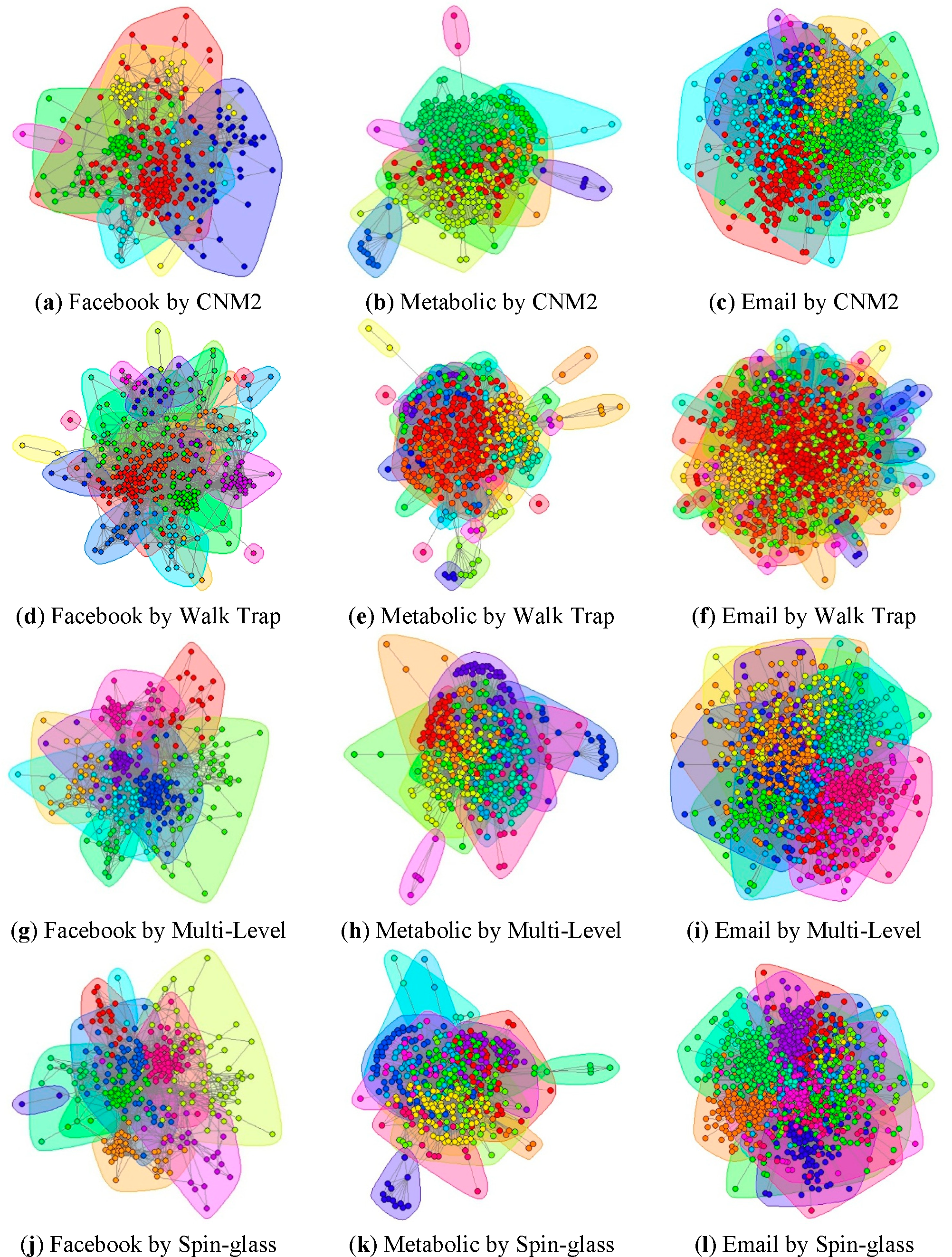
3.1. Evaluation With The SIR Model
For the investigation of the spreading experiment on the actual network, node i, which is used as the source of infection, can infect the scale of the other nodes to measure the impact of node i. When comparing the impact scale of the node as the initial infection source to other nodes in the network, the greater the number of nodes impacted, the greater the impact of the source node.
We adopt the SIR (Susceptible-Infected-Removed) model for the spreading experiment, i.e., a node infects its neighbors with the probability β, and it is assumed that each infected individual is changed to the “removed” status at a fixed rate γ. In this paper, it is assumed that γ = 1, i.e., in the process of each round of spreading, each infected node only has one chance to infect its neighbors with the probability β, and then the node is “removed”. The initial infection method is monophyletic, and the infection threshold β is set as small as possible; the purpose of this is to make the infection speed slow, and it can also make the selection of the infection source more meaningful. In addition, the number from the experimental result is the expectation value. Even when given two sets of identical conditions, the numbers of infected individuals from two groups of experiment are not the same as a result of the randomness and the small value of the infection probability β. Therefore, we need to regard each node as the initial infection source and take the arithmetic average of 100 independent experiments.
