Reducing False Arrhythmia Alarms Using Different Methods of Probability and Class Assignment in Random Forest Learning Methods
Abstract
1. Introduction
2. Related Works
2.1. Arrhythmia Detection
2.2. Random Forest as a Machine Learning Algorithm
3. State-of-the-Art Random Forest
3.1. Decision Tree as a Base Algorithm
| Algorithm 1: Decision tree algorithm pseudocode. |
| input: List of all explanatory variables (), training dataset () output: Decision tree () /1/ Start at the root node /2/ for each in do /3/ find the set S that minimizes the sum of the node impurities in the two child nodes in Equation (1) and choose the split that produces the minimum overall and /4/ end /5/ if stopping criterion is reached then do /6/ stop /7/ else do /8/ apply step 2 to each child node in turn /9/ end /10/ return DT |
3.2. Standard Random Forest Algorithm
| Algorithm 2: Random Forest algorithm pseudocode. |
| input: Number of Trees (), random subset of the features (), training dataset () output: random forest () /1/ is empty /2/ for each to do /3/ = Bootstrap Sample () /4/ = Random Decision Tree (, ) /5/ = /6/ end /7/ return |
4. Model Performance Measures
4.1. Confusion Matrix and Score Function
- True Positives (TP): the cases that predicted yes and the actual output was also yes,
- True Negatives (TN): the cases that predicted no and the actual output was no,
- False Positives (FP): the cases that predicted yes and the actual output was no, and
- False Negatives (FN): the cases that predicted no and the actual output was yes.
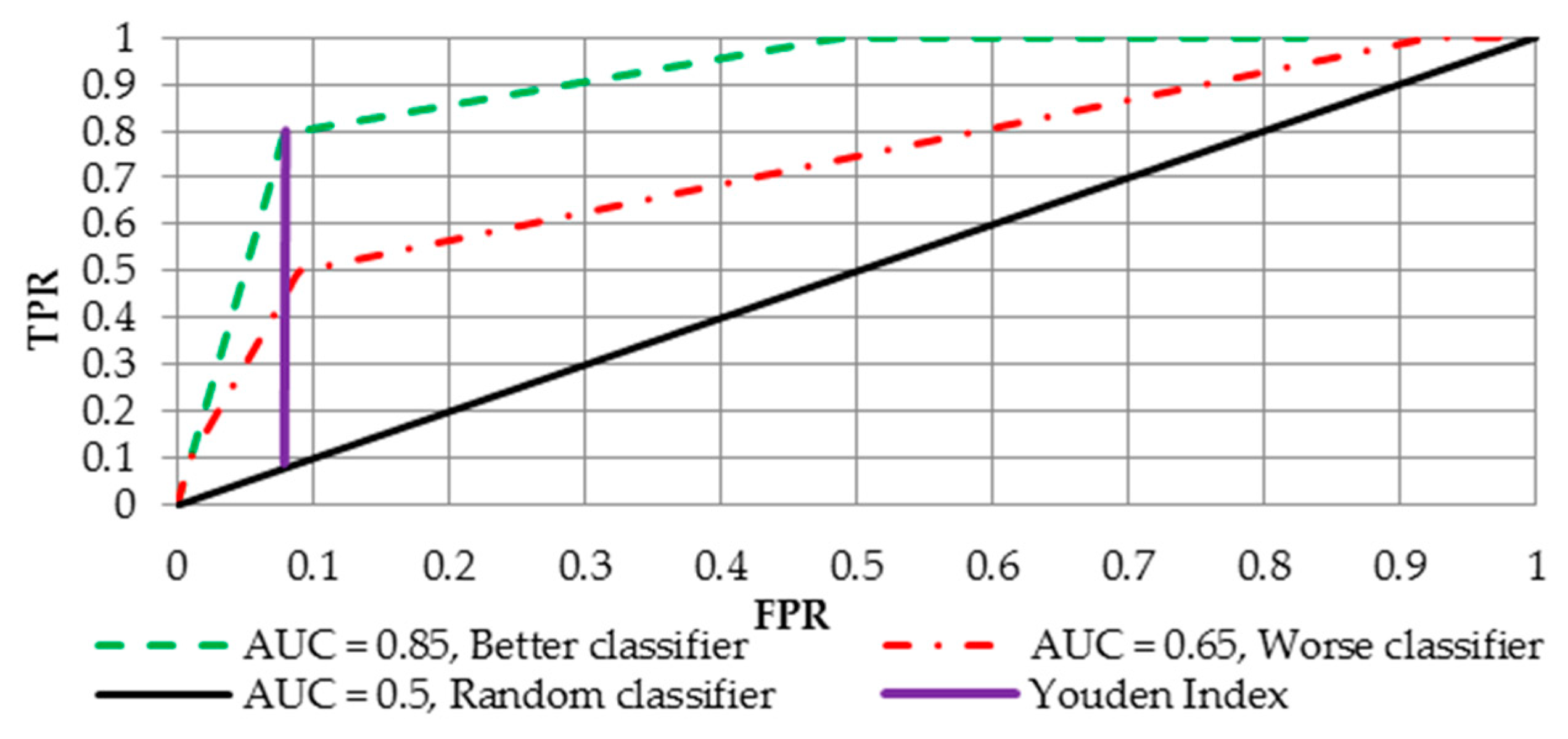
4.2. Receiver Operating Curve (ROC) and Area Under the ROC
4.3. Optimal Treshold for Class Determination
5. Methods of Probability and Class Determining
- (1)
- Probability: averaging all the probabilities from all the trees within the forest (Avg Prob; upper solid line table);
- (a)
- Class: cutoff (Youden Index) for class determination is derived based on the In Bag sample (abbreviation: Prob INB),
- (b)
- Class: cutoff (Youden Index) for class determination is derived based on the training sample (abbreviation: Prob Train),
- (c)
- Class: cutoff for class determination is set at 0.5 (abbreviation: Prob 0.5).
