the posterior samples, as the privacy cost under ABCDP is a function of the number of accepted posterior samples only.
- We theoretically analyze ABCDP by focusing on the effect of noisy posterior samples in terms of two quantities. The first quantity provides the probability of an output of ABCDP being different from that of ABC at any given time during inference. The second quantity provides the convergence rate, i.e., how fast the posterior integral using ABCDP’s noisy samples’ approaches that using non-private ABC’s samples. We write both quantities as a function of added noise for privacy to better understand the characteristics of ABCDP.
- We validate our theory in the experiments using several simulators. The results of these experiments are consistent with our theoretical findings on the flip probability and the average error induced by the noise addition for privacy.
2. Background
2.1. Approximate Bayesian Computation
Maximum Mean Discrepancy
2.2. Differential Privacy
2.3. AboveThreshold and Sparse Vector Technique
3. Problem Formulation
- Non-private step: The modeler draws a parameter sample ; then generates a pseudo-dataset , where for for a large T. We assume these parameter-pseudo-data pairs are publicly available (even to an adversary).
- Private step: the data owner takes the whole sequence of parameter-pseudo-data pairs and runs our ABCDP algorithm in order to output a set of differentially private binary indicators determining whether or not to accept each .
4. ABCDP
| Algorithm 1 Proposed c-sample ABCDP |
|
4.1. Effect of Noise Added to ABC
4.2. Convergence of Posterior Expectation of Rejection-ABCDP to Rejection-ABC
5. Related Work
6. Experiments
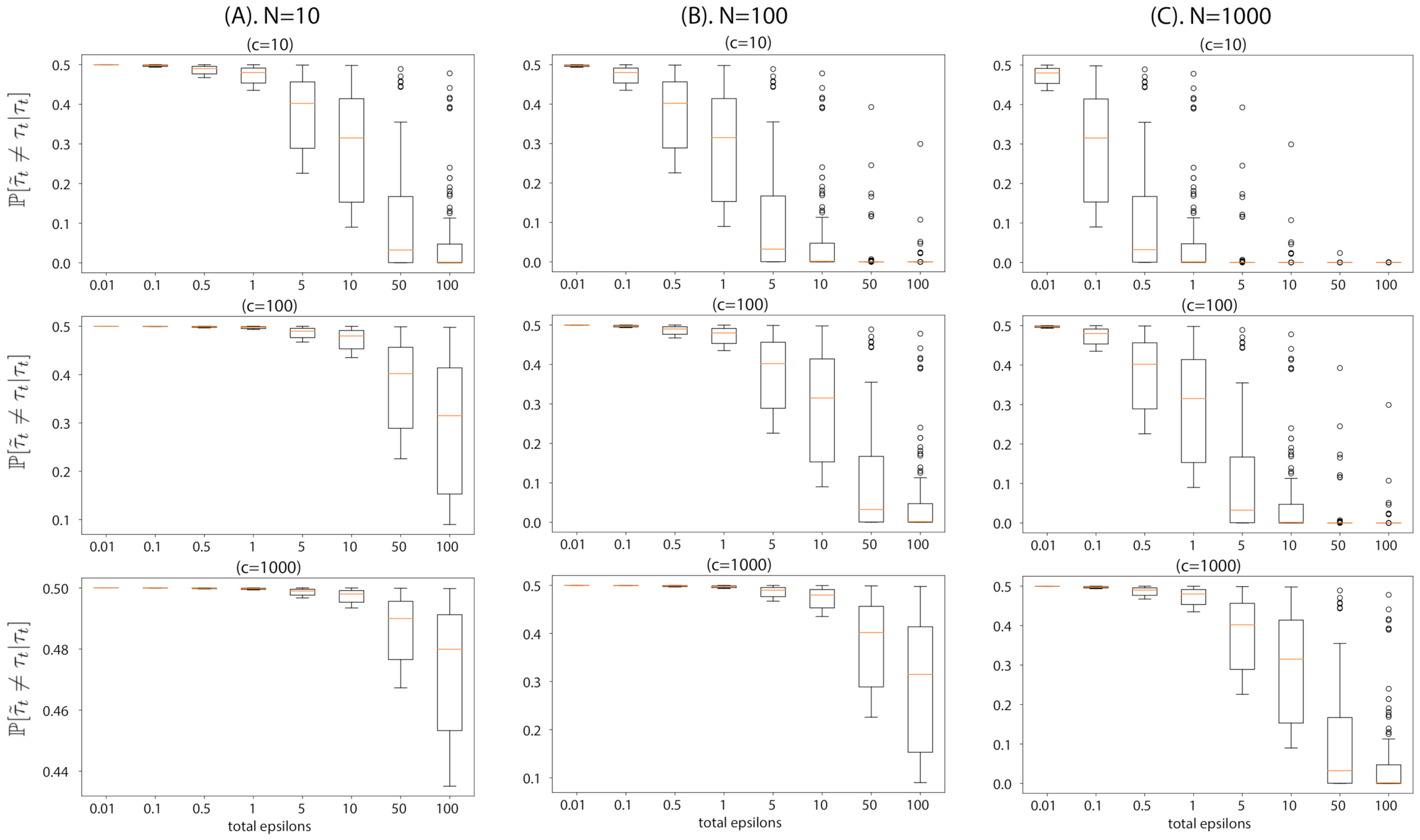
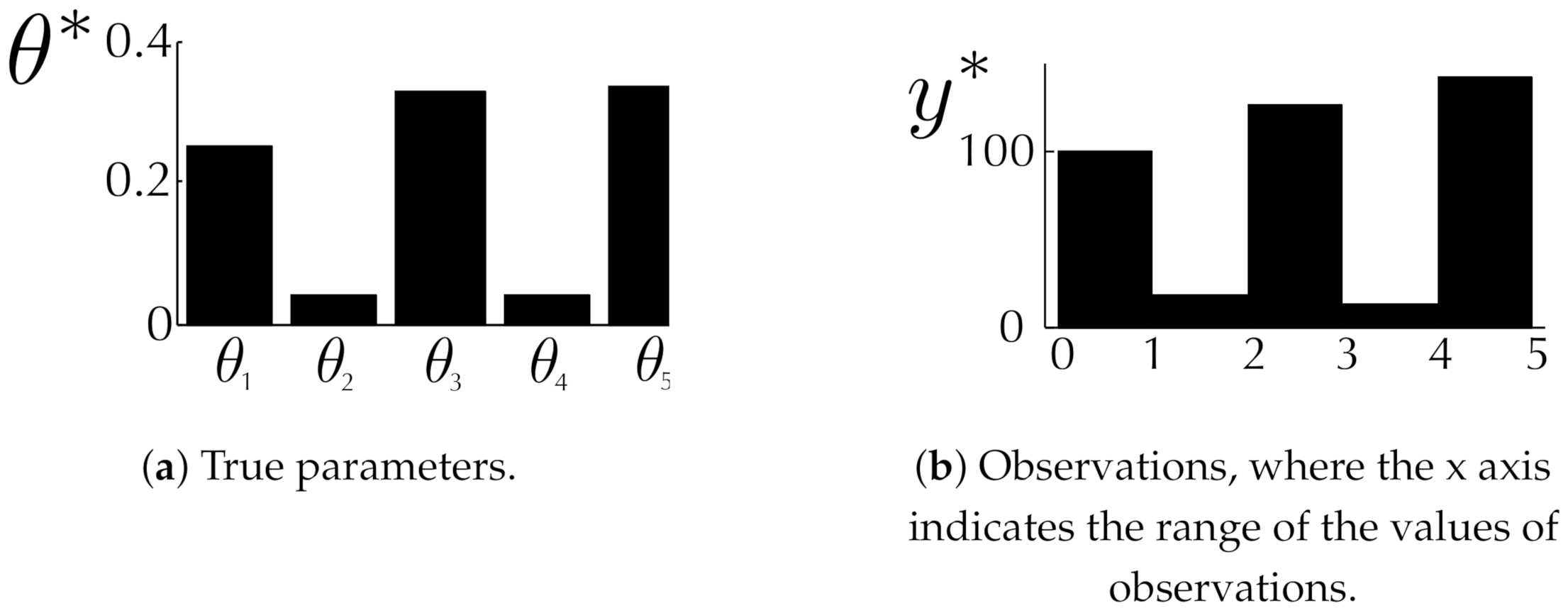
6.1. Toy Examples
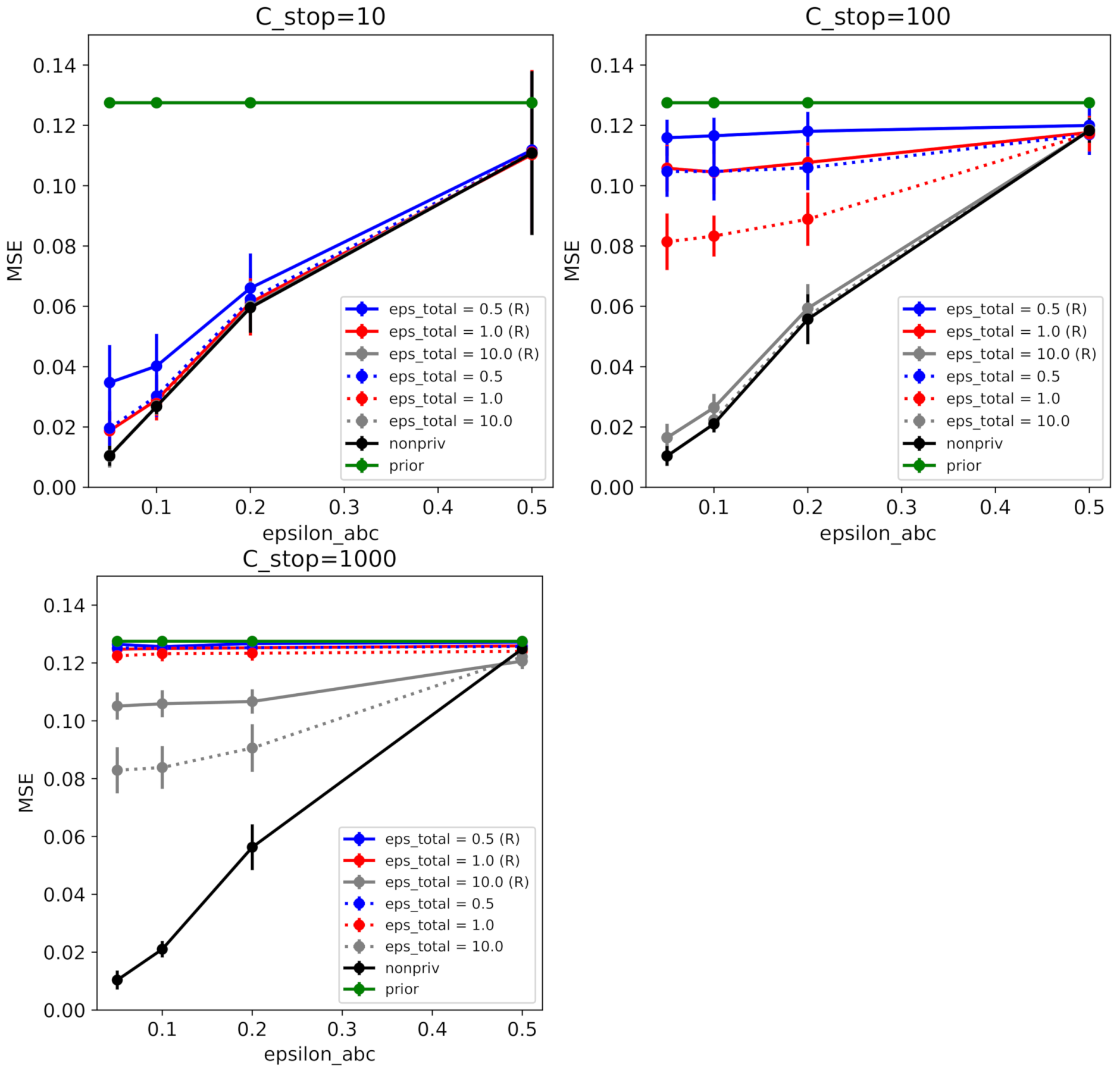
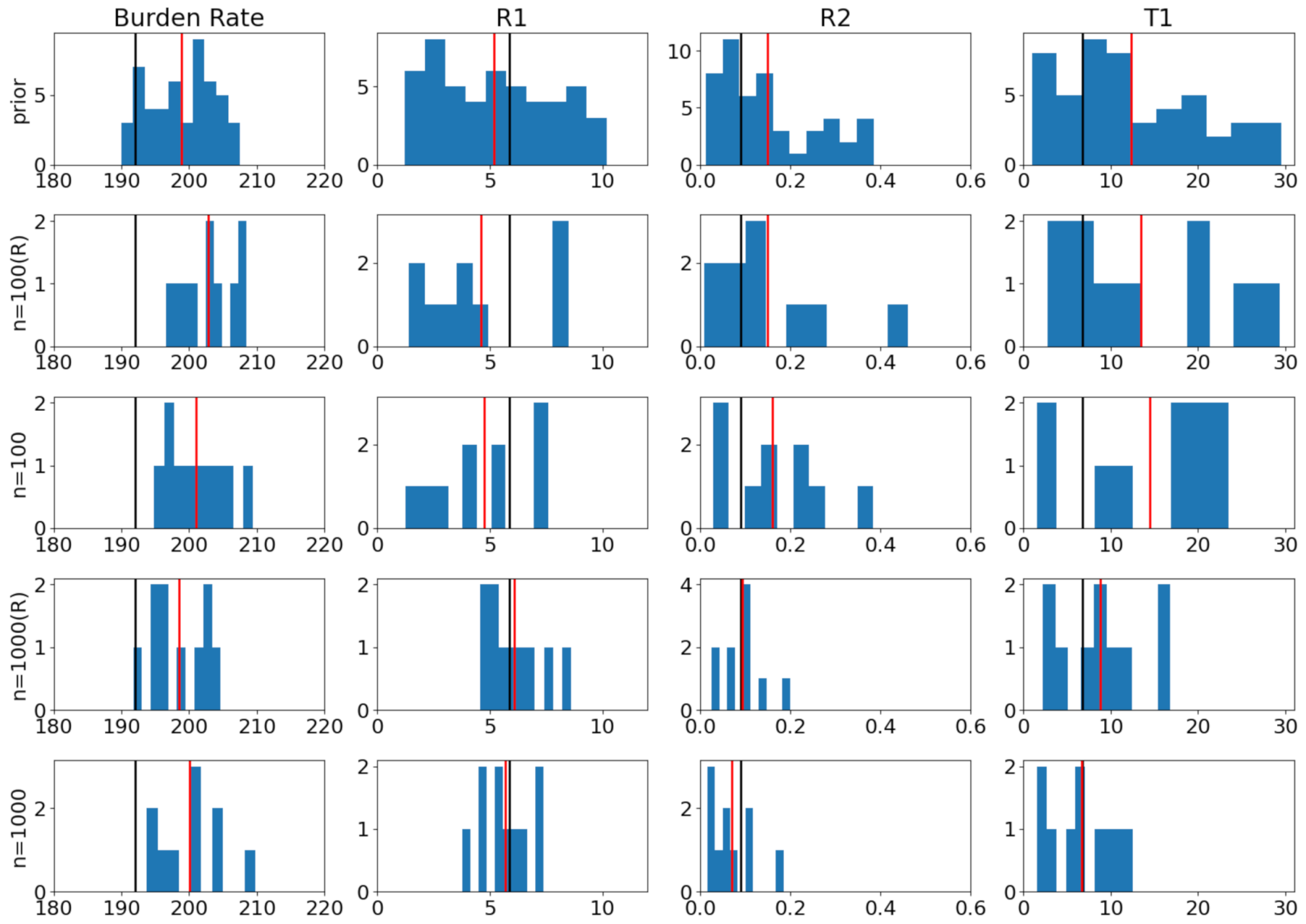
6.2. Coronavirus Outbreak Data
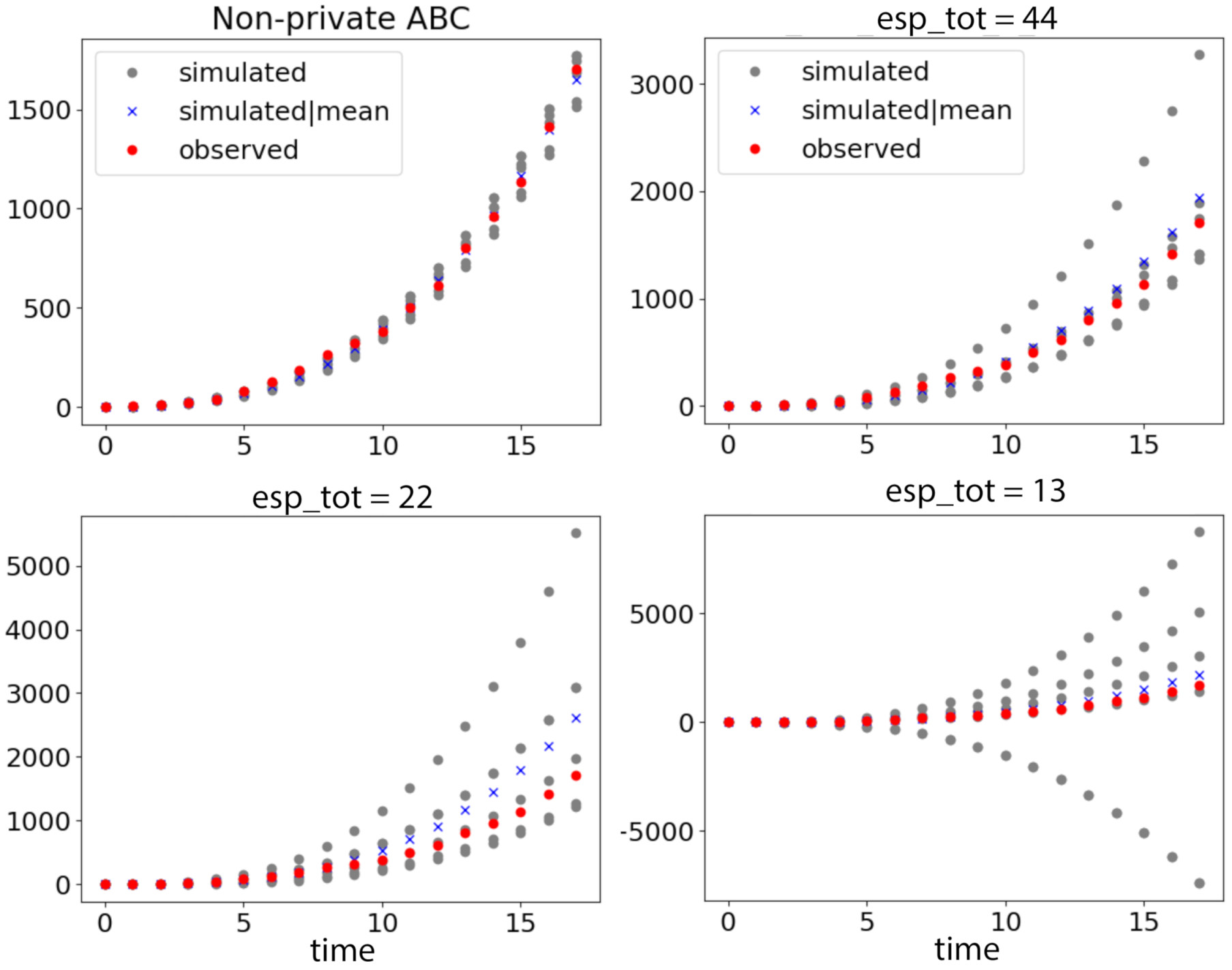
6.3. Modeling Tuberculosis (TB) Outbreak Using Stochastic Birth–Death Models
7. Summary and Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Note
Appendix A. Proof of Theorem 1
Appendix B. Proof of Lemma 1
Appendix C. Proof of Lemma 2
Appendix D. Proof of Proposition 1
Appendix E. Proof of Theorem 2
References
- Tavaré, S.; Balding, D.J.; Griffiths, R.C.; Donnelly, P. Inferring coalescence times from DNA sequence data. Genetics 1997, 145, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Ratmann, O.; Jørgensen, O.; Hinkley, T.; Stumpf, M.; Richardson, S.; Wiuf, C. Using Likelihood-Free Inference to Compare Evolutionary Dynamics of the Protein Networks of H. pylori and P. falciparum. PLoS Comput. Biol. 2007, 