For the experiment, select different values of
β according to the scale of different datasets; the selection is based on making the average infected network scale less than 20% and the maximal influence less than 50%. The infection threshold values of networks are provided in
Table 5. Because the power network is quite sparse and the average path length (18.989) is much longer than 6, the results of spreading are not as significant as others.
As shown in
Figure 4, the influence of every individual is presented by the size of the corresponding node in these topological graphs.
Take the spreading capability of the source node as a measurement standard of the node importance, and compare the importance index between the CbC and other nodes. The results of the experiment are described in a temperature figure in Section 3.2; the temperature’s corresponding value is the number of other nodes infected by the source node.
3.2. Experimental Analysis
With comparing the Pearson correlation coefficient between
CbCs (by CNM, Walk trap and Label Propagation) and other classic indexes (
Table 6), it is clearly reflected that the correlations between
CbCs and degree are the extremely strong correlations, so as to eigenvector. Meanwhile, the correlations between
CbCs and
Ks are strong ones.
The relation between the
CbC and degree of the node illustrated in
Figure 5 indicates their obviously positive correlation. The node with the larger degree has a higher
CbC. In the Email network, there are quite a number of nodes (as shown in the fourth quadrant of
Figure 5b) with high
CbC, but its degree is not large. It can be seen from the temperature that a node with large degree and high
CbC has strong spreading capability (e.g., the first quadrant). Compare the spreading capability of a node with the same quadrant; it can be seen that, when the degree of the node is close, the node with the high
CbC has strong spreading capability. Meanwhile, the insufficiency of using degree to measure the importance can also be seen; some nodes do not have large degrees but do have strong spreading capability, which is clearly shown in
Figure 5b. Because calculating
CbC (
i) requires traversing node
i’s neighborhood, the computational complexity of our algorithm is
O(
n<
k>), which grows linearly with the size of a sparse network. Compared with degree centrality (
O(
n), where
n is the number of nodes in the network),
CbC can better quantify the influence of nodes, but it has higher computational complexity.
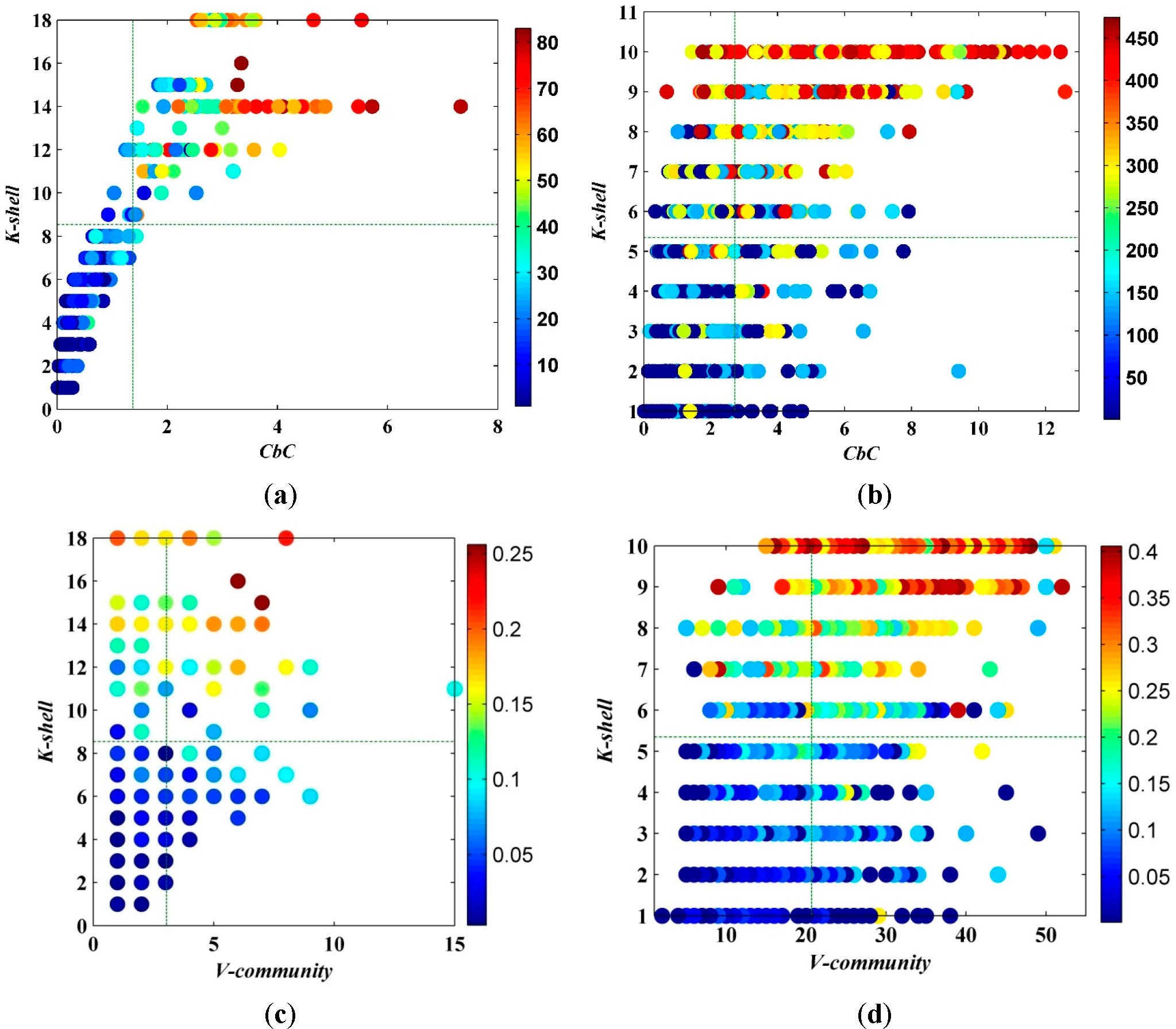
Compared with
V-community ((c) and (d) of
Figure 5 through
Figure 9 at the following part in this paper) presented by the reference [
33] in which the graphs in black and white were in the numbers from 6 to 10, the new measurement demonstrates a more statistically significant result.
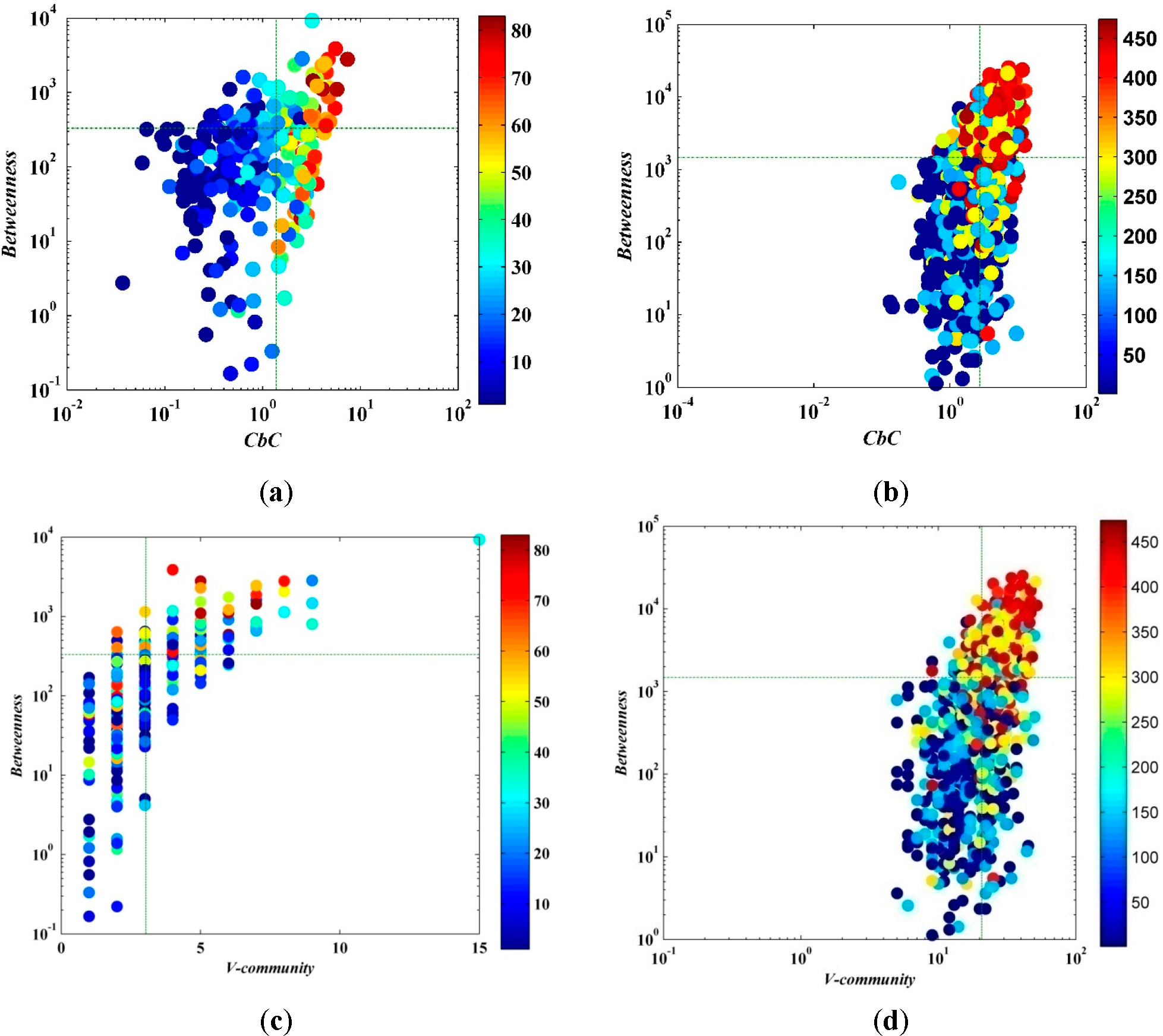
Betweenness is a measure of the centrality of a node in a network and is normally calculated as the fraction of the shortest paths between node pairs that pass through the node of interest. As shown in
