Role of Nitrate Reductase in NO Production in Photosynthetic Eukaryotes
Abstract
:1. Introduction
2. Nitrite: The Substrate for Reductive NO Production
3. Nitrate Reductase Is a Multidomain Protein
4. Does NR Catalyze Nitrite Reduction to NO?
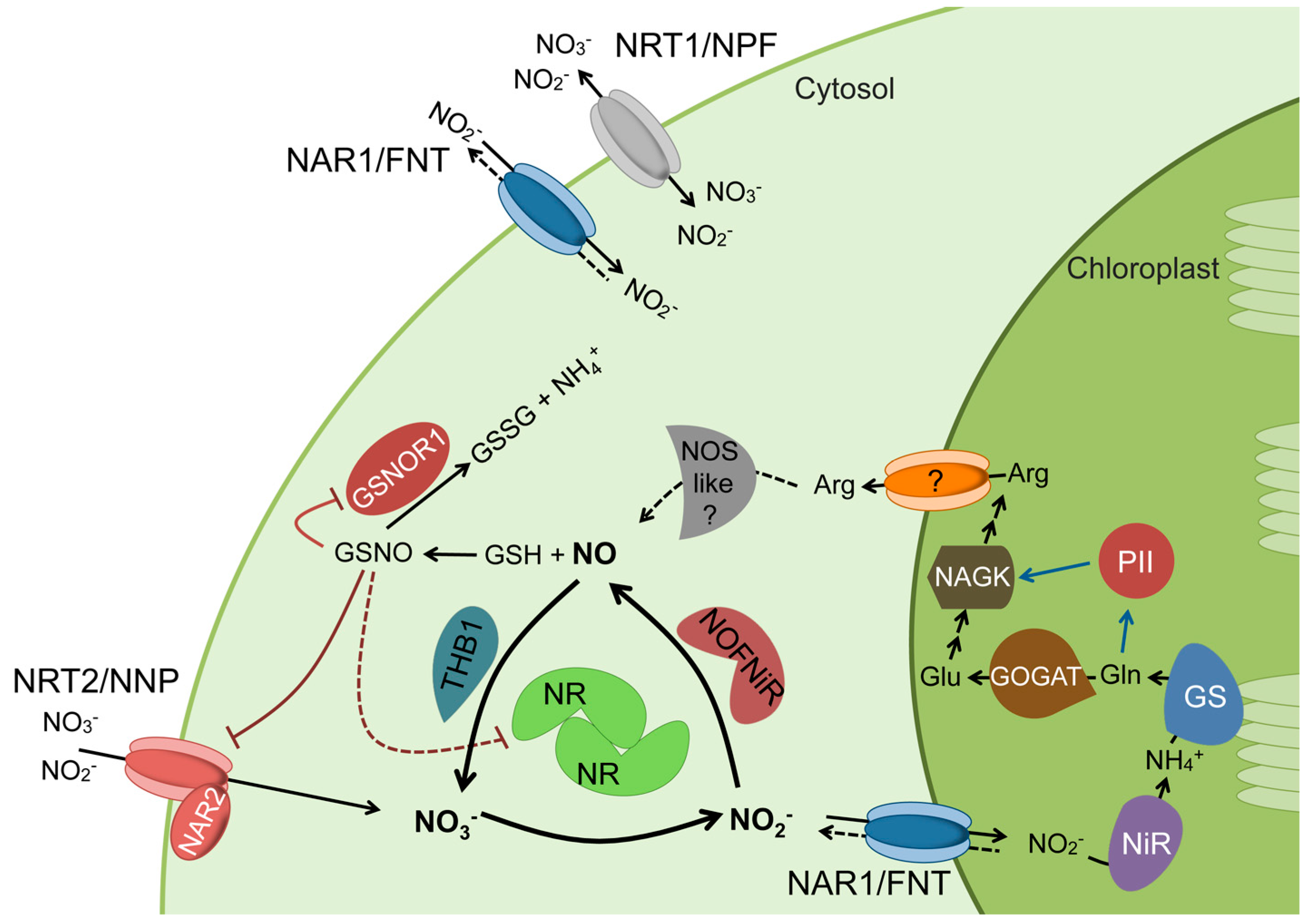
5. NR Does Not Catalyze In Vivo Nitrite Reduction to NO but Provides the Needed Electrons
6. NO Levels in the Cells Are Regulated
7. The NO Synthesis Systems Are Coordinated with Nitrogen Metabolism
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wendehenne, D.; Hancock, J.T. New frontiers in nitric oxide biology in plant. Plant Sci. 2011, 181, 507–508. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Leterrier, M.; Valderrama, R.; Airaki, M.; Chaki, M.; Palma, J.M.; Barroso, J.B. Nitric oxide imbalance provokes a nitrosative response in plants under abiotic stress. Plant Sci. 2011, 181, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.; Prats, E.; Pierre, S.; Hall, M.A.; Hebelstrup, K.H. Integrating nitric oxide into salicylic acid and jasmonic acid/ ethylene plant defense pathways. Front. Plant Sci. 2013, 4, 215. [Google Scholar] [CrossRef] [PubMed]
- Santolini, J.; Andre, F.; Jeandroz, S.; Wendehenne, D. Nitric oxide synthase in plants: Where do we stand? Nitric Oxide 2017, 63, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Astier, J.; Gross, I.; Durner, J. Nitric oxide production in plants: An update. J. Exp. Bot. 2018, 69, 3401–3411. [Google Scholar] [CrossRef] [PubMed]
- De Montaigu, A.; Sanz-Luque, E.; Galvan, A.; Fernandez, E. A soluble guanylate cyclase mediates negative signaling by ammonium on expression of nitrate reductase in Chlamydomonas. Plant Cell 2010, 22, 1532–1548. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Derrien, B.; Gautier, A.; Houille-Vernes, L.; Boulouis, A.; Saint-Marcoux, D.; Malnoe, A.; Rappaport, F.; de Vitry, C.; Vallon, O.; et al. Nitric oxide-triggered remodeling of chloroplast bioenergetics and thylakoid proteins upon nitrogen starvation in Chlamydomonas reinhardtii. Plant Cell 2014, 26, 353–372. [Google Scholar] [CrossRef] [PubMed]
- Yordanova, Z.P.; Iakimova, E.T.; Cristescu, S.M.; Harren, F.J.; Kapchina-Toteva, V.M.; Woltering, E.J. Involvement of ethylene and nitric oxide in cell death in mastoparan-treated unicellular alga Chlamydomonas reinhardtii. Cell Biol. Int. 2010, 34, 301–308. [Google Scholar] [PubMed]
- Hemschemeier, A.; Duner, M.; Casero, D.; Merchant, S.S.; Winkler, M.; Happe, T. Hypoxic survival requires a 2-on-2 hemoglobin in a process involving nitric oxide. Proc. Natl. Acad. Sci. USA 2013, 110, 10854–10859. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tian, D.; Kong, X.; Chen, Q.; Ef, A.A.; Hu, X.; Jia, A. The role of nitric oxide signalling in response to salt stress in Chlamydomonas reinhardtii. Planta 2016, 244, 651–669. [Google Scholar] [CrossRef] [PubMed]
