Understanding the Variability in Graph Data Sets through Statistical Modeling on the Stiefel Manifold
Abstract
1. Introduction
2. Background
2.1. Statistical Modeling for Graphs Data Sets
2.2. Models and Algorithms on the Stiefel Manifold
2.2.1. Compact Stiefel Manifolds of Orthonormal Frames
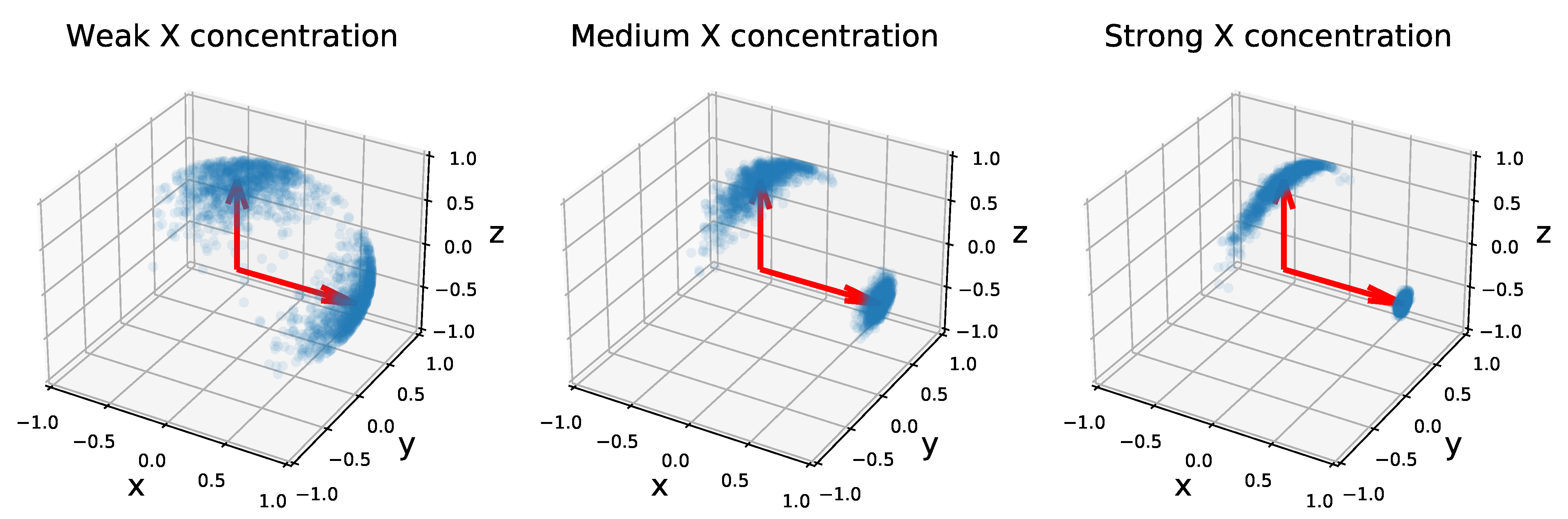
2.2.2. Von Mises–Fisher Distributions
2.2.3. Application to Network Modeling
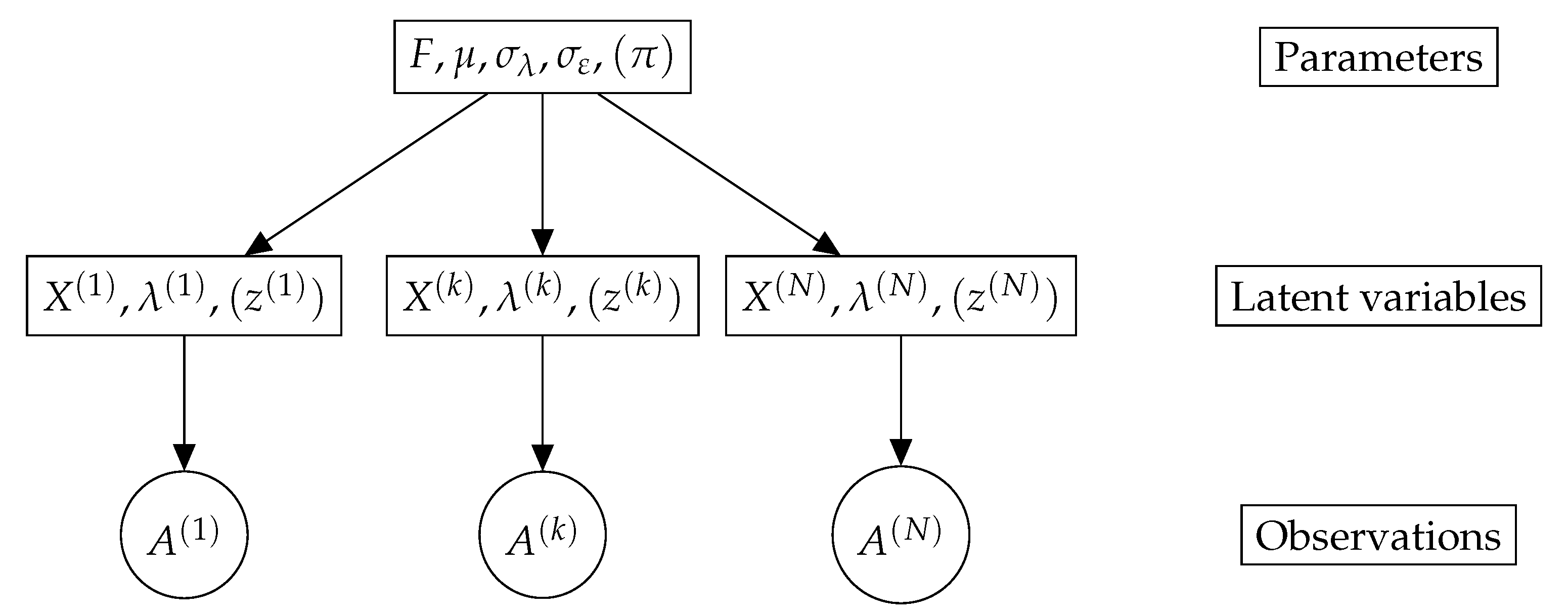
3. A Latent Variable Model for Graph Data Sets
3.1. Motivation
3.2. Model Description
3.3. Mixture Model
4. A Maximum Likelihood Estimation Algorithm
4.1. Maximum Likelihood Estimation for Exponential Models with the MCMC-SAEM Algorithm
- E-step: Using the current value of the parameter , compute the expectation
- M-step: Find .