Figure 6, it can be described from the trend that the
CbC and betweenness have a positive correlation. It can be seen from the figure that the node with larger betweenness and higher
CbC has stronger spreading capability. Moreover,
CbC can better measure the importance of a node compared to betweenness. When the node with higher
CbC (e.g., the first and the fourth quadrant), even if the betweenness is small (e.g., the fourth quadrant), the node can still have a spectacular range of influence. However, on the contrary, when the node has large betweenness but lower
CbC (e.g., the second quadrant), the number of nodes with strong spreading capability is significantly smaller. In addition, the computational complexity degree of
CbC is much lower than that of betweenness (calculating the shortest paths between all pairs of nodes in a network has the complexity
O(
n3) when using Floyd’s algorithm [
51]; for unweighted networks, calculating betweenness centrality requires
O(
nm) =
O(
n2<
k>) using Brandes’ algorithm [
52]).
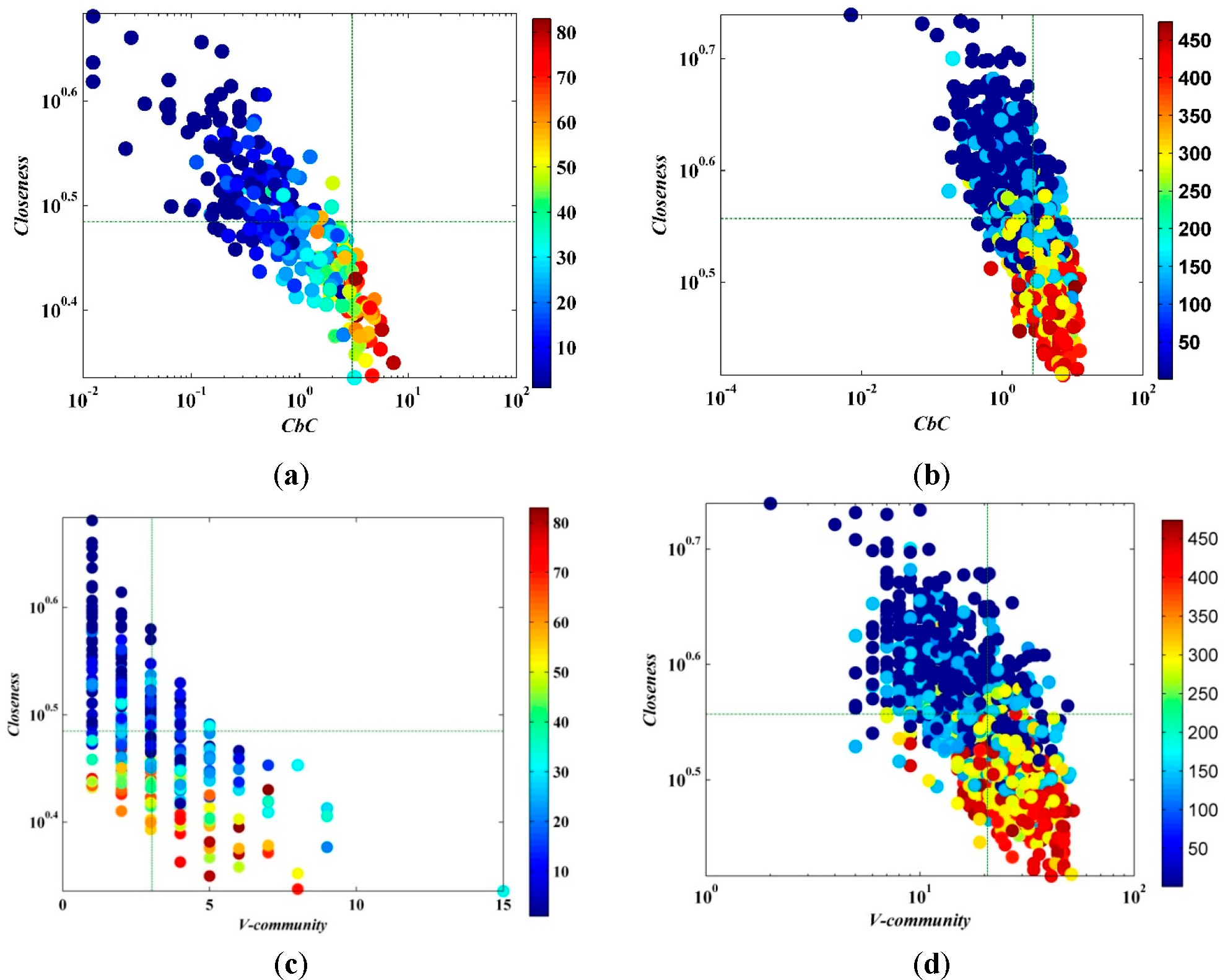
The value of closeness in the manuscript was calculated by Gephi0.8 and the formula was not the reciprocal of the sum on the geodesic distances on all other nodes in the network, but was calculated by the average distance from a given starting node to all other nodes in the network. Consequently, it can be considered as a measure of how long it will take to spread information from a given node to other reachable nodes in the network. The plots of