3, e230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazin, E.; Dawson, K.J.; Beaumont, M.A. Likelihood-Free Inference of Population Structure and Local Adaptation in a Bayesian Hierarchical Model. Genetics 2010, 185, 587–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schafer, C.M.; Freeman, P.E. Likelihood-Free Inference in Cosmology: Potential for the Estimation of Luminosity Functions. In Statistical Challenges in Modern Astronomy V; Springer: Berlin/Heidelberg, Germany, 2012; pp. 3–19. [Google Scholar]
- Pritchard, J.; Seielstad, M.; Perez-Lezaun, A.; Feldman, M. Population growth of human Y chromosomes: A study of Y chromosome microsatellites. Mol. Biol. Evol. 1999, 16, 1791–1798. [Google Scholar] [CrossRef] [Green Version]
- Fearnhead, P.; Prangle, D. Constructing summary statistics for approximate Bayesian computation: Semi-automatic approximate Bayesian computation. J. R. Stat. Soc. Ser. 2012, 74, 419–474. [Google Scholar] [CrossRef] [Green Version]
- Joyce, P.; Marjoram, P. Approximately Sufficient Statistics and Bayesian Computation. Stat. Appl. Genet. Molec. Biol. 2008, 7, 1544–6115. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.P.; Cornuet, J.; Marin, J.; Pillai, N.S. Lack of confidence in approximate Bayesian computation model choice. Proc. Natl. Acad. Sci. USA 2011, 108, 