- (2)
- Probability: majority class voting where the cutoff for class determining is derived based on the In Bag sample (upper-middle dashed line table; abbreviation: Voting Prob INB);
- (a)
- Class: cutoff (Youden Index) for class determining is derived based on In-Bag sample (abbreviation: Vote INB INB),
- (b)
- Class: cutoff for class determination is set to 0.5 (abbreviation: Vote INB 0.5).
- (3)
- Probability: majority class voting where the cutoff for class determining is based on the training sample (lower-middle dotted-dashed line table; abbreviation: Voting Prob Training);
- (a)
- Class: cutoff (Youden Index) for class determining is based on the training sample (abbreviation: Vote Train Train),
- (b)
- Class: cutoff for class determination is set to 0.5 (abbreviation: Vote Train 0.5).
- (4)
- Probability: majority class voting where the cutoff is set to 0.5 (bottom red line table, equivalent to the standard RF; abbreviation: Voting Prob 0.5);
- (a)
- Class: cutoff (Youden Index) for class determination is derived based on the In Bag sample (abbreviation: Vote 0.5 INB),
- (b)
- Class: cutoff (Youden Index) for class determination is derived based on the training sample (abbreviation: Vote 0.5 Train),
- (c)
- Class: cutoff for class determination is set to 0.5 (abbreviation: Vote 0.5 0.5).
6. Empirical Analysis
6.1. Feature Vector
6.2. Numerical Implementation
6.3. Detailed Results for AUC and Score for Various Samples
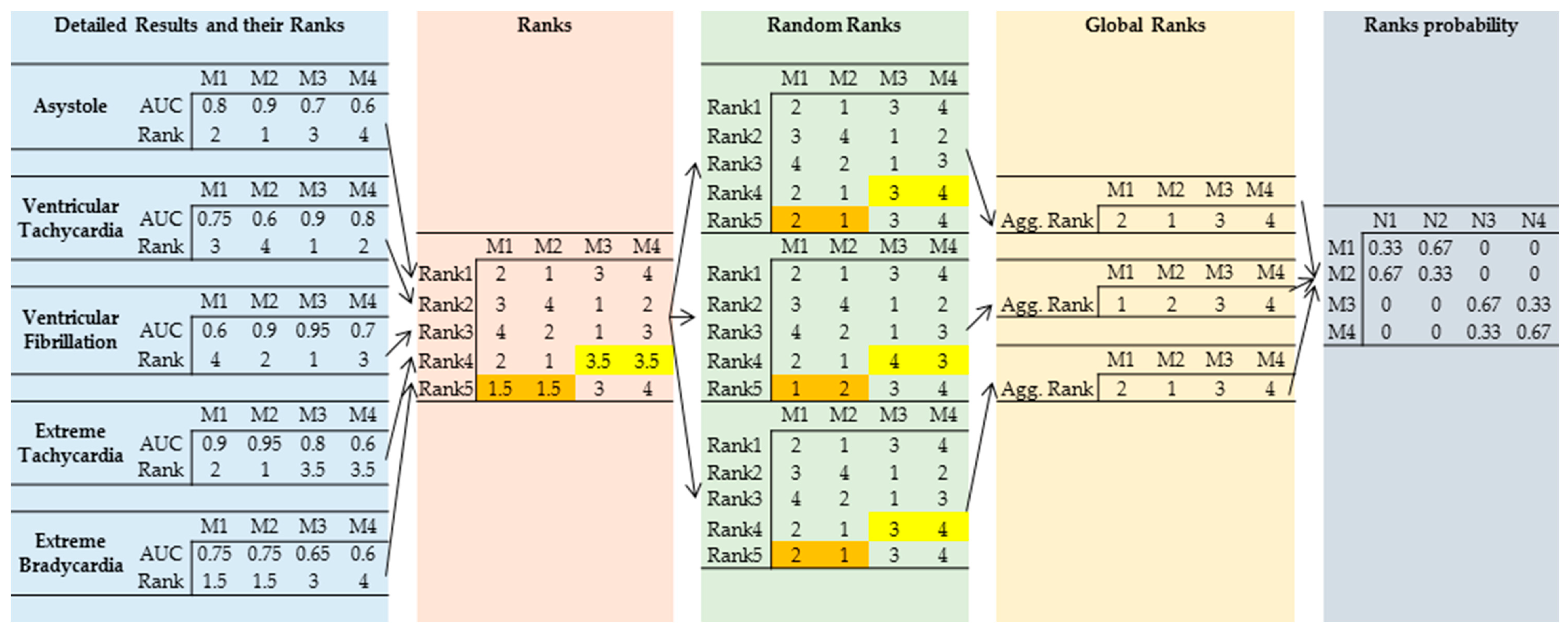
6.4. Aggregated Ranks for AUC and Score Based on the Validation Sample
7. Conclusions
- (1)
- Ventricular tachycardia is the arrhythmia for which the false alarms are the most difficult to detect since AUC values for both unseen samples (OOB and validation) have the lowest ranges, between 0.86 and 0.87;
- (2)
- Extreme tachycardia arrhythmia false alarms are by far the easiest to detect as the AUC values are close to 1 for both OOB and validation datasets;
- (3)
- The AUC value is the greatest for the OOB sample when the probability is selected using majority class voting where the cutoff for class assignment is derived based on the training sample;
- (4)
- In terms of the score measure, the validation dataset delivers better results for false alarms detection in comparison with Out of Bag; however, the results are biased with higher standard errors;
- (5)
- In terms of the score measure, ventricular tachycardia false alarms are difficult to capture as the scores are lower than 50, and slighly better scores are produced using the validaton dataset;
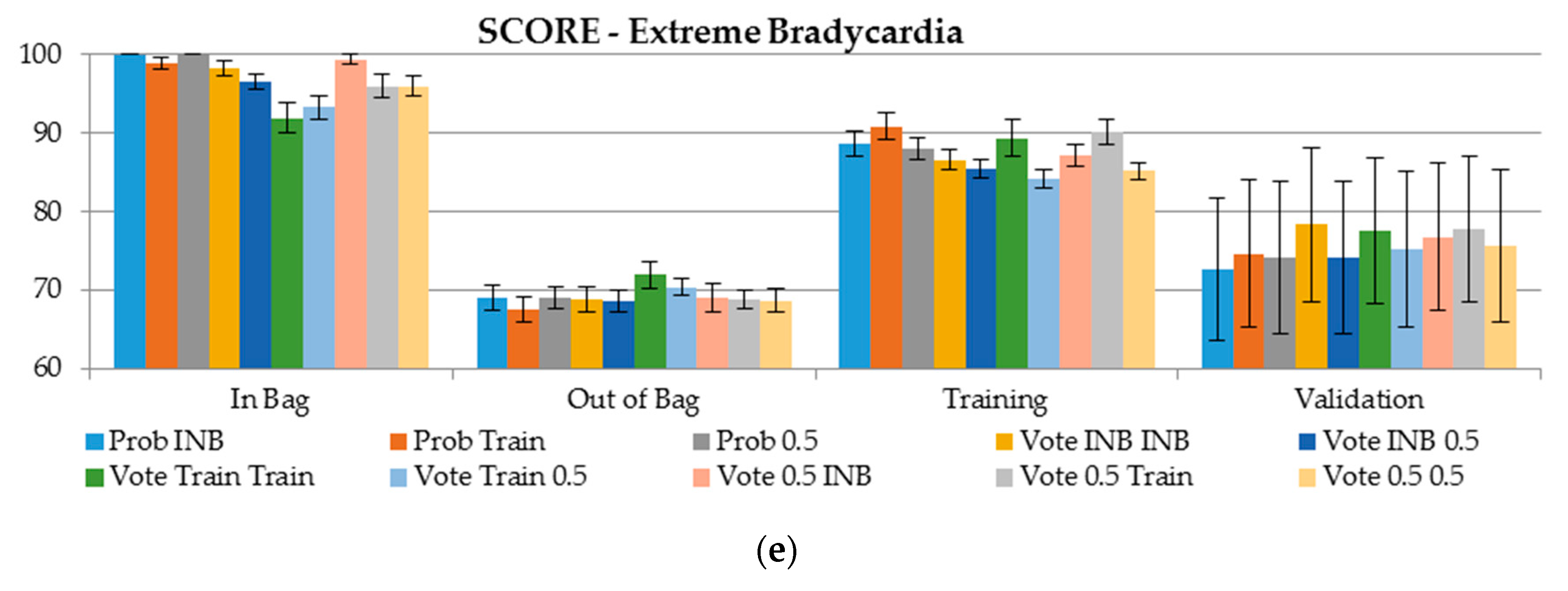
- (6)
- For ventricular fibrillation, the scores obtained on the validation dataset are 30 points better than using OOB (score 50 vs. 80, respectively). This was observed when the cutoff for the class assignment was derived based on the training sample, or the cutoff for class determining was set to 0.5 (Figure 4).
- (1)
- In 53.83% of the cases, the majority class voting with the cutoff set to 0.5 (Voting Prob 0.5) produced the highest AUC value (first ranking position: N1);