CbC and the closeness of nodes are given in
Figure 7;
CbC and closeness have a significantly negative correlation. Therefore, the closeness of neighbors with high
CbC is far less than for a node with low
CbC,
i.e., the node with high
CbC is mostly situated at the “bridge” of the communities, but it is not located in the central area of the community. It can be seen from the comparison of the spreading capacity of nodes that the node with higher
CbC has stronger spreading capability. The same as for betweenness centrality, calculating closeness centrality has a complexity of
O(
n3).
Based on the idea that an actor is more central if it has a relation to actors that are themselves central, it can be argued that the centrality of a node does not only depend on its number of adjacent nodes but also on their value of centrality. For example, Bonacichin [
53] defined the centrality of a node as positive multiple of the sum of adjacent centralities.
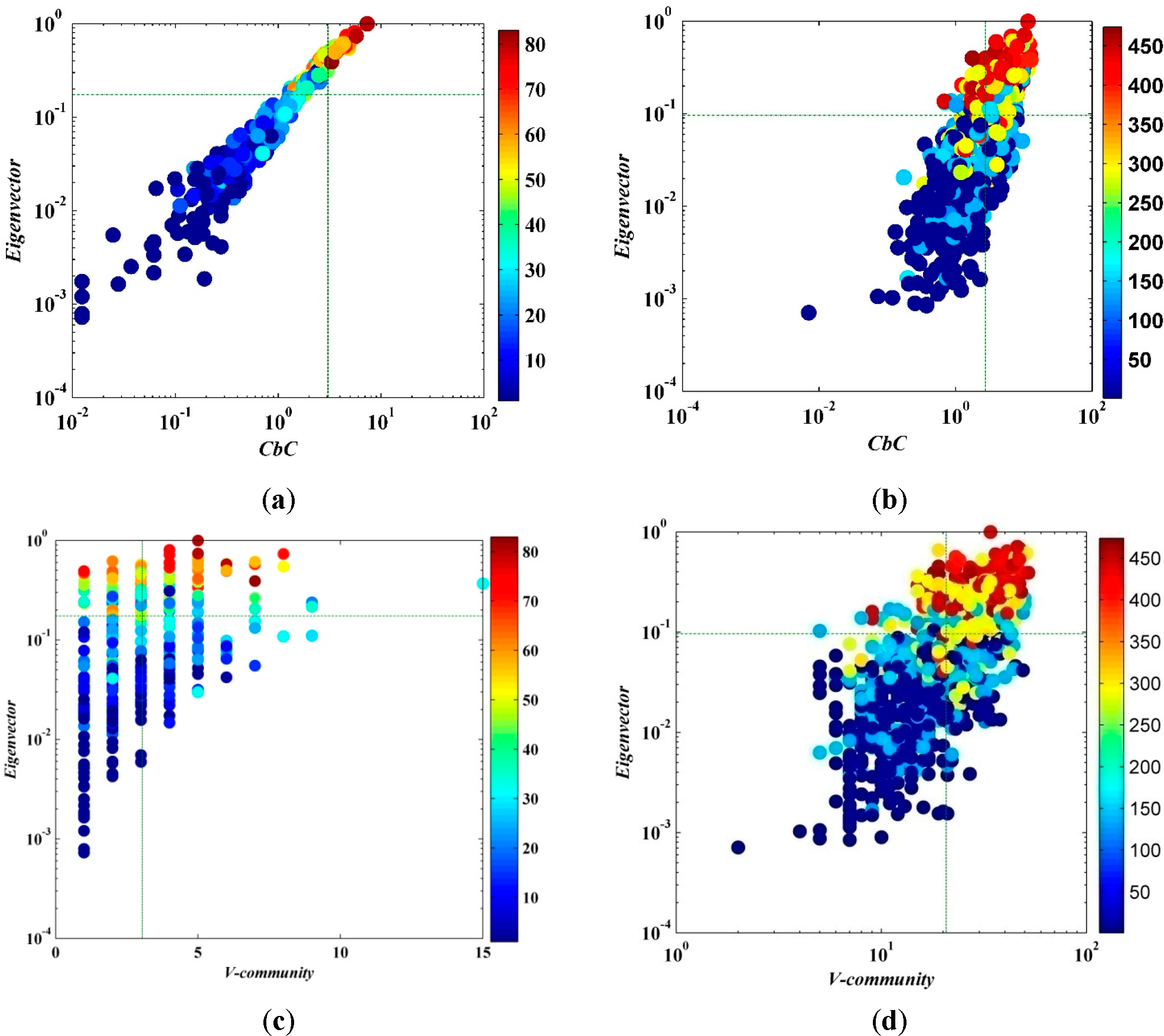
Figure 8 schematizes the correlation between
CbC and eigenvector centricity of a node in the network;
CbC and eigenvector centricity have significant positive correlation,
i.e., the node with higher
CbC has larger eigenvector centricity, and
vice versa. The computational complexity of eigenvector is
O(
n2), which is less than betweenness and closeness centrality but still larger than the algorithm we have proposed.