| Algorithm 1: The MCMC-SAEM Algorithm |
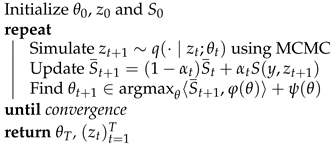 |
4.2. E-Step with Markov Chain Monte Carlo
4.2.1. Transition Kernel
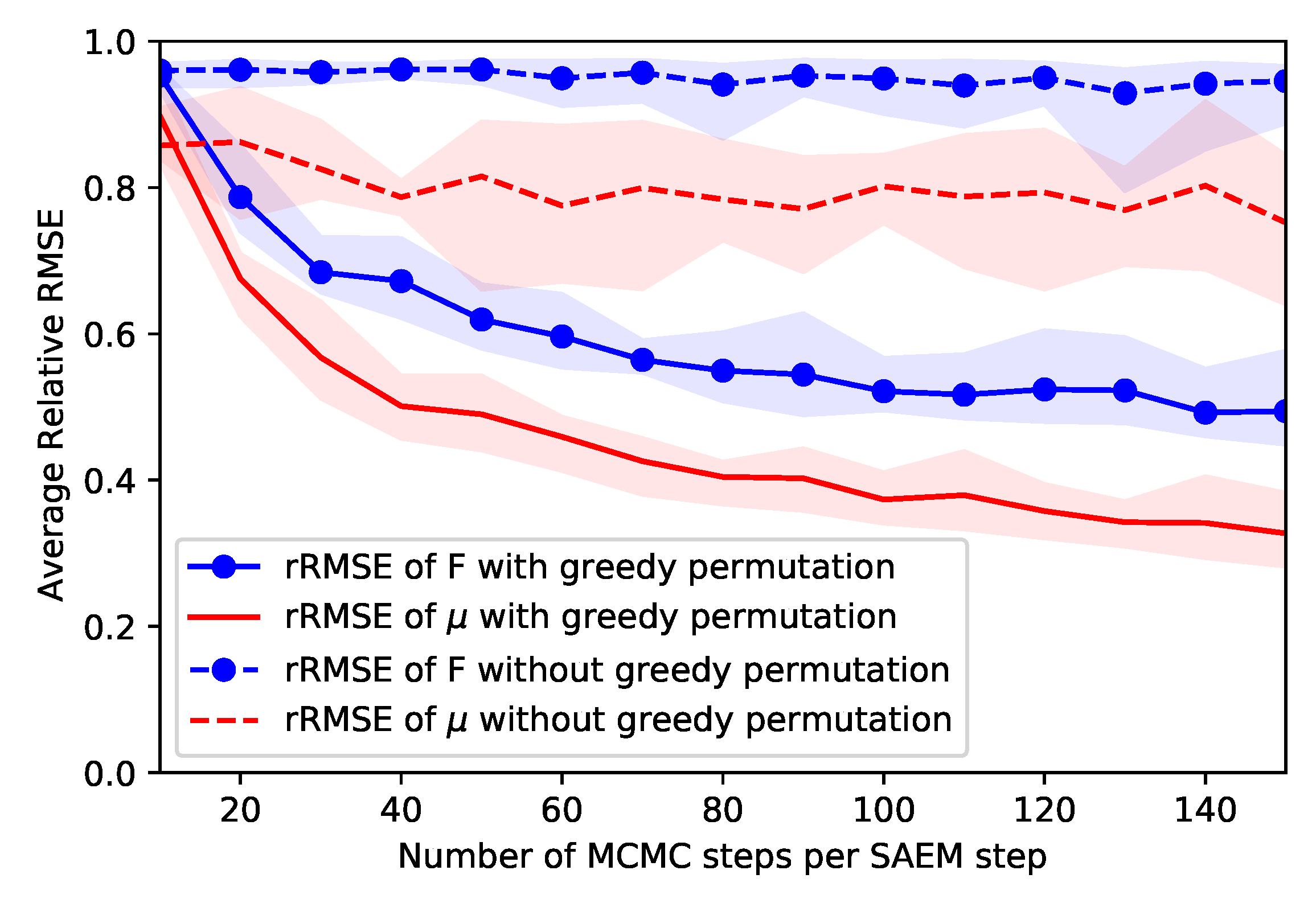
4.2.2. Permutation Invariance Problem
| Algorithm 2: Greedy column matching |
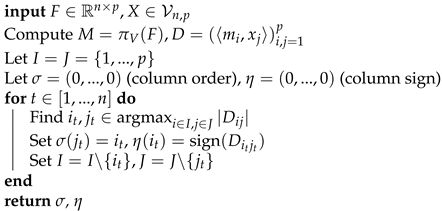 |
4.3. M-Step with Saddle-Point Approximations
4.3.1. Maximum Likelihood Estimation of Von Mises–Fisher Distributions