- (2)
- The best score (first ranking position: N1) was observed when the probability for each observation was determined using majority class voting with the cutoff for the class set to 0.5. The final class was determined using the cutoff derived based on the training sample (Vote 0.5 Train). This approach produces the highest score on the validation dataset.
Author Contributions
Funding
Conflicts of Interest
Appendix A
| TPR In Bag | TNR In Bag | TPR Out of Bag | TNR Out of Bag | TPR Training | TNR Training | TPR Validation | TNR Validation | |
|---|---|---|---|---|---|---|---|---|
| Prob INB | 100(±0) | 100(±0) | 77.2(±0.9) | 93.9(±0.3) | 89.9(±1.7) | 99(±0.1) | 75(±8.3) | 94(±2.2) |
| Prob Train | 100(±0) | 99.9(±0.1) | 77.2(±0.9) | 91.6(±0.4) | 100(±0) | 98.2(±0.3) | 80(±8.2) | 92(±2) |
| Prob 0.5 | 98.5(±0.8) | 100(±0) | 72.2(±1) | 96.9(±0.2) | 86.3(±0.8) | 100(±0) | 70(±8.2) | 97(±1.5) |
| Vote INB INB | 100(±0) | 100(±0) | 60.5(±3) | 97.7(±0.3) | 84.8(±1.3) | 100(±0) | 61.7(±6.6) | 97(±1.5) |
| Vote INB 0.5 | 100(±0) | 99(±0.2) | 78.3(±0.8) | 91.3(±0.4) | 100(±0) | 97.8(±0.3) | 75(±8.3) | 91(±2.3) |
| Vote Train Train | 100(±0) | 99.2(±0.2) | 77.3(±0.9) | 93.8(±0.7) | 100(±0) | 98.4(±0.2) | 75(±8.3) | 93(±2.1) |
| Vote Train 0.5 | 100(±0) | 99(±0.2) | 78.3(±0.7) | 93.1(±0.5) | 99.5(±0.5) | 97.9(±0.3) | 75(±8.3) | 91(±2.3) |
| Vote 0.5 INB | 100(±0) | 100(±0) | 77.2(±0.9) | 91.9(±0.3) | 92.9(±1.3) | 97.6(±0.3) | 80(±8.2) | 92(±2) |
| Vote 0.5 Train | 100(±0) | 99.9(±0.1) | 80.8(±1.5) | 89(±0.5) | 100(±0) | 97.1(±0.3) | 85(±7.6) | 90(±2.6) |
| Vote 0.5 0.5 | 87.3(±0.9) | 100(±0) | 72.2(±1) | 97.1(±0.2) | 79.8(±1.5) | 100(±0) | 70(±8.2) | 97(±1.5) |
| TPR In Bag | TNR In Bag | TPR Out of Bag | TNR Out of Bag | TPR Training | TNR Training | TPR Validation | TNR Validation | |
|---|---|---|---|---|---|---|---|---|
| Prob INB | 100(±0) | 100(±0) | 51.8(±1.1) | 91.1(±0.5) | 98(±0.3) | 99.6(±0.1) | 56.7(±6.3) | 90.1(±1.5) |
| Prob Train | 100 (±0) | 100(±0) | 58.9(±2.3) | 88.4(±0.8) | 99.6(±0.2) | 99.4(±0.1) | 65.6(±4.5) | 88.1(±1.9) |
| Prob 0.5 | 100(±0) | 100(±0) | 42.5(±0.9) | 94.4(±0.2) | 94(±0.4) | 100(±0) | 42.9(±5.7) | 94.5(±0.9) |
| Vote INB INB | 100(±0) | 100(±0) | 36.8(±1.4) | 95.4(±0.2) | 93.3(±1.1) | 100(±0) | 38.1(±5.2) | 95.3(±1.2) |
| Vote INB 0.5 | 100(±0) | 99.8(±0.1) | 64.2(±0.9) | 86.1(±0.4) | 100(±0) | 97.3(±0.2) | 68.9(±4.6) | 85.3(±1.6) |
| Vote Train Train | 100(±0) | 100(±0) | 53.4(±2.3) | 92.1(±0.5) | 99.1(±0.2) | 99.3(±0.1) | 55.6(±6.4) | 89.7(±1.5) |
| Vote Train 0.5 | 100(±0) | 99.6(±0.1) | 69.3(±0.6) | 86.1(±0.4) | 99.8(±0.2) | 96.7(±0.2) | 68.9(±4.6) | 85.3(±1.6) |
| Vote 0.5 INB | 100(±0) | 100(±0) | 58.7(±1.3) | 88.5(±0.5) | 98.3(±0.2) | 99.3(±0.1) | 63.3(±4.7) | 87.7(±1.8) |
| Vote 0.5 Train | 100(±0) | 100(±0) | 58.4(±2.4) | 88(±0.8) | 99.1(±0.2) | 99.4(±0.2) | 67.8(±5.4) | 88.9(±1.4) |
| Vote 0.5 0.5 | 99.5(±0.2) | 100(±0) | 39.5(±1.1) | 95.1(±0.2) | 90.5(±0.8) | 100(±0) | 39.3(±5.3) | 95.3(±1) |