- De Mia, M.; Lemaire, S.D.; Choquet, Y.; Wollman, F.A. Nitric oxide remodels the photosynthetic apparatus upon S-starvation in Chlamydomonas reinhardtii. Plant Physiol. 2019, 179, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Delledonne, M.; Xia, Y.; Dixon, R.A.; Lamb, C. Nitric oxide functions as a signal in plant disease resistance. Nature 1998, 394, 585–588. [Google Scholar] [PubMed]
- Durner, J.; Wendehenne, D.; Klessig, D.F. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 1998, 95, 10328–10333. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.; Mandon, J.; Persijn, S.; Cristescu, S.M.; Moshkov, I.E.; Novikova, G.V.; Hall, M.A.; Harren, F.J.; Hebelstrup, K.H.; Gupta, K.J. Nitric oxide in plants: An assessment of the current state of knowledge. AoB Plants 2013, 5, pls052. [Google Scholar] [CrossRef] [PubMed]
- Maia, L.B.; Moura, J.J.G. Putting xanthine oxidoreductase and aldehyde oxidase on the NO metabolism map: Nitrite reduction by molybdoenzymes. Redox Biol. 2018, 19, 274–289. [Google Scholar] [CrossRef] [PubMed]
- Foresi, N.; Correa-Aragunde, N.; Parisi, G.; Calo, G.; Salerno, G.; Lamattina, L. Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 2010, 22, 3816–3830. [Google Scholar] [CrossRef] [PubMed]
- Astier, J.; Jeandroz, S.; Wendehenne, D. Nitric oxide synthase in plants: The surprise from algae. Plant Sci. 2018, 268, 64–66. [Google Scholar] [CrossRef] [PubMed]
- Jeandroz, S.; Wipf, D.; Stuehr, D.J.; Lamattina, L.; Melkonian, M.; Tian, Z.; Zhu, Y.; Carpenter, E.J.; Wong, G.K.; Wendehenne, D. Occurrence, structure, and evolution of nitric oxide synthase-like proteins in the plant kingdom. Sci. Signal 2016, 9, re2. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, E.; Galvan, A. Nitrate assimilation in Chlamydomonas. Eukaryot. Cell 2008, 7, 555–559. [Google Scholar] [PubMed]
- Crawford, N.M. Nitrate: Nutrient and signal for plant growth. Plant Cell 1995, 7, 859–868. [Google Scholar] [PubMed]
- Dechorgnat, J.; Nguyen, C.T.; Armengaud, P.; Jossier, M.; Diatloff, E.; Filleur, S.; Daniel-Vedele, F. From the soil to the seeds: The long journey of nitrate in plants. J. Exp. Bot. 2011, 62, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Tsay, Y.F. Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 2011, 23, 1945–1957. [Google Scholar] [CrossRef] [PubMed]
- Krapp, A.; David, L.C.; Chardin, C.; Girin, T.; Marmagne, A.; Leprince, A.S.; Chaillou, S.; Ferrario-Mery, S.; Meyer, C.; Daniel-Vedele, F. Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 789–798. [Google Scholar] [CrossRef] [PubMed]
- O’brien, J.A.; Vega, A.; Onore Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G.; Gutié Rrez, R.A. Nitrate Transport, Sensing, and Responses in Plants. Mol. Plant 2016, 9, 837–856. [Google Scholar] [CrossRef] [PubMed]
- Calatrava, V.; Chamizo-Ampudia, A.; Sanz-Luque, E.; Ocana-Calahorro, F.; Llamas, A.; Fernandez, E.; Galvan, A. How Chlamydomonas handles nitrate and the nitric oxide cycle. J. Exp. Bot. 2017, 68, 2593–2602. [Google Scholar] [CrossRef] [PubMed]