4.3.2. Application to the Proposed Model
4.4. Algorithm for the Mixture Model
4.5. Numerical Implementation Details
| Algorithm 3: Maximum Likelihood Estimation algorithm for |
 |
5. Experiments
5.1. Experiments on Synthetic Data
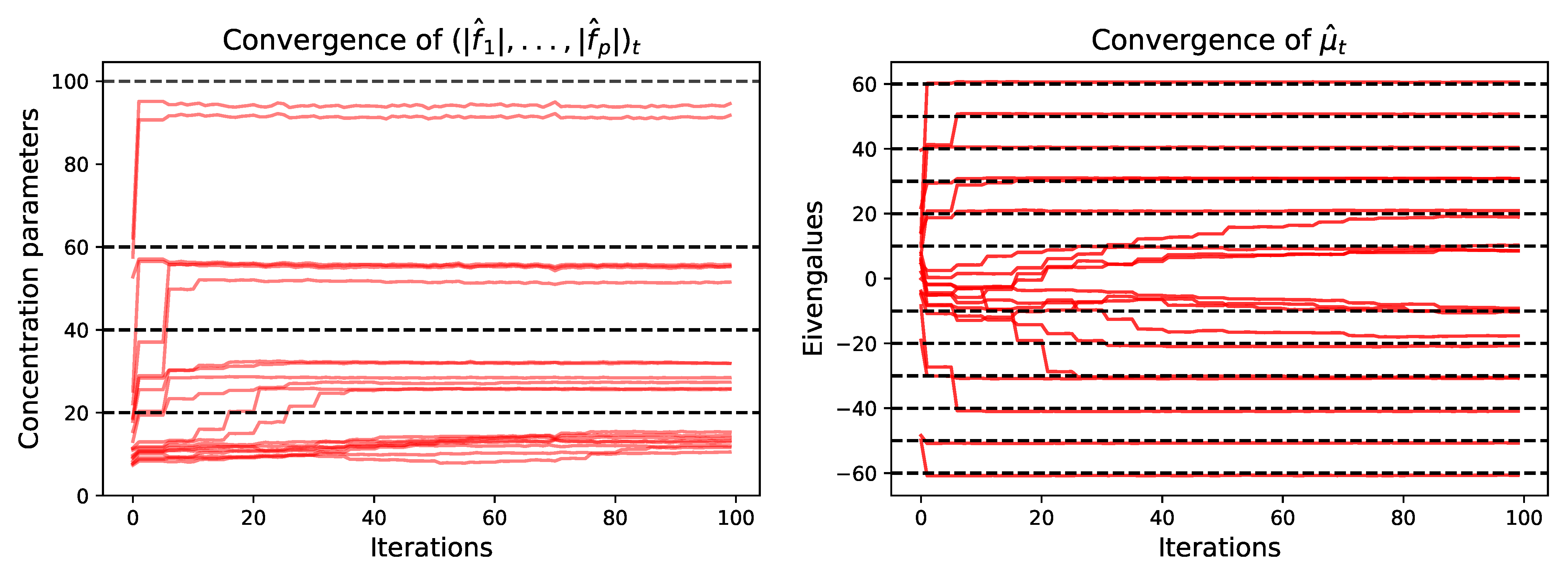
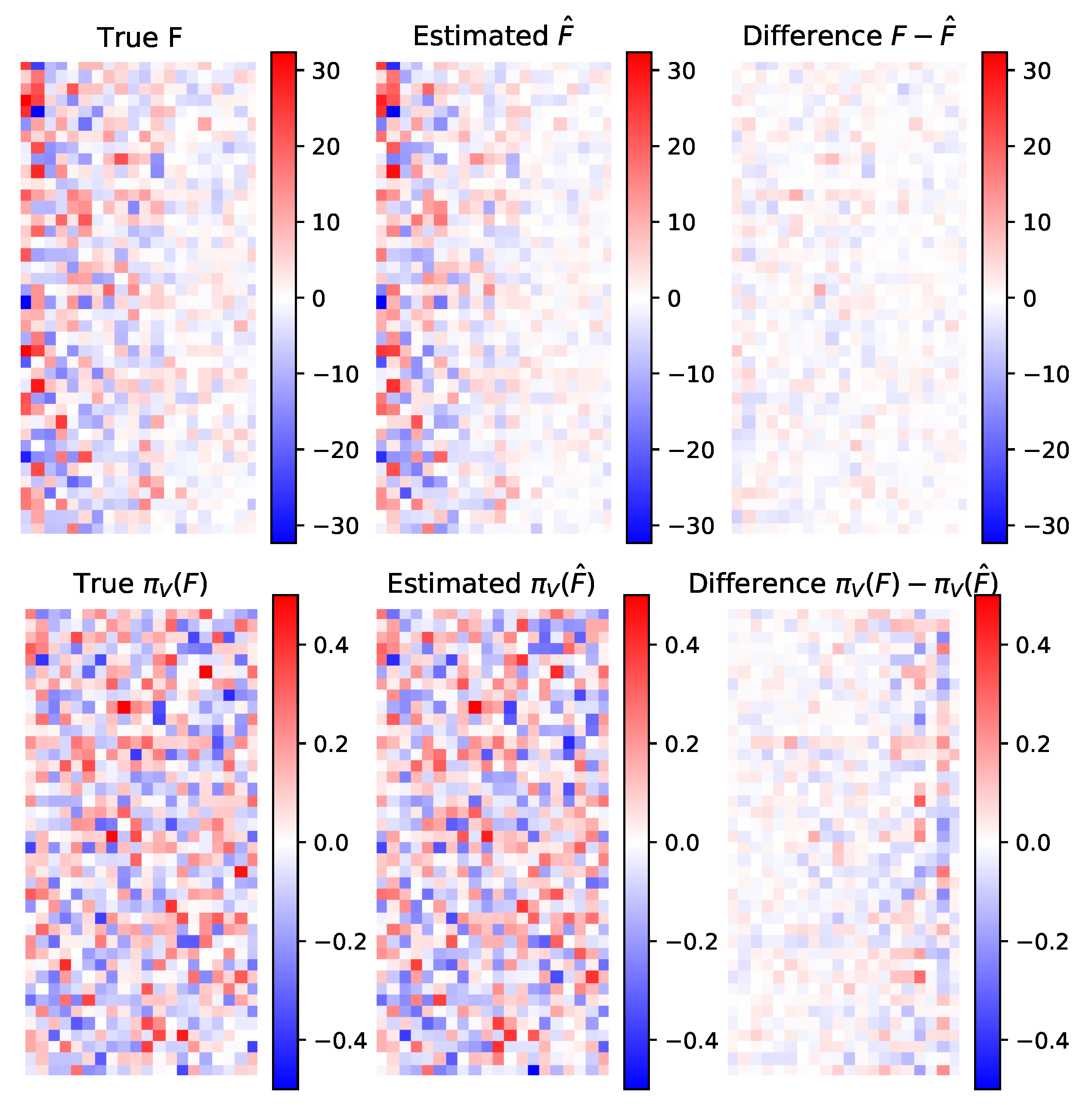
5.1.1. Parameters Estimation Performance
Small Dimension
High Dimension
Model Selection