| TPR In Bag | TNR In Bag | TPR Out of Bag | TNR Out of Bag | TPR Training | TNR Training | TPR Validation | TNR Validation | |
|---|---|---|---|---|---|---|---|---|
| Prob INB | 100(±0) | 100(±0) | 23.3(±9.5) | 96.2(±0.5) | 66.7(±6.7) | 98.9(±0.8) | 16.7(±16.7) | 98.2(±1.9) |
| Prob Train | 100(±0) | 96.6(±0.5) | 56.7(±8) | 95.4(±0.6) | 100(±0) | 96.2(±0.5) | 83.3(±16.7) | 94.2(±2.6) |
| Prob 0.5 | 86.7(±6.7) | 100(±0) | 13.3(±4.2) | 97.7(±0.8) | 56.7(±6.1) | 99.6(±0.4) | 16.7(±16.7) | 98.2(±1.9) |
| Vote INB INB | 100(±0) | 100(±0) | 0(±0) | 99.6(±0.4) | 13.3(±8.4) | 100(±0) | 0(±0) | 100(±0) |
| Vote INB 0.5 | 100(±0) | 95.4(±0.6) | 70(±6.8) | 94.2(±1) | 100(±0) | 95.4(±0.6) | 83.3(±16.7) | 92.4(±3.8) |
| Vote Train Train | 100(±0) | 96.2(±0.5) | 56.7(±3.3) | 96.2(±0.5) | 100(±0) | 96.2(±0.5) | 83.3(±16.7) | 96.3(±2.3) |
| Vote Train 0.5 | 100(±0) | 95.4(±0.6) | 70(±6.8) | 94.2(±1) | 100(±0) | 95.4(±0.6) | 83.3(±16.7) | 92.4(±3.8) |
| Vote 0.5 INB | 100(±0) | 100(±0) | 33.3(±9.9) | 95.4(±0.6) | 83.3(±6.1) | 98.1(±0.9) | 50(±22.4) | 98.2(±1.9) |
| Vote 0.5 Train | 100(±0) | 98.1(±0.9) | 43.3(±10.9) | 93.1(±1.2) | 100(±0) | 96.2(±0.5) | 83.3(±16.7) | 94.2(±2.6) |
| Vote 0.5 0.5 | 73.3(±6.7) | 100(±0) | 10(±4.5) | 98.9(±0.8) | 50(±4.5) | 100(±0) | 16.7(±16.7) | 100(±0) |
| TPR In Bag | TNR In Bag | TPR Out of Bag | TNR Out of Bag | TPR Training | TNR Training | TPR Validation | TNR Validation | |
|---|---|---|---|---|---|---|---|---|
| Prob INB | 100(±0) | 100(±0) | 99.2(±0.1) | 69.4(±4.5) | 100(±0) | 98.6(±1.5) | 99.2(±0.8) | 77.8(±15.6) |
| Prob Train | 100(±0) | 100(±0) | 99.2(±0.1) | 66.7(±4.9) | 100(±0) | 100(±0) | 99.2(±0.8) | 77.8(±15.6) |
| Prob 0.5 | 100(±0) | 100(±0) | 99.2(±0.1) | 76.4(±1.5) | 99.3(±0.1) | 100(±0) | 99.2(±0.8) | 77.8(±15.6) |
| Vote INB INB | 100(±0) | 100(±0) | 99.7(±0.2) | 54.2(±4.9) | 100(±0) | 86.1(±4.1) | 99.2(±0.8) | 66.7(±17.7) |
| Vote INB 0.5 | 99.2(±0.1) | 100(±0) | 98.8(±0.2) | 83.3(±3.1) | 99.2(±0.1) | 100(±0) | 98.5(±1.1) | 88.9(±11.8) |
| Vote Train Train | 100(±0) | 100(±0) | 99.2(±0.1) | 62.5(±3.8) | 100(±0) | 100(±0) | 99.2(±0.8) | 66.7(±17.7) |
| Vote Train 0.5 | 99.2(±0.1) | 100(±0) | 98.8(±0.2) | 83.3(±3.1) | 99.2(±0.1) | 100(±0) | 98.5(±1.1) | 88.9(±11.8) |
| Vote 0.5 INB | 100(±0) | 100(±0) | 99.2(±0.1) | 66.7(±5.4) | 100(±0) | 93.1(±3.9) | 99.2(±0.8) | 66.7(±17.7) |
| Vote 0.5 Train | 100(±0) | 100(±0) | 99.2(±0.1) | 70.8(±3.8) | 100(±0) | 100(±0) | 99.2(±0.8) | 77.8(±15.6) |
| Vote 0.5 0.5 | 99.4(±0.2) | 100(±0) | 99.1(±0) | 79.2(±2.2) | 99.2(±0.1) | 100(±0) | 99.2(±0.8) | 77.8(±15.6) |
| TPR In Bag | TNR In Bag | TPR Out of Bag | TNR Out of Bag | TPR Training | TNR Training | TPR Validation | TNR Validation | |
|---|---|---|---|---|---|---|---|---|
| Prob INB | 100(±0) | 100(±0) | 86.7(±0.8) | 87.9(±1.3) | 96.1(±0.7) | 94.6(±0.6) | 82.5(±8.7) | 87(±4.7) |
| Prob Train | 99.5(±0.3) | 100(±0) | 85.5(±1.1) | 88.1(±1.6) | 96.4(±1) | 97.9(±0.8) | 82.5(±8.7) | 91(±3.7) |
| Prob 0.5 | 100(±0) | 100(±0) | 86.5(±0.7) | 88.9(±1.2) | 95.4(±0.8) | 96.6(±0.7) | 82.5(±8.7) | 88.5(±3.9) |