- Monachello, D.; Allot, M.; Oliva, S.; Krapp, A.; Daniel-Vedele, F.; Barbier-Brygoo, H.; Ephritikhine, G. Two anion transporters AtClCa and AtClCe fulfil interconnecting but not redundant roles in nitrate assimilation pathways. New Phytol. 2009, 183, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, M.; Georgescu, M.N.; Takahashi, M. A nitrite transporter associated with nitrite uptake by higher plant chloroplasts. Plant Cell Physiol. 2007, 48, 1022–1035. [Google Scholar] [CrossRef] [PubMed]
- Tsay, Y.-F.; Chiu, C.-C.; Tsai, C.-B.; Ho, C.-H.; Hsu, P.-K. Nitrate transporters and peptide transporters. FEBS Lett. 2007, 581, 2290–2300. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.; Takahashi, M.; Arimura, G.; Isayama, T.; Irifune, K.; Goshima, N.; Morikawa, H. Determination of levels of nitrate, nitrite and ammonium ions in leaves of ions in leaves of various plants by capillary electrophoresis. Plant Cell Physiol. 1996, 37, 878–880. [Google Scholar] [CrossRef]
- Ferrario-Mery, S.; Bouvet, M.; Leleu, O.; Savino, G.; Hodges, M.; Meyer, C. Physiological characterisation of Arabidopsis mutants affected in the expression of the putative regulatory protein PII. Planta 2005, 223, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Camargo, A.; Llamas, A.; Schnell, R.A.; Higuera, J.J.; Gonzalez-Ballester, D.; Lefebvre, P.A.; Fernandez, E.; Galvan, A. Nitrate signaling by the regulatory gene NIT2 in Chlamydomonas. Plant Cell 2007, 19, 3491–3503. [Google Scholar] [CrossRef] [PubMed]
- Castaings, L.; Camargo, A.; Pocholle, D.; Gaudon, V.; Texier, Y.; Boutet-Mercey, S.; Taconnat, L.; Renou, J.P.; Daniel-Vedele, F.; Fernandez, E.; et al. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009, 57, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 2013, 4, 1617. [Google Scholar] [PubMed]
- Marchive, C.; Roudier, F.; Castaings, L.; Bréhaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 2013, 4, 1713. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.G.; Lea, P.J. Glutamate in plants: Metabolism, regulation, and signalling. J. Exp. Bot. 2007, 58, 2339–2358. [Google Scholar] [CrossRef] [PubMed]
- Van der leij, M.; Smith, S.; Miller, J. Remobilisation of vacuolar stored nitrate in barley root cells. Planta 1998, 205, 64–72. [Google Scholar] [CrossRef]
- Cookson, S.J.; Williams, L.E.; Miller, A.J. Light-dark changes in cytosolic nitrate pools depend on nitrate reductase activity in Arabidopsis leaf cells. Plant Physiol. 2005, 138, 1097–1105. [Google Scholar] [PubMed]
- Miller, A.J.; Smith, S.J. Cytosolic nitrate ion homeostasis: Could it have a role in sensing nitrogen status? Ann. Bot. 2008, 101, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.J.; Fan, X.; Orsel, M.; Smith, S.J.; Wells, D.M. Nitrate transport and signalling. J. Exp. Bot. 2007, 58, 2297–2306. [Google Scholar] [PubMed]