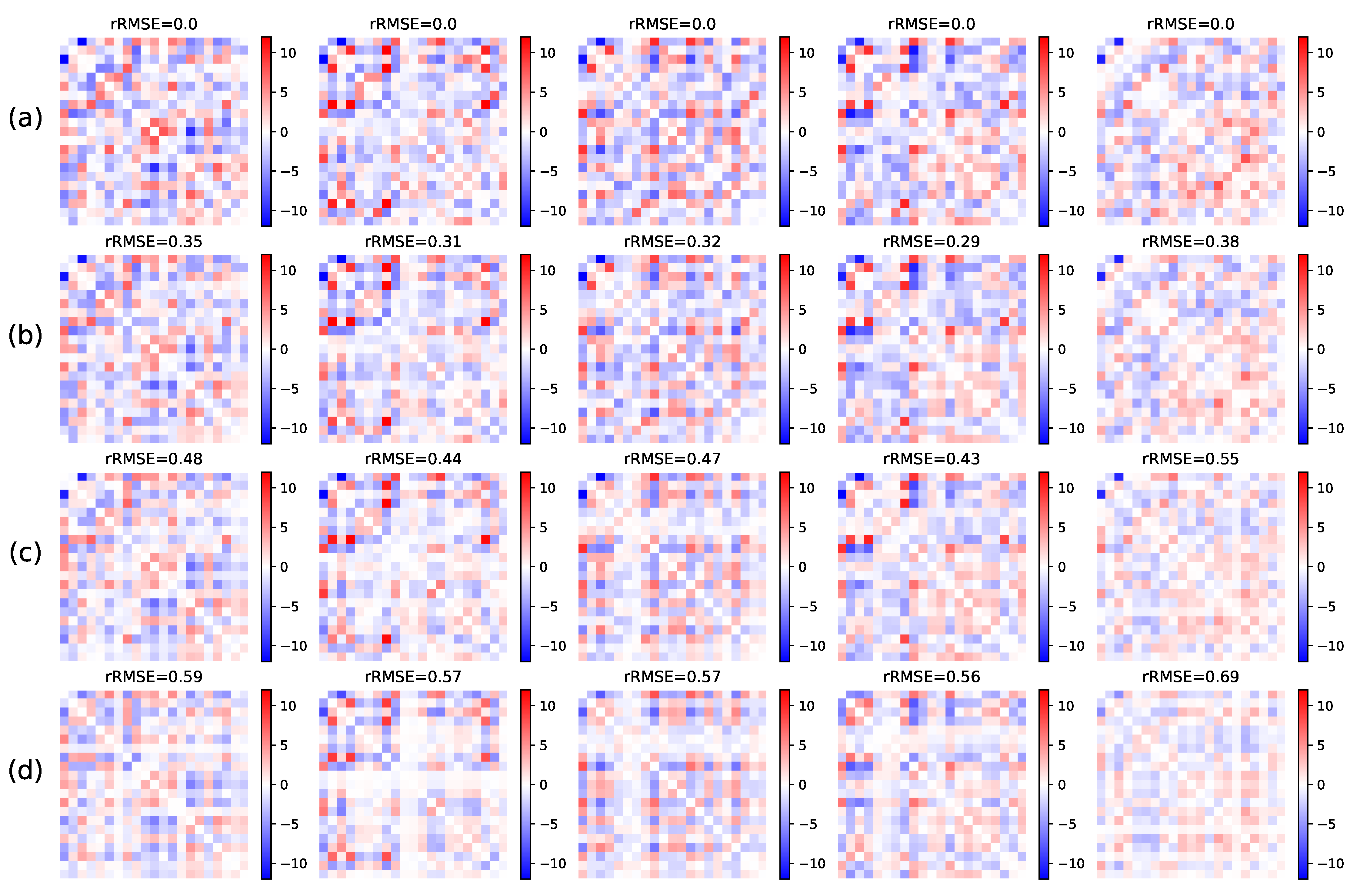
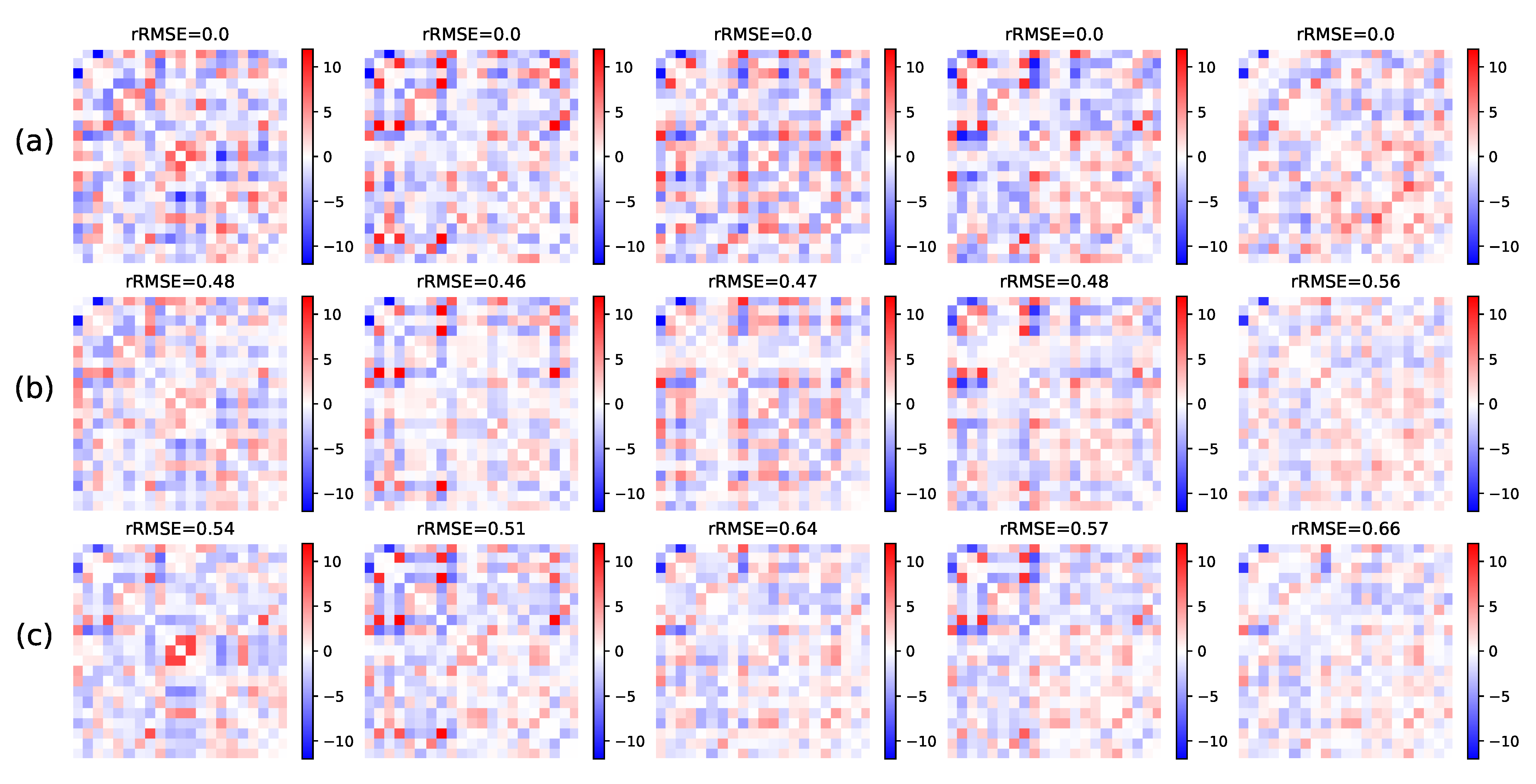
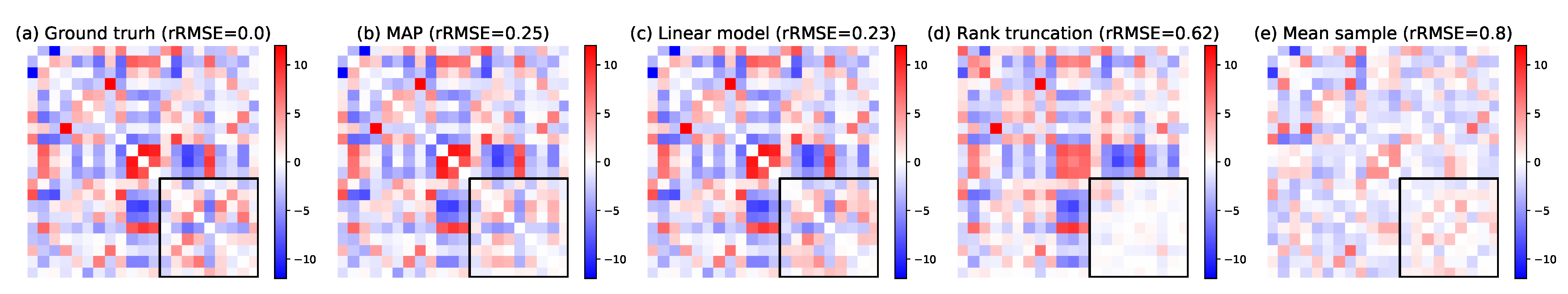
5.1.2. Missing Links Imputation
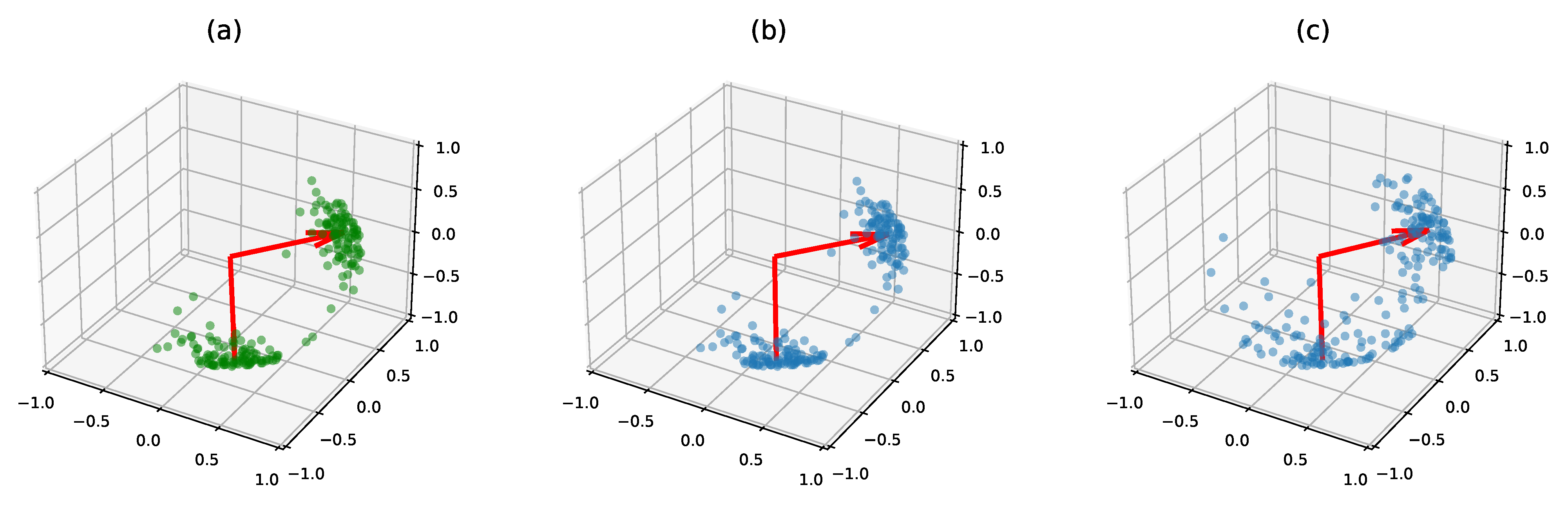
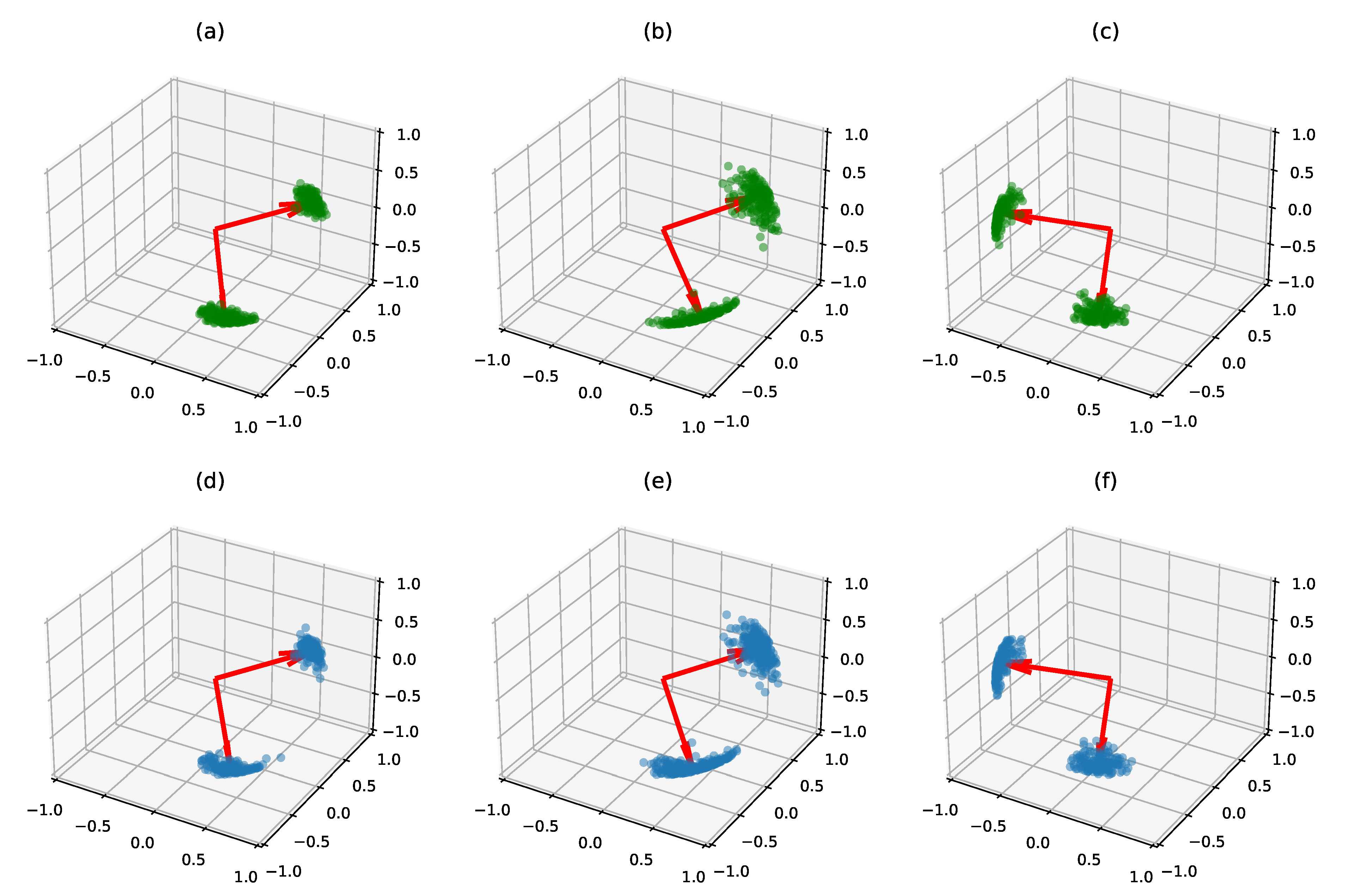
5.1.3. Clustering on Synthetic Data
Small Dimension
Larger Dimension
Model Selection
5.2. Experiments on Brain Connectivity Networks
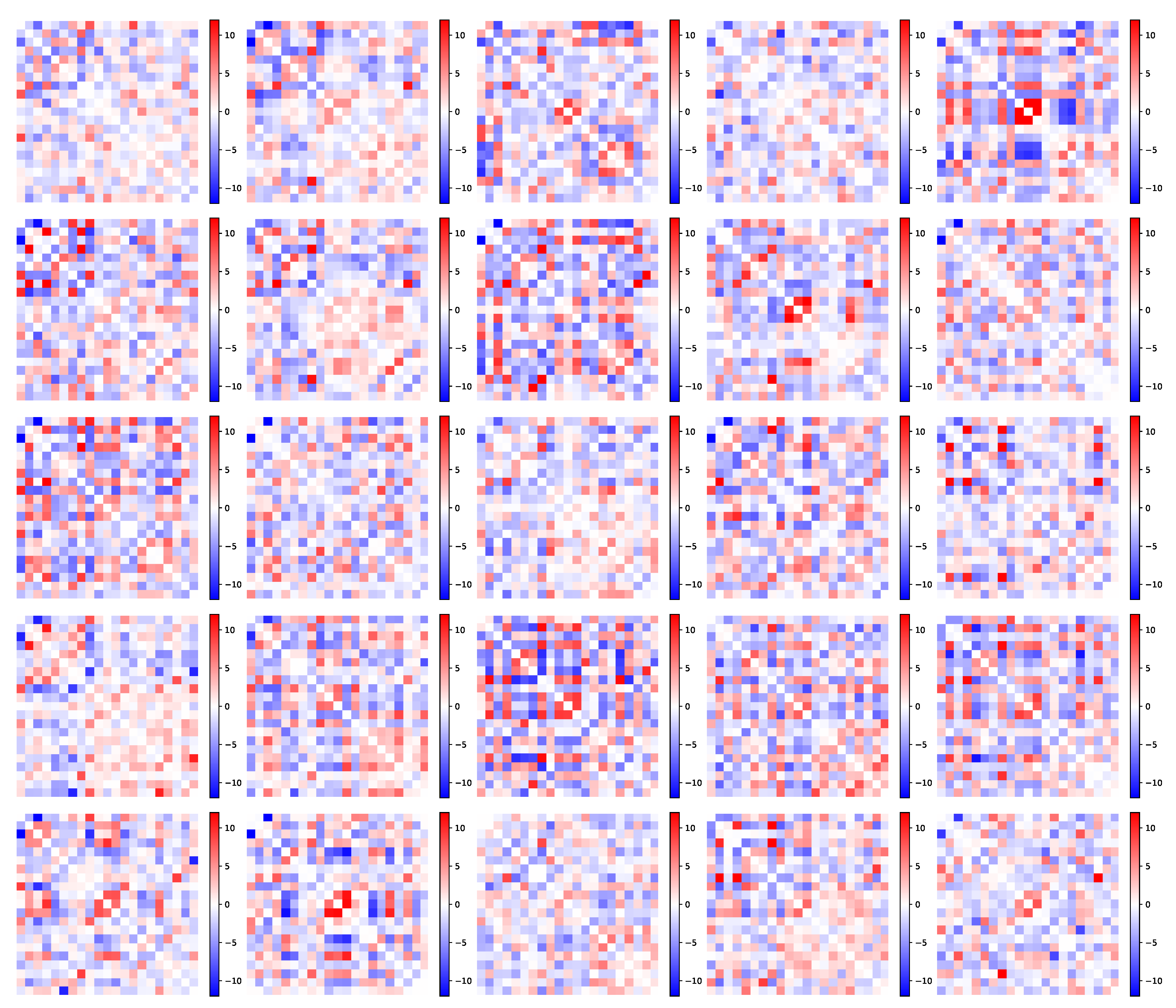
5.2.1. Parameter Estimation