15112–15117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunes, M.; Balding, D. On Optimal Selection of Summary Statistics for Approximate Bayesian Computation. Stat. Appl. Genet. Molec. Biol. 2010, 9. [Google Scholar] [CrossRef] [PubMed]
- Aeschbacher, S.; Beaumont, M.A.; Futschik, A. A Novel Approach for Choosing Summary Statistics in Approximate Bayesian Computation. Genetics 2012, 192, 1027–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drovandi, C.; Pettitt, A.; Lee, A. Bayesian Indirect Inference Using a Parametric Auxiliary Model. Statist. Sci. 2015, 30, 72–95. [Google Scholar] [CrossRef] [Green Version]
- Homer, N.; Szelinger, S.; Redman, M.; Duggan, D.; Tembe, W.; Muehling, J.; Pearson, J.V.; Stephan, D.A.; Nelson, S.F.; Craig, D.W. Resolving Individuals Contributing Trace Amounts of DNA to Highly Complex Mixtures Using High-Density SNP Genotyping Microarrays. PLoS Genet. 2008, 4, e1000167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, A.; Shmatikov, V. Privacy-preserving Data Exploration in Genome-wide Association Studies. In Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Chicago, IL, USA, 11–14 August 2013; pp. 1079–1087. [Google Scholar]