In contrast to common belief, there are plausible circumstances where the best spreaders do not correspond to the most highly connected or the most central people. Thus, [
19] suggested that the position of the node relative to the organization of the network determines its spreading influence more than a local property of a node and defined
K-shell. The relation between
CbC and
K-shell value
Ks is presented in
Figure 9. As
Ks in the network is distributed centrally, the positive correlation of
CbC and
Ks is not very obvious. However, as the
Ks values of a large number of nodes in the network are the same, it is deficient to use as an index to measure the importance of nodes. The experiment conducted in this paper will correct to “0.0001” for the
CbC of a node. Therefore, it can avoid repetition to the greatest extent. Through the spreading capability of a node,
Figure 9 shows that, if the
Ks values of the nodes are the same, the node with the higher
CbC has the stronger spreading capability.
Furthermore, compared with
V-community in
Figure 9c,d, the measurement we presented is no longer a discrete variable. Precisely because of the continuity, the ranking of importance can be described by
CbC more appropriately in a sense. The details of its advantages will be provided in the next section.
3.3. The Advantage of CbC
Firstly, we compared the Pearson correlation coefficient (
r) between spreading capabilities (in the same spreading probability
β = 0.05) and
CbCs calculated by CNM, Walk Trap and Label Propagation, and further compared the Pearson correlation coefficient (
r) between spreading capabilities of nodes and other classical centrality indicators (
Table 7).
By comparing the experimental results in
Table 7, the correlations between
CbCs and spreading capabilities are all the extremely strong correlations. Furthermore, the results show that the Pearson Correlation Coefficient (
r) for
CbC is almost larger than that for other centrality measures except the eigenvector. The values of Pearson correlation coefficient on eigenvector centralities are consistently higher than others. Furthermore, in our current research on effects of spreading capability depending on diverse probabilities of propagation, the fascinating results shows that the Pearson correlation coefficient on eigenvector centrality in Email network is declining with the increase of propagation probability. However, the value of the Coefficients are no lower than 0.8, namely, the correlations are all extremely strong ones between eigenvector and those influence ranking results by varying propagation probabilities.