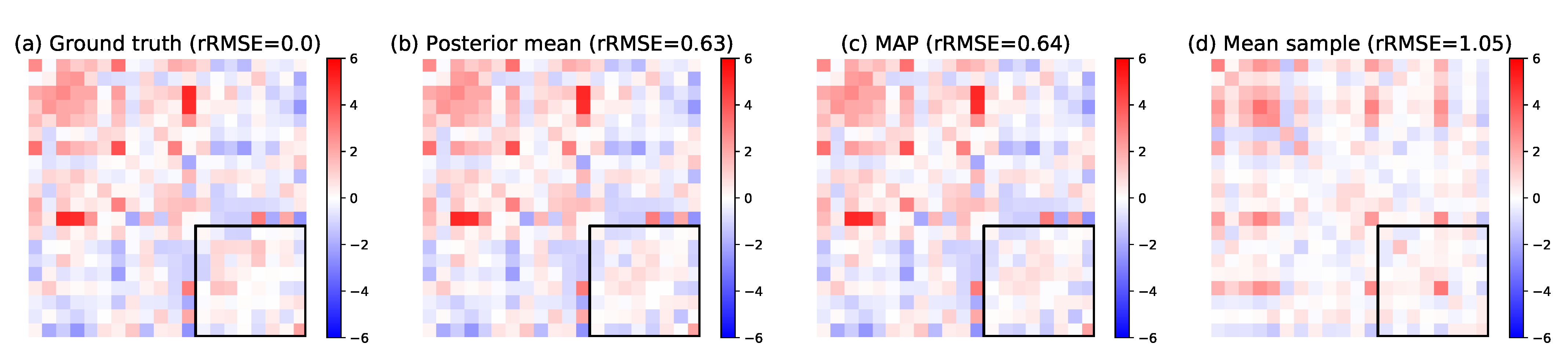
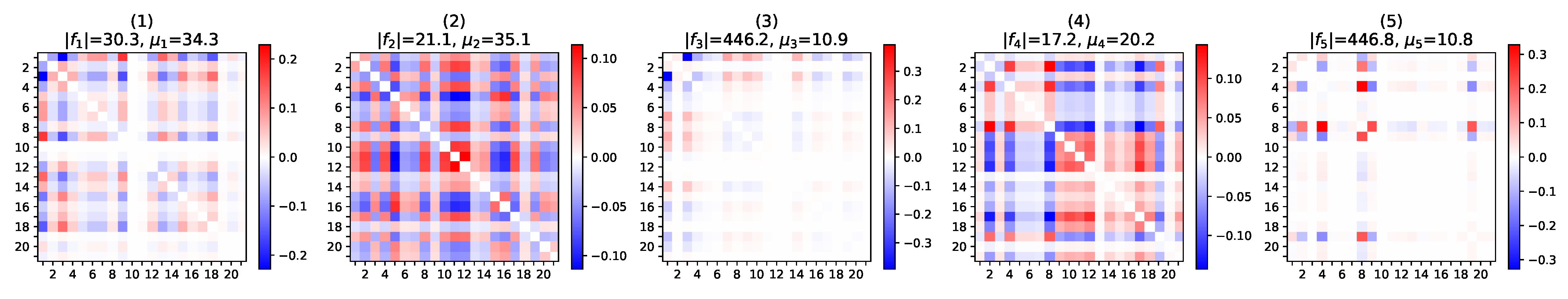
5.2.2. Pattern Interpretation
5.2.3. Link Prediction
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. SAEM Maximization Step
Appendix A.1. Maximum Likelihood Estimates for , ,
Appendix A.2. Saddle-Point Approximation of
- . The matrix is composed of blocks. Block is the identity and all the other blocks are set to zero.