| Vote INB INB | 99.3(±0.4) | 100(±0) | 86.7(±0.9) | 87.1(±1.4) | 94.9(±0.6) | 95.6(±0.7) | 85(±8.9) | 88.5(±3.9) |
| Vote INB 0.5 | 98.6(±0.4) | 100(±0) | 86(±0.7) | 89.4(±1.1) | 94(±0.6) | 97.4(±0.4) | 82.5(±8.7) | 88.5(±3.9) |
| Vote Train Train | 96.9(±1) | 97.9(±0.8) | 89.1(±1.5) | 85.2(±2.4) | 95.9(±1.3) | 96.6(±1.2) | 85(±8.9) | 88.5(±3.9) |
| Vote Train 0.5 | 97.1(±0.7) | 100(±0) | 86.7(±0.5) | 91.2(±0.9) | 93.5(±0.6) | 96.6(±0.6) | 82.5(±8.7) | 90.5(±3.9) |
| Vote 0.5 INB | 99.8(±0.2) | 100(±0) | 87(±1) | 86.6(±1.3) | 95.4(±0.7) | 94.8(±0.9) | 85(±8.9) | 87(±4.7) |
| Vote 0.5 Train | 98.3(±0.7) | 99.7(±0.3) | 86.7(±0.7) | 87.1(±1.8) | 96.1(±0.8) | 97.7(±0.6) | 84.5(±8.9) | 91(±3.7) |
| Vote 0.5 0.5 | 98.3(±0.5) | 100(±0) | 85.7(±0.9) | 90.4(±0.9) | 93.7(±0.5) | 97.9(±0.3) | 82.5(±8.7) | 93(±3.6) |
| Method | Asystole | Ventricular Tachycardia | Ventricular Flutter or Fibrillation | Extreme Tachycardia | Extreme Bradycardia | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| TPR | TNR | TPR | TNR | TPR | TNR | TPR | TNR | TPR | TNR | |
| Gajowniczek et al. (Vote 0.5 training) | 100 | 97 | 99 | 99 | 100 | 96 | 100 | 100 | 96 | 98 |
| Plesinger et al. [3] | 100 | 68 | 80 | 80 | 100 | 85 | 99 | 78 | 91 | 60 |
| Krasteva et al. [3] | 100 | 99 | 81 | 77 | 100 | 96 | 100 | 100 | 98 | 91 |
| Kalidas et al. [3] | 100 | 86 | 84 | 85 | 100 | 65 | 100 | 89 | 100 | 93 |
| Hoog Antink et al. [3] | 100 | 89 | 93 | 85 | 100 | 92 | 100 | 100 | 100 | 77 |
| Eerikäinen et al. [3] | 90 | 90 | 89 | 63 | 67 | 94 | 98 | 88 | 93 | 83 |
| Asadi et al. [22] | 39 | 49 | 68 | 85 | 78 | 89 | 87 | 82 | 56 | 60 |
| Srivastava et al. [23] | 100 | 89 | 100 | 67 | 83 | 88 | 100 | 67 | 100 | 72 |
Appendix B
References
- Aboukhalil, A.; Nielsen, L.; Saeed, M.; Mark, R.G.; Clifford, G.D. Reducing false alarm rates for critical arrhythmias using the arterial blood pressure waveform. J. Biomed. Inform. 2008, 41, 442–451. [Google Scholar] [CrossRef]
- Drew, B.J.; Harris, P.; Zègre-Hemsey, J.K.; Mammone, T.; Schindler, D.; Salas-Boni, R.; Bai, Y.; Tinoco, A.; Ding, Q.; Hu, X. Insights into the problem of alarm fatigue with physiologic monitor devices: A comprehensive observational study of consecutive intensive care unit patients. PLoS ONE 2014, 9, e110274. [Google Scholar] [CrossRef] [PubMed]
- Clifford, G.D.; Silva, I.; Moody, B.; Li, Q.; Kella, D.; Shahin, A.; Kooistra, T.L.; Perry, D.; Mark, R.G. The PhysioNet/computing in cardiology challenge 2015: Reducing false arrhythmia alarms in the ICU. In Proceedings of the 2015 Computing in Cardiology Conference (CinC), Nice, France, 6–9 September 2015; pp. 273–276. [Google Scholar] [CrossRef]
- Pan, J.; Tompkins, W.J. A Real-Time QRS Detection Algorithm. IEEE Trans. Biomed. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B. ECG signal classification and parameter estimation using multiwavelet transform. Biomed. Res. 2017, 28. Available online: http://www.biomedres.info/biomedical-research/ecg-signal-classification-and-parameter-estimation-using-multiwavelet-transform.html (accessed on 29 March 2019).