- Segonzac, C.; Boyer, J.-C.; Ipotesi, E.; Szponarski, W.; Tillard, P.; Touraine, B.; Sommerer, N.; Rossignol, M.; Gibrat, R. Nitrate efflux at the root plasma membrane: Identification of an Arabidopsis excretion transporter. Plant Cell 2007, 19, 3760–3777. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Gordon-Weeks, R.; Shen, Q.; Miller, A.J. Glutamine transport and feedback regulation of nitrate reductase activity in barley roots leads to changes in cytosolic nitrate pools. J. Exp. Bot. 2006, 57, 1333–1340. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, E.; Gonzalez-Montelongo, R.; Giraldez, T.; Alvarez de la Rosa, D.; Siverio, J.M. Molecular components of nitrate and nitrite efflux in yeast. Eukaryot. Cell 2014, 13, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Campbell, W.H. Nitrate reductase structure, function and regulation: Bridging the Gap between Biochemistry and Physiology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 277–303. [Google Scholar] [CrossRef] [PubMed]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Galvan, A.; Fernandez, E. Nitrate Reductase Regulates Plant Nitric Oxide Homeostasis. Trends Plant Sci. 2017, 22, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Barbier, G.G.; Hecht, H.J.; Mendel, R.R.; Campbell, W.H.; Schwarz, G. Structural basis of eukaryotic nitrate reduction: Crystal structures of the nitrate reductase active site. Plant Cell 2005, 17, 1167–1179. [Google Scholar] [CrossRef] [PubMed]
- Dean, J.V.; Harper, J.E. The Conversion of Nitrite to Nitrogen Oxide(s) by the Constitutive NAD(P)H-Nitrate Reductase Enzyme from Soybean. Plant Physiol. 1988, 88, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Wildt, J.; Kley, D.; Rockel, A.; Rockel, P.; Segscheneider, H.J. Emission of NO from several higher plant species. J. Geophys. Res. 1997, 102, 5919–5927. [Google Scholar] [CrossRef]
- Dean, J.V.; Harper, J.E. Nitric Oxide and Nitrous Oxide Production by Soybean and Winged Bean during the in Vivo Nitrate Reductase Assay. Plant Physiol. 1986, 82, 718–723. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, H.; Sakihama, Y. Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: In vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett. 2000, 468, 89–92. [Google Scholar] [CrossRef]
- Rockel, P.; Strube, F.; Rockel, A.; Wildt, J.; Kaiser, W.M. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 2002, 53, 103–110. [Google Scholar] [PubMed]
- Meyer, C.; Lea, U.S.; Provan, F.; Kaiser, W.M.; Lillo, C. Is nitrate reductase a major player in the plant NO (nitric oxide) game? Photosynth. Res. 2005, 83, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Llamas, A.; Kalakoutskii, K.; Fernandez, E. Molybdenum cofactor amounts in Chlamydomonas reinhardtii depend on the Nit5 gene function related to molybdate transport. Plant Cell Environ. 2000, 23, 1247–1255. [Google Scholar] [CrossRef]
- Walker-Simmons, M.; Kudrna, D.A.; Warner, R.L. Reduced Accumulation of ABA during Water Stress in a Molybdenum Cofactor Mutant of Barley. Plant Physiol. 1989, 90, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, J.Q.; Crawford, N.M. Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol. Gen. Genet. 1993, 239, 289–297. [Google Scholar] [PubMed]