- . In this formula, is the diagonal matrix with diagonal p singular values of F. The function is the cumulant generating function.
- is the unique solution of the so-called saddle-point equation . It has the explicit expression , with
- is given by and by .
- can be computed explicitly:
- The parameter T is defined in Equation (8) of [49] and computed in the supplementary material of the paper in the case of vMF distributions. In first approximation, T can be considered zero.
Appendix B. Gradient Formulas
Appendix B.1. Model with Gaussian Perturbation
Appendix B.2. Binary Model
Appendix C. Algorithm for the Clustering Model
| Algorithm A1: Maximum Likelihood Estimation of for the mixture model |
 |
Appendix D. Symmetry of Von Mises–Fisher Distributions
Appendix E. Additional Details on the UK Biobank Experiment
Appendix E.1. Impact of the Number p of Patterns
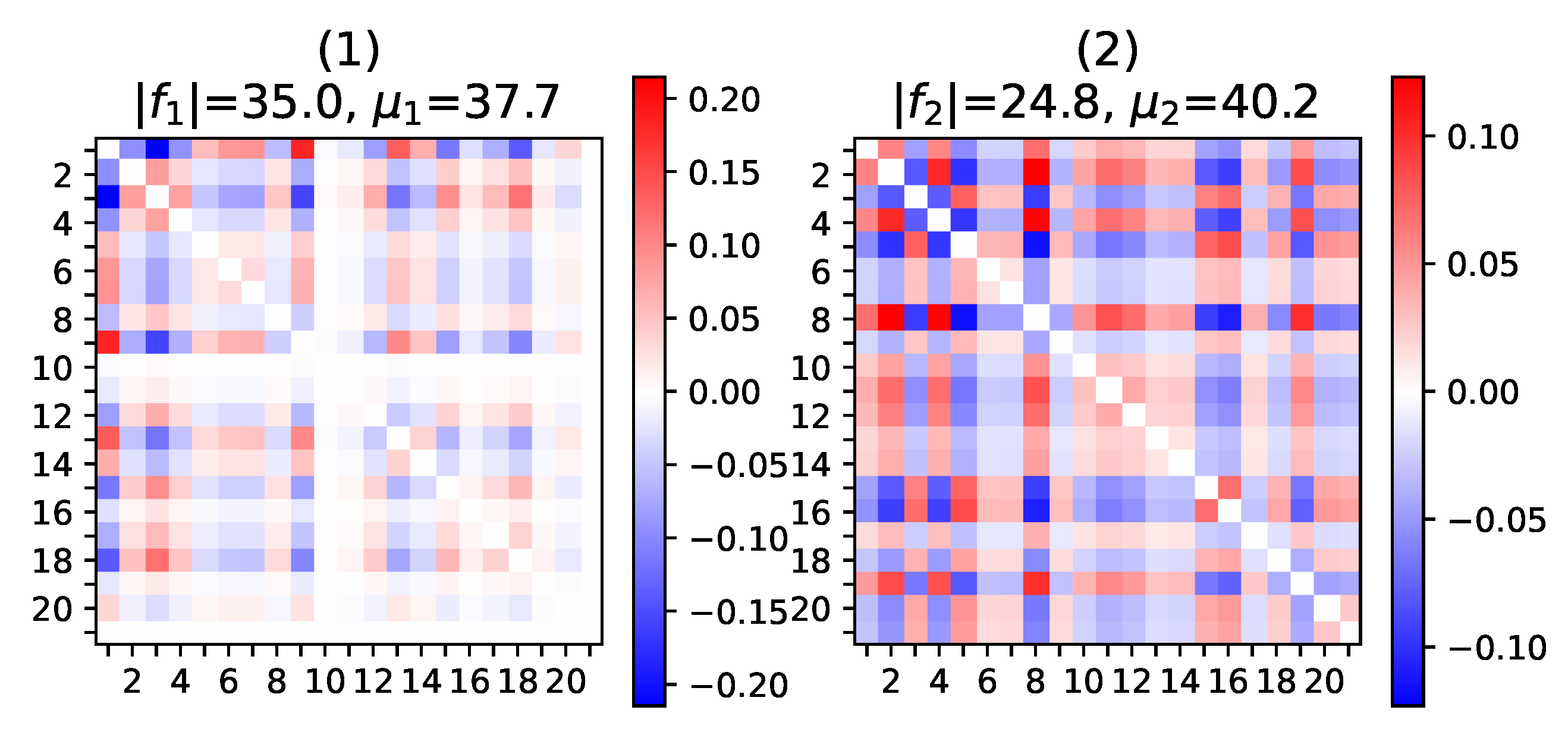
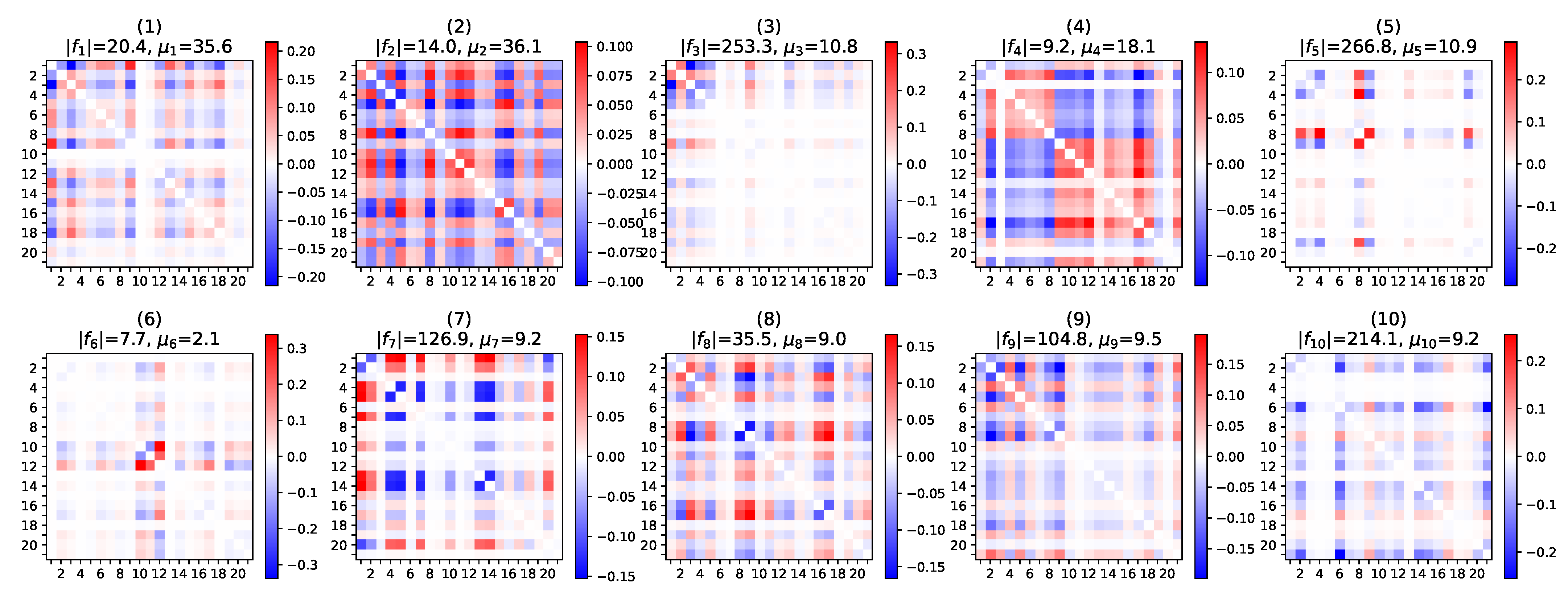
Appendix E.2. Brain Regions of the UK Biobank fMRI Correlation Networks

References
- Newman, M.E.J. Networks—An Introduction; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Ni, C.C.; Lin, Y.Y.; Luo, F.; Gao, J. Community Detection on Networks with Ricci Flow. Sci. Rep. 2019, 9, 9984. [Google Scholar] [CrossRef]
- Martínez, V.; Berzal, F.; Cubero, J.C. A Survey of Link Prediction in Complex Networks. ACM Comput. Surv. 2016, 49, 1–33. [Google Scholar] [CrossRef]
- Shen, X.; Finn, E.S.; Scheinost, D.; Rosenberg, M.D.; Chun, M.M.; Papademetris, X.; Constable, R.T. Using Connectome-Based Predictive Modeling to Predict Individual Behavior from Brain Connectivity. Nat. Protoc. 2017, 12, 506–518. [Google Scholar] [CrossRef]
- Banks, D.; Carley, K. Metric Inference for Social Networks. J. Classif. 1994, 11, 121–149. [Google Scholar] [CrossRef]
- Rubinov, M.; Sporns, O. Complex Network Measures of Brain Connectivity: Uses and Interpretations. NeuroImage 2010, 52, 1059–1069. [Google Scholar] [CrossRef]
- Simonovsky, M.; Komodakis, N. GraphVAE: Towards Generation of Small Graphs Using Variational Autoencoders. In Artificial Neural Networks and Machine Learning—ICANN 2018; Lecture Notes in Computer Science; Kůrková, V., Manolopoulos, Y., Hammer, B., Iliadis, L., Maglogiannis, I., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 412–422. [Google Scholar] [CrossRef]
- Pozzi, F.A.; Fersini, E.; Messina, E.; Liu, B. Sentiment Analysis in Social Networks; Morgan Kaufmann: Burlington, MA, USA, 2016. [Google Scholar]
- Monti, F.; Bronstein, M.; Bresson, X. Geometric Matrix Completion with Recurrent Multi-Graph Neural Networks. Adv. Neural Inf. Process. Syst. 2017, 30, 3697–3707. [Google Scholar]
- Narayanan, T.; Subramaniam, S. Community Structure Analysis of Gene Interaction Networks in Duchenne Muscular Dystrophy. PLoS ONE 2013, 8, e67237. [Google Scholar] [CrossRef] [PubMed][Green Version]
- He, Y.; Evans, A. Graph Theoretical Modeling of Brain Connectivity. Curr. Opin. Neurol. 2010, 23, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Duvenaud, D.K.; Maclaurin, D.; Iparraguirre, J.; Bombarell, R.; Hirzel, T.; Aspuru-Guzik, A.; Adams, R.P. Convolutional Networks on Graphs for Learning Molecular Fingerprints. Adv. Neural Inf. Process. Syst. 2015, 28, 2224–2232. [Google Scholar]
- Lovász, L. Large Networks and Graph Limits; Colloquium Publications; American Mathematical Society: Providence, RI, USA, 2012; Volume 60. [Google Scholar] [CrossRef]
- Hanneke, S.; Fu, W.; Xing, E.P. Discrete Temporal Models of Social Networks. Electron. J. Stat. 2010, 4, 585–605. [Google Scholar] [CrossRef]
- Fornito, A.; Zalesky, A.; Bullmore, E. Fundamentals of Brain Network Analysis; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Zheng, W.; Yao, Z.; Li, Y.; Zhang, Y.; Hu, B.; Wu, D.; Alzheimer’s Disease Neuroimaging Initiative. Brain Connectivity Based Prediction of Alzheimer’s Disease in Patients With Mild Cognitive Impairment Based on Multi-Modal Images. Front. Hum. Neurosci. 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Das, N.; Gonçalves, T.; Quaresma, P.; Kundu, M. The Journey of Graph Kernels through Two Decades. Comput. Sci. Rev. 2018, 27, 88–111. [Google Scholar] [CrossRef]
- Damoiseaux, J.S. Effects of Aging on Functional and Structural Brain Connectivity. NeuroImage 2017, 160, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Chikuse, Y. Statistics on Special Manifolds; Lecture Notes in Statistics; Springer: New York, NY, USA, 2003. [Google Scholar] [CrossRef]
- Harris, J.K. An Introduction to Exponential Random Graph Modeling; Number 173 in Quantitative Applications in the Social Sciences; SAGE: Thousand Oaks, CA, USA, 2014. [Google Scholar]
- Erdos, P.; Rényi, A. On Random Graphs. Publ. Math. 1959, 6, 290–297. [Google Scholar]
- Peixoto, T.P. Bayesian Stochastic Blockmodeling. In Advances in Network Clustering and Blockmodeling; Doreian, P., Batagelj, V., Ferligoj, A., Eds.; Wiley Series in Computational and Quantitative Social Science; Wiley: New York, NY, USA, 2020; pp. 289–332. Available online: http://arxiv.org/abs/1705.10225 (accessed on 19 April 2021).