- Christov, I.I. Real time electrocardiogram QRS detection using combined adaptive threshold. Biomed. Eng. Online 2004, 3, 28. [Google Scholar] [CrossRef] [PubMed]
- Arzeno, N.M.; Deng, Z.-D.; Poon, C.-S. Analysis of First-Derivative Based QRS Detection Algorithms. IEEE Trans. Biomed. Eng. 2008, 55, 478–484. [Google Scholar] [CrossRef]
- Silva, I.; Moody, B.; Behar, J.; Johnson, A.; Oster, J.; Clifford, G.D.; Moody, G.B. Robust detection of heart beats in multimodal data. Physiol. Meas. 2015, 36, 1629–1644. [Google Scholar] [CrossRef]
- Gierałtowski, J.; Ciuchciński, K.; Grzegorczyk, I.; Kośna, K.; Soliński, M.; Podziemski, P. RS slope detection algorithm for extraction of heart rate from noisy, multimodal recordings. Physiol. Meas. 2015, 36, 1743–1761. [Google Scholar] [CrossRef]
- Antink, C.H.; Leonhardt, S.; Walter, M. Reducing false alarms in the ICU by quantifying self-similarity of multimodal biosignals. Physiol. Meas. 2016, 37, 1233–1252. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Thakor, N.V.; Mower, M.M. Ventricular fibrillation detection by a regression test on the autocorrelation function. Med Biol. Eng. Comput. 1987, 25, 241–249. [Google Scholar] [CrossRef]
- Balasundaram, K.; Masse, S.; Nair, K.; Farid, T.; Nanthakumar, K.; Umapathy, K. Wavelet-based features for characterizing ventricular arrhythmias in optimizing treatment options. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011. [Google Scholar] [CrossRef]
- Khadra, L.; Al-Fahoum, A.S.; Al-Nashash, H. Detection of life-threatening cardiac arrhythmias using the wavelet transformation. Med. Biol. Eng. Comput. 1997, 35, 626–632. [Google Scholar] [CrossRef]
- Li, H.; Han, W.; Hu, C.; Meng, M.Q.-H. Detecting ventricular fibrillation by fast algorithm of dynamic sample entropy. In Proceedings of the 2009 IEEE International Conference on Robotics and Biomimetics (ROBIO), Guilin, China, 19–23 December 2009. [Google Scholar] [CrossRef]
- Alonso-Atienza, F.; Morgado, E.; Fernandez-Martinez, L.; Garcia-Alberola, A.; Rojo-Alvarez, J.L. Detection of Life-Threatening Arrhythmias Using Feature Selection and Support Vector Machines. IEEE Trans. Biomed. Eng. 2014, 61, 832–840. [Google Scholar] [CrossRef] [PubMed]
- Mjahad, A.; Rosado-Muñoz, A.; Bataller-Mompeán, M.; Francés-Víllora, J.V.; Guerrero-Martínez, J.F. Ventricular Fibrillation and Tachycardia detection from surface ECG using time-frequency representation images as input dataset for machine learning. Comput. Methods Programs Biomed. 2017, 141, 119–127. [Google Scholar] [CrossRef]
- Anas, E.; Lee, S.Y.; Hasan, M.K. Sequential algorithm for life threatening cardiac pathologies detection based on mean signal strength and EMD functions. Biomed. Eng. Online 2010, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Prabhakararao, E.; Manikandan, M.S. Efficient and robust ventricular tachycardia and fibrillation detection method for wearable cardiac health monitoring devices. Healthc. Technol. Lett. 2016, 3, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhao, L.; Tang, H.; Li, Q.; Wei, S.; Li, J. Life-threatening false alarm rejection in ICU: Using the rule-based and multi-channel information fusion method. Physiol. Meas. 2016, 37, 1298–1312. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.; Couto, P. Detection of false arrhythmia alarms with emphasis on ventricular tachycardia. Physiol. Meas. 2016, 37, 1326–1339. [Google Scholar] [CrossRef]
- Plesinger, F.; Klimes, P.; Halamek, J.; Jurak, P. Taming of the monitors: Reducing false alarms in intensive care units. Physiol. Meas. 2016, 37, 1313–1325. [Google Scholar] [CrossRef]
- Asadi, F.; Mollakazemi, M.J.; Ghiasi, S.; Sadati, S.H. Enhancement of life-threatening arrhythmia discrimination in the intensive care unit with morphological features and interval feature extraction via random forest classifier. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016; pp. 57–60. [Google Scholar] [CrossRef]
- Srivastava, C.; Sharma, S.; Jalali, A. A novel algorithm for reducing false arrhythmia alarms in intensive care units. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 2525–2528. [Google Scholar] [CrossRef]
- Saxena, A.; Prasad, M.; Gupta, A.; Bharill, N.; Patel, O.P.; Tiwari, A.; Er, M.J.; Ding, W.; Lin, C.-T.; Lin, C.-T. A review of clustering techniques and developments. Neurocomputing 2017, 267, 664–681. [Google Scholar] [CrossRef]
- Zabkowski, T.; Gajowniczek, K.; Szupiluk, R. Grade analysis for energy usage patterns segmentation based on smart meter data. In Proceedings of the 2015 IEEE 2nd International Conference on Cybernetics (CYBCONF), Gdynia, Poland, 24–26 June 2015. [Google Scholar] [CrossRef]
- Sorzano, C.O.S.; Vargas, J.; Montano, A.P. A survey of dimensionality reduction techniques. arXiv, 2014; arXiv:1403.2877. [Google Scholar]