- Schwarz, G.; Mendel, R.R.; Ribbe, M.W. Molybdenum cofactors, enzymes and pathways. Nature 2009, 460, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Keceli, G.; Cao, R.; Su, J.; Mi, Z. Molybdenum-containing nitrite reductases: Spectroscopic characterization and redox mechanism. Redox Rep. 2017, 22, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Krizowski, S.; Fischer-Schrader, K.; Niks, D.; Tejero, J.; Sparacino-Watkins, C.; Wang, L.; Ragireddy, V.; Frizzell, S.; Kelley, E.E.; et al. Sulfite Oxidase Catalyzes Single-Electron Transfer at Molybdenum Domain to Reduce Nitrite to Nitric Oxide. Antioxid. Redox Signal 2015, 23, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Moche, M.; Stremlau, S.; Hecht, L.; Gobel, C.; Feussner, I.; Stohr, C. Effect of nitrate supply and mycorrhizal inoculation on characteristics of tobacco root plasma membrane vesicles. Planta 2010, 231, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Tischner, R.; Planchet, E.; Kaiser, W.M. Mitochondrial electron transport as a source for nitric oxide in the unicellular green alga Chlorella sorokiniana. FEBS Lett. 2004, 576, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Planchet, E.; Jagadis Gupta, K.; Sonoda, M.; Kaiser, W.M. Nitric oxide emission from tobacco leaves and cell suspensions: Rate limiting factors and evidence for the involvement of mitochondrial electron transport. Plant J. 2005, 41, 732–743. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Stoimenova, M.; Kaiser, W.M. In higher plants, only root mitochondria, but not leaf mitochondria reduce nitrite to NO, in vitro and in situ. J. Exp. Bot. 2005, 56, 2601–2609. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Kaiser, W.M. Production and scavenging of nitric oxide by barley root mitochondria. Plant Cell Physiol. 2010, 51, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Igamberdiev, A.U. The anoxic plant mitochondrion as a nitrite: NO reductase. Mitochondrion 2011, 11, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, A.; Kumari, A.; Mur, L.A.J.; Gupta, K.J. A discrete role for alternative oxidase under hypoxia to increase nitric oxide and drive energy production. Free Radic. Biol. Med. 2018, 122, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Plouviez, M.; Wheeler, D.; Shilton, A.; Packer, M.A.; McLenachan, P.A.; Sanz-Luque, E.; Ocana-Calahorro, F.; Fernandez, E.; Guieysse, B. The biosynthesis of nitrous oxide in the green alga Chlamydomonas reinhardtii. Plant J. 2017, 91, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Havemeyer, A.; Bittner, F.; Wollers, S.; Mendel, R.; Kunze, T.; Clement, B. Identification of the missing component in the mitochondrial benzamidoxime prodrug-converting system as a novel molybdenum enzyme. J. Biol. Chem. 2006, 281, 34796–34802. [Google Scholar] [CrossRef] [PubMed]