- Chandna, S.; Maugis, P.A. Nonparametric Regression for Multiple Heterogeneous Networks. arXiv 2020, arXiv:2001.04938. [Google Scholar]
- Zhang, Z.; Cui, P.; Zhu, W. Deep Learning on Graphs: A Survey. arXiv 2020, arXiv:1812.04202. [Google Scholar] [CrossRef]
- Banka, A.; Rekik, I. Adversarial Connectome Embedding for Mild Cognitive Impairment Identification Using Cortical Morphological Networks. In Connectomics in NeuroImaging; Lecture Notes in Computer Science; Schirmer, M.D., Venkataraman, A., Rekik, I., Kim, M., Chung, A.W., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 74–82. [Google Scholar] [CrossRef]
- Ma, J.; Zhu, X.; Yang, D.; Chen, J.; Wu, G. Attention-Guided Deep Graph Neural Network for Longitudinal Alzheimer’s Disease Analysis. In Medical Image Computing and Computer Assisted Intervention —MICCAI 2020; Martel, A.L., Abolmaesumi, P., Stoyanov, D., Mateus, D., Zuluaga, M.A., Zhou, S.K., Racoceanu, D., Joskowicz, L., Eds.; Springer International Publishing: Cham, Switzerland, 2020; Volume 12267, pp. 387–396. [Google Scholar] [CrossRef]
- Westveld, A.H.; Hoff, P.D. A Mixed Effects Model for Longitudinal Relational and Network Data, with Applications to International Trade and Conflict. Ann. Appl. Stat. 2011, 5, 843–872. [Google Scholar] [CrossRef]
- D’Souza, N.S.; Nebel, M.B.; Wymbs, N.; Mostofsky, S.; Venkataraman, A. Integrating Neural Networks and Dictionary Learning for Multidimensional Clinical Characterizations from Functional Connectomics Data. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.T., Khan, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 11766, pp. 709–717. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, Z.; Dunson, D.B. Auto-Encoding Graph-Valued Data with Applications to Brain Connectomes. arXiv 2019, arXiv:1911.02728. [Google Scholar]
- Edelman, A.; Arias, T.A.; Smith, S.T. The Geometry of Algorithms with Orthogonality Constraints. SIAM J. Matrix Anal. Appl. 1998, 20, 303–353. [Google Scholar] [CrossRef]
- Zimmermann, R. A Matrix-Algebraic Algorithm for the Riemannian Logarithm on the Stiefel Manifold under the Canonical Metric. SIAM J. Matrix Anal. Appl. 2017, 38, 322–342. [Google Scholar] [CrossRef]
- Khatri, C.G.; Mardia, K.V. The von Mises–Fisher Matrix Distribution in Orientation Statistics. J. R. Stat. Soc. Ser. B (Methodol.) 1977, 39, 95–106. [Google Scholar] [CrossRef]
- Karush, W. Minima of Functions of Several Variables with Inequalities as Side Constraints. Masters’s Thesis, Department of Mathematics, University of Chicago, Chicago, IL, USA, 1939. [Google Scholar]
- Kuhn, E.; Lavielle, M. Coupling a Stochastic Approximation Version of EM with an MCMC Procedure. ESAIM Probab. Stat. 2004, 8, 115–131. [Google Scholar] [CrossRef]
- Hoff, P.D.; Ward, M.D. Modeling Dependencies in International Relations Networks. Political Anal. 2004, 12, 160–175. [Google Scholar] [CrossRef]
- Hoff, P.D. Modeling Homophily and Stochastic Equivalence in Symmetric Relational Data. In Proceedings of the 20th International Conference on Neural Information Processing Systems; NIPS’07; Curran Associates Inc.: Red Hook, NY, USA, 2007; pp. 657–664. [Google Scholar]
- Hoff, P.D. Model Averaging and Dimension Selection for the Singular Value Decomposition. J. Am. Stat. Assoc. 2007, 102, 674–685. [Google Scholar] [CrossRef]
- Hoff, P.D. Simulation of the Matrix Bingham—von Mises—Fisher Distribution, With Applications to Multivariate and Relational Data. J. Comput. Graph. Stat. 2009, 18, 438–456. [Google Scholar] [CrossRef]
- Chen, J.; Han, G.; Cai, H.; Ma, J.; Kim, M.; Laurienti, P.; Wu, G. Estimating Common Harmonic Waves of Brain Networks on Stiefel Manifold. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2020; Lecture Notes in Computer Science; Martel, A.L., Abolmaesumi, P., Stoyanov, D., Mateus, D., Zuluaga, M.A., Zhou, S.K., Racoceanu, D., Joskowicz, L., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 367–376. [Google Scholar] [CrossRef]
- Allassonnière, S.; Younes, L. A Stochastic Algorithm for Probabilistic Independent Component Analysis. Ann. Appl. Stat. 2012, 6, 125–160. [Google Scholar] [CrossRef]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum Likelihood from Incomplete Data Via the EM Algorithm. J. R. Stat. Soc. Ser. B (Methodol.) 1977, 39, 1–22. [Google Scholar] [CrossRef]
- Allassonnière, S.; Kuhn, E.; Trouvé, A. Construction of Bayesian Deformable Models via a Stochastic Approximation Algorithm: A Convergence Study. Bernoulli 2010, 16, 641–678. [Google Scholar] [CrossRef]
- Debavelaere, V.; Durrleman, S.; Allassonnière, S. On the Convergence of Stochastic Approximations under a Subgeometric Ergodic Markov Dynamic. Electron. J. Stat. 2021, 15, 1583–1609. [Google Scholar]
- Hastings, W.K. Monte Carlo Sampling Methods Using Markov Chains and Their Applications. Biometrika 1970, 57, 97–109. [Google Scholar] [CrossRef]
- Robert, C.P.; Casella, G. Monte Carlo Statistical Methods; Springer: New York, NY, USA, 2010. [Google Scholar]
- Li, J.; Fuxin, L.; Todorovic, S. Efficient Riemannian Optimization On The Stiefel Manifold Via The Cayley Transform. arXiv 2020, arXiv:2002.01113. [Google Scholar]