- Tanaka, M.M.; Francis, A.R.; Luciani, F.; Sisson, S.A. Using approximate Bayesian computation to estimate tuberculosis transmission parameters from genotype data. Genetics 2006, 173, 1511–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwork, C.; McSherry, F.; Nissim, K.; Smith, A. Calibrating noise to sensitivity in private data analysis. In Proceedings of the TCC, New York, NY, USA, 4–7 March 2006; Springer: Berlin/Heidelberg, Germany, 2006; Volume 3876, pp. 265–284. [Google Scholar]
- Chaudhuri, K.; Monteleoni, C.; Sarwate, A.D. Differentially Private Empirical Risk Minimization. J. Mach. Learn. Res. 2011, 12, 1069–1109. [Google Scholar] [PubMed]
- Dwork, C.; Roth, A. The Algorithmic Foundations of Differential Privacy. Found. Trends Theor. Comput. Sci. 2014, 9, 211–407. [Google Scholar] [CrossRef]
- Park, M.; Jitkrittum, W.; Sejdinovic, D. K2-ABC: Approximate Bayesian Computation with Infinite Dimensional Summary Statistics via Kernel Embeddings. In Proceedings of the AISTATS, Cadiz, Spain, 9–11 May 2016. [Google Scholar]
- Nakagome, S.; Fukumizu, K.; Mano, S. Kernel approximate Bayesian computation in population genetic inferences. Stat. Appl. Genet. Mol. Biol. 2013, 12, 667–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gleim, A.; Pigorsch, C. Approximate Bayesian Computation with Indirect Summary Statistics; University of Bonn: Bonn, Germany, 2013. [Google Scholar]
- Gretton, A.; Borgwardt, K.; Rasch, M.; Schölkopf, B.; Smola, A. A Kernel Two-Sample Test. J. Mach. Learn. Res. 2012, 13, 723–773. [Google Scholar]