- Kozmin, S.G.; Leroy, P.; Pavlov, Y.I.; Schaaper, R.M. YcbX and yiiM, two novel determinants for resistance of Escherichia coli to N-hydroxylated base analogues. Mol. Microbiol. 2008, 68, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Chamizo-Ampudia, A.; Galvan, A.; Fernandez, E.; Llamas, A. The Chlamydomonas reinhardtii molybdenum cofactor enzyme crARC has a Zn-dependent activity and protein partners similar to those of its human homologue. Eukaryot. Cell 2011, 10, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Tejada-Jimenez, M.; Chamizo-Ampudia, A.; Calatrava, V.; Galvan, A.; Fernandez, E.; Llamas, A. From the Eukaryotic Molybdenum Cofactor Biosynthesis to the Moonlighting Enzyme mARC. Molecules 2018, 23, 3287. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Giles, L.J.; Ruppelt, C.; Mendel, R.R.; Bittner, F.; Kirk, M.L. Oxyl and hydroxyl radical transfer in mitochondrial amidoxime reducing component-catalyzed nitrite reduction. J. Am. Chem. Soc. 2015, 137, 5276–5279. [Google Scholar] [CrossRef] [PubMed]
- Chamizo-Ampudia, A.; Galvan, A.; Fernandez, E.; Llamas, A. Study of Different Variants of Mo Enzyme crARC and the Interaction with Its Partners crCytb5-R and crCytb5-1. Int. J. Mol. Sci. 2017, 18, 670. [Google Scholar] [CrossRef] [PubMed]
- Llamas, A.; Chamizo-Ampudia, A.; Tejada-Jimenez, M.; Galvan, A.; Fernandez, E. The molybdenum cofactor enzyme mARC: Moonlighting or promiscuous enzyme? Biofactors 2017, 43, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Kubitza, C.; Bittner, F.; Ginsel, C.; Havemeyer, A.; Clement, B.; Scheidig, A.J. Crystal structure of human mARC1 reveals its exceptional position among eukaryotic molybdenum enzymes. Proc. Natl. Acad. Sci. USA 2018, 115, 11958–11963. [Google Scholar] [CrossRef] [PubMed]
- Islinger, M.; Luers, G.H.; Li, K.W.; Loos, M.; Volkl, A. Rat liver peroxisomes after fibrate treatment. A survey using quantitative mass spectrometry. J. Biol. Chem. 2007, 282, 23055–23069. [Google Scholar] [CrossRef] [PubMed]
- Kotthaus, J.; Wahl, B.; Havemeyer, A.; Kotthaus, J.; Schade, D.; Garbe-Schonberg, D.; Mendel, R.; Bittner, F.; Clement, B. Reduction of N(omega)-hydroxy-l-arginine by the mitochondrial amidoxime reducing component (mARC). Biochem. J. 2011, 433, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Sparacino-Watkins, C.E.; Tejero, J.; Sun, B.; Gauthier, M.C.; Thomas, J.; Ragireddy, V.; Merchant, B.A.; Wang, J.; Azarov, I.; Basu, P.; et al. Nitrite reductase and nitric-oxide synthase activity of the mitochondrial molybdopterin enzymes mARC1 and mARC2. J. Biol. Chem. 2014, 289, 10345–10358. [Google Scholar] [CrossRef] [PubMed]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Ocana-Calahorro, F.; Mariscal, V.; Carreras, A.; Barroso, J.B.; Galvan, A.; Fernandez, E. A dual system formed by the ARC and NR molybdoenzymes mediates nitrite-dependent NO production in Chlamydomonas. Plant Cell Environ. 2016, 39, 2097–2107. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Luque, E.; Ocana-Calahorro, F.; de Montaigu, A.; Chamizo-Ampudia, A.; Llamas, A.; Galvan, A.; Fernandez, E. THB1, a truncated hemoglobin, modulates nitric oxide levels and nitrate reductase activity. Plant J. 2015, 81, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Hebelstrup, K.H.; Mur, L.A.; Igamberdiev, A.U. Plant hemoglobins: Important players at the crossroads between oxygen and nitric oxide. FEBS Lett. 2011, 585, 3843–3849. [Google Scholar] [CrossRef] [PubMed]