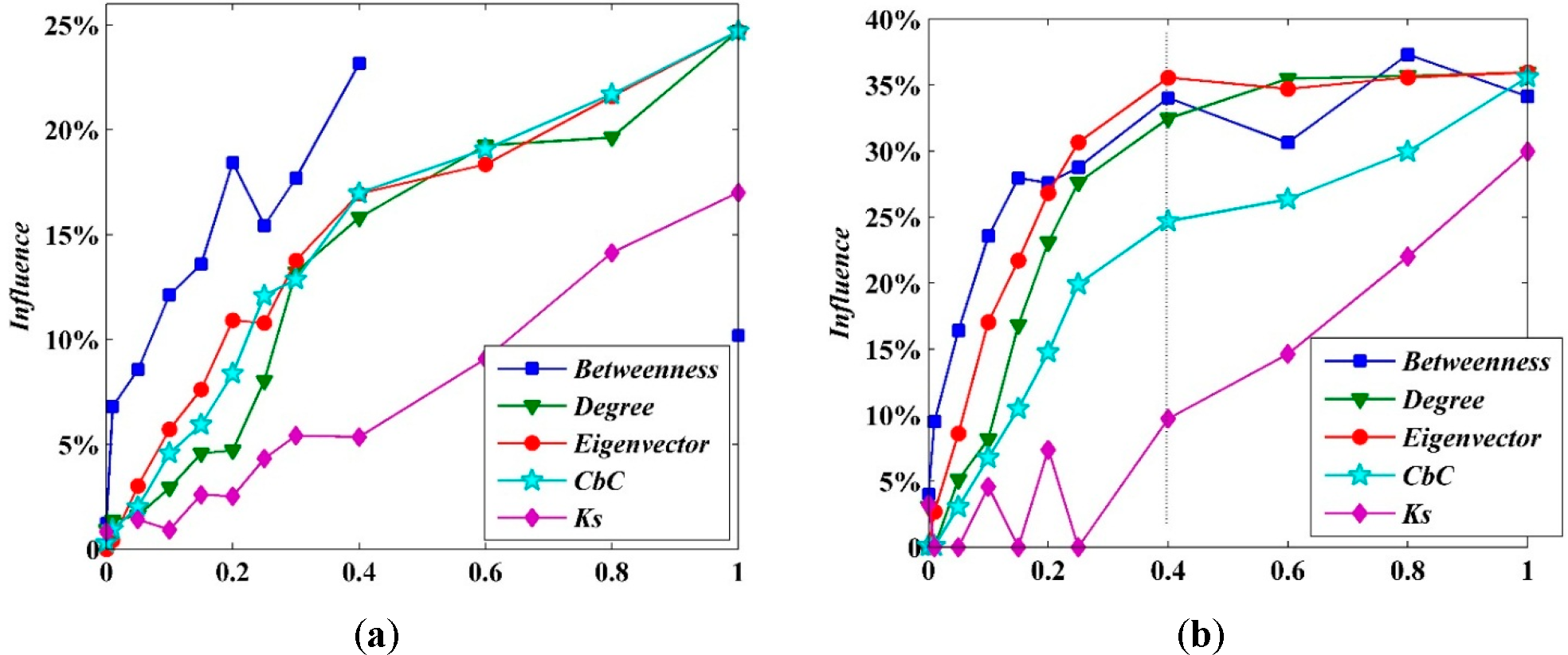
For convenience in computation, one may normalize the variables above by dividing them by
N (the number of nodes), and, in doing so, they also have the meaning of “ranking”. The normalization of the indicators (betweenness, degree, eigenvector,
CbC and
K-shell) is shown in
Figure 10. The ordinate axis is the average influence of the infection source. According to the curves, the spreading capability of the sources and the ranking of selected indicators almost have positive correlations. Totally depending on the experimental data results without considering the possible error bars for points, the
CbC is the only index that the influence of the source is monotone increasing. Meanwhile, the competitors, degree and eigenvector, both exist in fluctuation; the average influence of larger indexes is lower than that of smaller ones, especially when the indicator has a high level (ranking results of more than 0.4). Nevertheless, the differentials are not significant enough to support such a definitive conclusion, even worse, they are probably caused by possible error. Furthermore,
Figure 10b represents that the influence measured by
CbC is increasing at a much slower pace. Accordingly, the influence of the source can be ranked by
CbC more steadily and homogeneously. Comparing between
Figure 10a and
Figure 10b, it can be seen clearly that, the larger the network, the more obvious this performance.
In
Figure 10, the average influence of sources with level = 0.4 as ranked by
CbC is 24%, compared to 35% for those ranked by eigenvector. It is interesting to note that, when the indicators (betweenness and degree) are at a lower level (<0.4), the average influence decreases by approximately 30% to 35%. Some nodes with ordinary index values have distinguished spreading capability. In other words, these “critical influential spreaders” would be neglected by ranking with betweenness, degree or eigenvector. Identifying the influential spreaders, especially these “critical nodes”, is the very advantage of the
CbC method.
The influences of the top 1% of nodes are listed in
Table 8 (the top four nodes in the Facebook network with 324 nodes) and
Table 9 (the top 12 nodes in the Email network with 1133 nodes).
Table 8 illustrates that node 211 was identified only by
CbC and was neglected by the other indicators. However, the average influence of node 211 is more than 22%, which is much larger than the average value of the entire network (0.077437, as discussed in Section 3.1 above). It is definitely an influential spreader and is 14th when ranked by influential capability. Meanwhile, when ranking by degree or betweenness, node 186 is in the top 1%, but it is in 105th place for influential capability, with only a 10% influential range. Even worse, node 153 is third highest ranking by betweenness but has no more than a 6.5% influential result, even less than the average influence of this network. In addition, the influence of node 33 is marginally lower than that of node 211. Actually, although node 33 is not in the top 1%, it is ranked 8th place by
CbC. As expected, with the increase of the network size, the advantages of identifying critical influential spreaders by
CbC are more obvious. The results of the Email network are shown in
Table 9.