- Yazgana, P.; Kusakci, A.O. A Literature Survey on Association Rule Mining Algorithms. Southeast Eur. J. Soft Comput. 2016, 5. [Google Scholar] [CrossRef]
- Fabris, F.; Magalhães, J.P.; de Freitas, A.A. A review of supervised machine learning applied to ageing research. Biogerontology 2017, 18, 171–188. [Google Scholar] [CrossRef]
- Kostopoulos, G.; Karlos, S.; Kotsiantis, S.; Ragos, O. Semi-supervised regression: A recent review. J. Intell. Fuzzy Syst. 2018, 35, 1483–1500. [Google Scholar] [CrossRef]
- Bakir, G.; Hofmann, T.; Schölkopf, B.; Smola, A.J.; Taskar, B.; Vishwanathan, S.V.N. (Eds.) Predicting Structured Data; MIT Press: Cambridge, MA, USA, 2007. [Google Scholar]
- Ren, Y.; Zhang, L.; Suganthan, P.N. Ensemble Classification and Regression-Recent Developments, Applications and Future Directions. IEEE Comput. Intell. Mag. 2016, 11, 41–53. [Google Scholar] [CrossRef]
- Tripoliti, E.E.; Fotiadis, D.I.; Manis, G. Dynamic construction of Random Forests: Evaluation using biomedical engineering problems. In Proceedings of the 10th IEEE International Conference on Information Technology and Applications in Biomedicine, Corfu, Greece, 2–5 November 2010. [Google Scholar] [CrossRef]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. Experiments with a new boosting algorithm. In Proceedings of the ICML’96 Proceedings of the Thirteenth International Conference on International Conference on Machine Learning, Bari, Italy, 3–6 July 1996; pp. 148–156. [Google Scholar]
- Goel, E.; Abhilasha, E. Random forest: A review. Int. J. Adv. Res. Comput. Sci. Softw. Eng. 2017, 7, 251–257. [Google Scholar] [CrossRef]
- Kuncheva, L. Diversity in multiple classifier systems. Inf. Fusion. 2004, 6, 3–4. [Google Scholar] [CrossRef]
- Boinee, P.; De Angelis, A.; Foresti, G.L. Meta random forests. Int. J. Comput. Intell. 2005, 2, 138–147. [Google Scholar]
- Robnik-Šikonja, M. Improving Random Forests. Lect. Notes Comput. Sci. 2004, 359–370. [Google Scholar] [CrossRef]
- Tsymbal, A.; Pechenizkiy, M.; Cunningham, P. Dynamic Integration with Random Forests. Mach. Learn. ECML 2006, 801–808. [Google Scholar] [CrossRef]
- Bernard, S.; Adam, S.; Heutte, L. Dynamic Random Forests. Pattern Recognit. Lett. 2012, 33, 1580–1586. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, H. Search for the smallest random forest. Stat. Interface 2009, 2, 381–388. [Google Scholar] [CrossRef]
- Wu, X.; Chen, Z. Toward dynamic ensembles: The BAGA apprach. In Proceedings of the 3rd ACS/IEEE International Conference onComputer Systems and Applications, Cairo, Egypt, 3–6 January 2005. [Google Scholar] [CrossRef]
- Gajowniczek, K.; Karpio, K.; Łukasiewicz, P.; Orłowski, A.; Ząbkowski, T. Q-Entropy Approach to Selecting High Income Households. Acta Phys. Pol. A 2015, 127, A-38–A-44. [Google Scholar] [CrossRef]
- Breiman, L. Classification and Regression Trees; Routledge: New York, NA, USA, 2017. [Google Scholar] [CrossRef]
- Gastwirth, J.L. The Estimation of the Lorenz Curve and Gini Index. Rev. Econ. Stat. 1972, 54, 306. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 261–277. [Google Scholar] [CrossRef]
- Gajowniczek, K.; Orłowski, A.; Ząbkowski, T. Entropy Based Trees to Support Decision Making for Customer Churn Management. Acta Phys. Pol. A 2016, 129, 971–979. [Google Scholar] [CrossRef]
- Gajowniczek, K.; Ząbkowski, T.; Orłowski, A. Comparison of Decision Trees with Rényi and Tsallis Entropy Applied for Imbalanced Churn Dataset. In Proceedings of the 2015 Federated Conference on Computer Science and Information Systems, Łódź, Poland, 13–16 September 2015. [Google Scholar] [CrossRef]
- Youden, W.J. An index for rating diagnostic tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef]
- Unal, I. Defining an optimal cut-point value in roc analysis: An alternative approach. Comput. Math. Methods Med. 2017, 2017. [Google Scholar] [CrossRef]
- Eerikäinen, L.M.; Vanschoren, J.; Rooijakkers, M.J.; Vullings, R.; Aarts, R.M. Reduction of false arrhythmia alarms using signal selection and machine learning. Physiol. Meas. 2016, 37, 1204–1216. [Google Scholar] [CrossRef] [PubMed]
- Rooijakkers, M.J.; Rabotti, C.; Oei, S.G.; Mischi, M. Low-complexity R-peak detection for ambulatory fetal monitoring. Physiol. Meas. 2012, 33, 1135–1150. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.; Heldt, T.; Moody, G.B.; Mark, R.G. An open-source algorithm to detect onset of arterial blood pressure pulses. Comput. Cardiol. 2003, 30, 259–262. [Google Scholar] [CrossRef]
- R: A Language and Environment for Statistical Computing. Available online: https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computing (accessed on 29 March 2019).