- Smola, A.; Gretton, A.; Song, L.; Schölkopf, D. A Hilbert space embedding for distributions. In Algorithmic Learning Theory, Proceedings of the 18th International Conference, Sendai, Japan, 1–4 October 2007; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Sriperumbudur, B.; Fukumizu, K.; Lanckriet, G. Universality, characteristic kernels and RKHS embedding of measures. J. Mach. Learn. Res. 2011, 12, 2389–2410. [Google Scholar]
- Dwork, C.; Kenthapadi, K.; McSherry, F.; Mironov, I.; Naor, M. Our Data, Ourselves: Privacy Via Distributed Noise Generation. In Advances in Cryptology—EUROCRYPT 2006, Proceedings of the 24th Annual International Conference on the Theory and Applications of Cryptographic Techniques, St. Petersburg, Russia, 28 May–1 June 2006; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4004, pp. 486–503. [Google Scholar]
- Mironov, I. Rényi Differential Privacy. In Proceedings of the 30th IEEE Computer Security Foundations Symposium (CSF), Santa Barbara, CA, USA, 21–25 August 2017; pp. 263–275. [Google Scholar]
- Lyu, M.; Su, D.; Li, N. Understanding the Sparse Vector Technique for Differential Privacy. Proc. VLDB Endow. 2017, 10, 637–648. [Google Scholar] [CrossRef] [Green Version]
- Gong, R. Exact Inference with Approximate Computation for Differentially Private Data via Perturbations. arXiv 2019, arXiv:stat.CO/1909.12237. [Google Scholar]