These data depict that nodes 134, 205, 219, 206, 198, 201 and 140 are in the top 1% level ranking by CbC but are ignored by all other indicators. However, the influences of these seven nodes are all significantly greater than the average of the entire network (0.149706), and the influence of node 198 is third highest of all 1133 nodes in the Email network.
Furthermore, as shown in
Table 8 and
Table 9, the very top 1% nodes ranking by eigenvector centrality, unfortunately, more or less miss some “critical nodes”, which can be explored by
CbC. It seems that, although the ranking result by eigenvector centrality of entirety is satisfied, at the top level it is not precise.
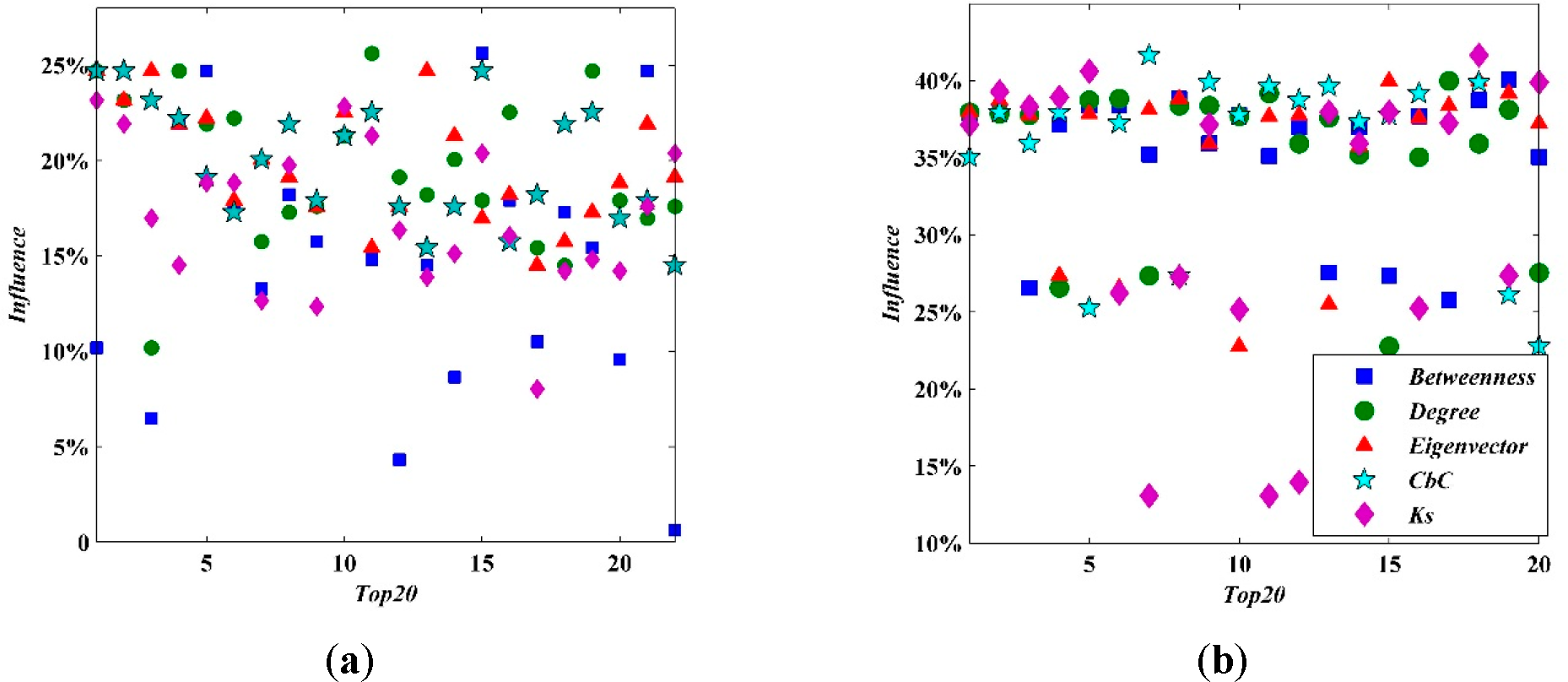
Above all,
CbC plays an important role in identifying the critical influential spreaders. However, the influence of source cannot be strictly ranked by any ordered indicators. Given are two scatter diagrams plotting out the influence of the top 20 nodes as ranked by the chosen indicators (
Figure 11). Even more, the first and second spreaders are not found by the indicators above. In the Facebook network, the two most influential spreaders are node 16 and node 219 (influences of 0.256173 and 0.253086, respectively). In the Email network, the most influential spreaders are node 396 and node 358 (both have influences of 0.418358). Our research indicates that
CbC can help to identify critical spreaders; however, in order to strictly rank these spreaders, further research is needed, which is exactly what we plan to focus on for future work.