- Pal, S.; Sengupta, S.; Mitra, R.; Banerjee, A. Conjugate Priors and Posterior Inference for the Matrix Langevin Distribution on the Stiefel Manifold. Bayesian Anal. 2020, 15, 871–908. [Google Scholar] [CrossRef]
- Jupp, P.E.; Mardia, K.V. Maximum Likelihood Estimators for the Matrix Von Mises-Fisher and Bingham Distributions. Ann. Stat. 1979, 7, 599–606. [Google Scholar] [CrossRef]
- Kume, A.; Preston, S.P.; Wood, A.T.A. Saddlepoint Approximations for the Normalizing Constant of Fisher–Bingham Distributions on Products of Spheres and Stiefel Manifolds. Biometrika 2013, 100, 971–984. [Google Scholar] [CrossRef]
- Ali, M.; Gao, J. Classification of Matrix-Variate Fisher–Bingham Distribution via Maximum Likelihood Estimation Using Manifold Valued Data. Neurocomputing 2018, 295, 72–85. [Google Scholar] [CrossRef]
- Butler, R.W. Saddlepoint Approximations with Applications; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Debavelaere, V.; Durrleman, S.; Allassonnière, S.; Alzheimer’s Disease Neuroimaging Initiative. Learning the Clustering of Longitudinal Shape Data Sets into a Mixture of Independent or Branching Trajectories. Int. J. Comput. Vis. 2020, 128, 2794–2809. [Google Scholar] [CrossRef]
- Lam, S.K.; Pitrou, A.; Seibert, S. Numba: A LLVM-Based Python JIT Compiler. In Proceedings of the Second Workshop on the LLVM Compiler Infrastructure in HPC; LLVM ’15; Association for Computing Machinery: New York, NY, USA, 2015; pp. 1–6. [Google Scholar] [CrossRef]
- Kaneko, T.; Fiori, S.; Tanaka, T. Empirical Arithmetic Averaging Over the Compact Stiefel Manifold. IEEE Trans. Signal Process. 2013, 61, 883–894. [Google Scholar] [CrossRef]
- Lu, L.; Zhou, T. Link Prediction in Complex Networks: A Survey. Phys. A Stat. Mech. Its Appl. 2011, 390, 1150–1170. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, Y. Link Prediction Based on Graph Neural Networks. Adv. Neural Inf. Process. Syst. 2018, 31, 5171–5181. [Google Scholar]
- Celeux, G.; Frühwirth-Schnatter, S.; Robert, C.P. Model Selection for Mixture Models—Perspectives and Strategies. In Handbook of Mixture Analysis, 1st ed.; Frühwirth-Schnatter, S., Celeux, G., Robert, C.P., Eds.; Chapman and Hall: London, UK; CRC Press: Boca Raton, FL, USA, 2019; pp. 117–154. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef] [PubMed]
- Kiviniemi, V.; Kantola, J.H.; Jauhiainen, J.; Hyvärinen, A.; Tervonen, O. Independent Component Analysis of Nondeterministic fMRI Signal Sources. NeuroImage 2003, 19, 253–260. [Google Scholar] [CrossRef]
- Horn, A.; Ostwald, D.; Reisert, M.; Blankenburg, F. The Structural–Functional Connectome and the Default Mode Network of the Human Brain. NeuroImage 2014, 102, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Eysenck, M.W. Cognitive Psychology: A Student’s Handbook; Psychology Press: New York, NY, USA, 2010. [Google Scholar]
- Purves, D.; Augustine, G.J.; Fitzpatrick, D.; Hall, W.C.; LaMantia, A.S.; Mooney, R.D.; Platt, M.L.; White, L.E. (Eds.) Neuroscience, 6th ed.; Sinauer Associates is an Imprint of Oxford University Press: New York, NY, USA, 2017. [Google Scholar]
- Nguyen, L.T.; Kim, J.; Shim, B. Low-Rank Matrix Completion: A Contemporary Survey. IEEE Access 2019, 7, 94215–94237. [Google Scholar] [CrossRef]
- Kume, A.; Sei, T. On the Exact Maximum Likelihood Inference of Fisher–Bingham Distributions Using an Adjusted Holonomic Gradient Method. Stat. Comput. 2018, 28, 835–847. [Google Scholar] [CrossRef]
- Schiratti, J.B.; Allassonniere, S.; Colliot, O.; Durrleman, S. Learning Spatiotemporal Trajectories from Manifold-Valued Longitudinal Data. Adv. Neural Inf. Process. Syst. 2015, 28, 2404–2412. [Google Scholar]
- Hammond, D.K.; Vandergheynst, P.; Gribonval, R. Wavelets on Graphs via Spectral Graph Theory. Appl. Comput. Harmon. Anal. 2011, 30, 129–150. [Google Scholar] [CrossRef]
- Atasoy, S.; Donnelly, I.; Pearson, J. Human Brain Networks Function in Connectome-Specific Harmonic Waves. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-Regression: A Basic Tool of Chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mantoux, C.; Couvy-Duchesne, B.; Cacciamani, F.; Epelbaum, S.; Durrleman, S.; Allassonnière, S. Understanding the Variability in Graph Data Sets through Statistical Modeling on the Stiefel Manifold. Entropy 2021, 23, 490. https://doi.org/10.3390/e23040490
Mantoux C, Couvy-Duchesne B, Cacciamani F, Epelbaum S, Durrleman S, Allassonnière S. Understanding the Variability in Graph Data Sets through Statistical Modeling on the Stiefel Manifold. Entropy. 2021; 23(4):490. https://doi.org/10.3390/e23040490
Chicago/Turabian StyleMantoux, Clément, Baptiste Couvy-Duchesne, Federica Cacciamani, Stéphane Epelbaum, Stanley Durrleman, and Stéphanie Allassonnière. 2021. "Understanding the Variability in Graph Data Sets through Statistical Modeling on the Stiefel Manifold" Entropy 23, no. 4: 490. https://doi.org/10.3390/e23040490
APA StyleMantoux, C., Couvy-Duchesne, B., Cacciamani, F., Epelbaum, S., Durrleman, S., & Allassonnière, S. (2021). Understanding the Variability in Graph Data Sets through Statistical Modeling on the Stiefel Manifold. Entropy, 23(4), 490. https://doi.org/10.3390/e23040490