- Lintusaari, J.; Blomstedt, P.; Rose, B.; Sivula, T.; Gutmann, M.; Kaski, S.; Corander, J. Resolving outbreak dynamics using approximate Bayesian computation for stochastic birth?death models [version 2; peer review: 2 approved]. Wellcome Open Res. 2019, 4. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Wang, Y.X. Improving Sparse Vector Technique with Renyi Differential Privacy. In Proceedings of the 2020 Conference on Neural Information Processing Systems, Virtual, 6–12 December 2020; Volume 33. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, M.; Vinaroz, M.; Jitkrittum, W. ABCDP: Approximate Bayesian Computation with Differential Privacy. Entropy 2021, 23, 961. https://doi.org/10.3390/e23080961
Park M, Vinaroz M, Jitkrittum W. ABCDP: Approximate Bayesian Computation with Differential Privacy. Entropy. 2021; 23(8):961. https://doi.org/10.3390/e23080961
Chicago/Turabian StylePark, Mijung, Margarita Vinaroz, and Wittawat Jitkrittum. 2021. "ABCDP: Approximate Bayesian Computation with Differential Privacy" Entropy 23, no. 8: 961. https://doi.org/10.3390/e23080961
APA StylePark, M., Vinaroz, M., & Jitkrittum, W. (2021). ABCDP: Approximate Bayesian Computation with Differential Privacy. Entropy, 23(8), 961. https://doi.org/10.3390/e23080961