- Wright, M.N.; Ziegler, A. Ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Softw. 2017, 77. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Building Predictive Models inRUsing thecaretPackage. J. Stat. Softw. 2008, 28. [Google Scholar] [CrossRef]
- Pihur, V.; Datta, S.; Datta, S. RankAggreg, an R package for weighted rank aggregation. BMC Bioinform. 2009, 10, 62. [Google Scholar] [CrossRef] [PubMed]



| Predicted Value | |||
|---|---|---|---|
| Positive (P) | Negative (N) | ||
| Real value | Positive (P) | True Positive (TP) | False Negative (FN) |
| Negative (N) | False Positive (FP) | True Negative (TN) | |
| No. | Tree1 | Tree2 | Tree3 | Tree4 | Avg Prob INB | Avg Prob OOB | Avg Prob Train |
|---|---|---|---|---|---|---|---|
| 1 | 0.6(1) | 0.7(0) | 0.8(1) | 0.2(0) | 0.70 | 0.45 | 0.58 |
| 2 | 0.5(1) | 0.7(1) | 0.1(0) | 0.1(0) | 0.50 | 0.10 | 0.35 |
| 3 | 0.1(0) | 0.9(1) | 0.4(1) | 0.8(0) | 0.65 | 0.45 | 0.55 |
| No. | Tree1 | Tree2 | Tree3 | Tree4 | Vote Prob INB | Vote Prob OOB | Vote Prob Train |
|---|---|---|---|---|---|---|---|
| 1 | YES (1) | YES (0) | YES (1) | NO (0) | 1.00 | 0.50 | 0.75 |
| 2 | NO (1) | YES (1) | NO (0) | NO (0) | 0.50 | 0.00 | 0.25 |
| 3 | NO (0) | YES (1) | NO (1) | YES (0) | 0.50 | 0.50 | 0.50 |
| Arrhythmia Type | NO | YES |
|---|---|---|
| Ventricular Tachycardia | 252 | 89 |
| Asystole | 100 | 22 |
| Extreme Tachycardia | 9 | 131 |
| Ventricular Fibrillation or Flutter | 52 | 6 |
| Extreme Bradycardia | 43 | 46 |
| Approach | Ranks | |||
|---|---|---|---|---|
| N1 | N2 | N3 | N4 | |
| Avg Prob | 27.86% | 51.25% | 15.38% | 5.51% |
| Voting Prob INB | 9.03% | 13.14% | 22.77% | 55.06% |
| Voting Prob Train | 9.28% | 13.80% | 51.89% | 25.03% |
| Voting Prob 0.5 | 53.83% | 21.81% | 9.96% | 14.40% |
| Approach | Rank | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N1 | N2 | N3 | N4 | N5 | N6 | N7 | N8 | N9 | N10 | |
| Prob INB | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 53.06% | 23.73% | 5.25% | 17.95% | 0.00% |
| Prob Train | 0.00% | 25.04% | 74.26% | 0.70% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% |
| Prob 0.5 | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 61.91% | 38.09% | 0.00% |
| Vote INB INB | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 100.00% |
| Vote INB 0.5 | 0.00% | 74.96% | 23.82% | 0.00% | 0.61% | 0.61% | 0.00% | 0.00% | 0.00% | 0.00% |
| Vote Train Train | 0.00% | 0.00% | 0.35% | 0.00% | 7.97% | 38.62% | 53.06% | 0.00% | 0.00% | 0.00% |
| Vote Train 0.5 | 0.00% | 0.00% | 1.58% | 59.19% | 37.74% | 1.49% | 0.00% | 0.00% | 0.00% | 0.00% |
| Vote 0.5 INB | 0.00% | 0.00% | 0.00% | 40.11% | 53.68% | 6.22% | 0.00% | 0.00% | 0.00% | 0.00% |
| Vote 0.5 Train | 100.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% |
| Vote 0.5 0.5 | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 23.20% | 32.84% | 43.96% | 0.00% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gajowniczek, K.; Grzegorczyk, I.; Ząbkowski, T. Reducing False Arrhythmia Alarms Using Different Methods of Probability and Class Assignment in Random Forest Learning Methods. Sensors 2019, 19, 1588. https://doi.org/10.3390/s19071588
Gajowniczek K, Grzegorczyk I, Ząbkowski T. Reducing False Arrhythmia Alarms Using Different Methods of Probability and Class Assignment in Random Forest Learning Methods. Sensors. 2019; 19(7):1588. https://doi.org/10.3390/s19071588
Chicago/Turabian StyleGajowniczek, Krzysztof, Iga Grzegorczyk, and Tomasz Ząbkowski. 2019. "Reducing False Arrhythmia Alarms Using Different Methods of Probability and Class Assignment in Random Forest Learning Methods" Sensors 19, no. 7: 1588. https://doi.org/10.3390/s19071588
APA StyleGajowniczek, K., Grzegorczyk, I., & Ząbkowski, T. (2019). Reducing False Arrhythmia Alarms Using Different Methods of Probability and Class Assignment in Random Forest Learning Methods. Sensors, 19(7), 1588. https://doi.org/10.3390/s19071588