- Ohwaki, Y.; Kawagishi-Kobayashi, M.; Wakasa, K.; Fujihara, S.; Yoneyama, T. Induction of class-1 non-symbiotic hemoglobin genes by nitrate, nitrite and nitric oxide in cultured rice cells. Plant Cell Physiol. 2005, 46, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.D. Non-symbiotic haemoglobins-What’s happening beyond nitric oxide scavenging? AoB Plants 2012, 2012, pls004. [Google Scholar] [CrossRef] [PubMed]
- Trevisan, S.; Manoli, A.; Begheldo, M.; Nonis, A.; Enna, M.; Vaccaro, S.; Caporale, G.; Ruperti, B.; Quaggiotti, S. Transcriptome analysis reveals coordinated spatiotemporal regulation of hemoglobin and nitrate reductase in response to nitrate in maize roots. New Phytol. 2011, 192, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Luque, E.; Ocana-Calahorro, F.; Galvan, A.; Fernandez, E. THB1 regulates nitrate reductase activity and THB1 and THB2 transcription differentially respond to NO and the nitrate/ammonium balance in Chlamydomonas. Plant Signal Behav. 2015, 10, e1042638. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, I.; Lindermayr, C. Nitric oxide-based protein modification: Formation and site-specificity of protein S-nitrosylation. Front. Plant Sci. 2013, 4, 137. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Galisteo, A.P.; Rodriguez-Serrano, M.; Pazmino, D.M.; Gupta, D.K.; Sandalio, L.M.; Romero-Puertas, M.C. S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: Changes under abiotic stress. J. Exp. Bot. 2012, 63, 2089–2103. [Google Scholar] [CrossRef] [PubMed]
- Frungillo, L.; Skelly, M.J.; Loake, G.J.; Spoel, S.H.; Salgado, I. S-nitrosothiols regulate nitric oxide production and storage in plants through the nitrogen assimilation pathway. Nat. Commun. 2014, 5, 5401. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Luque, E.; Ocana-Calahorro, F.; Llamas, A.; Galvan, A.; Fernandez, E. Nitric oxide controls nitrate and ammonium assimilation in Chlamydomonas reinhardtii. J. Exp. Bot. 2013, 64, 3373–3383. [Google Scholar] [CrossRef] [PubMed]
- Gow, A.J.; Farkouh, C.R.; Munson, D.A.; Posencheg, M.A.; Ischiropoulos, H. Biological significance of nitric oxide-mediated protein modifications. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004, 287, L262–L268. [Google Scholar] [CrossRef] [PubMed]
- Deeb, R.S.; Nuriel, T.; Cheung, C.; Summers, B.; Lamon, B.D.; Gross, S.S.; Hajjar, D.P. Characterization of a cellular denitrase activity that reverses nitration of cyclooxygenase. Am. J. Physiol. Heart Circ. Physiol. 2013, 305, H687–H698. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef] [PubMed]
- Farnese, F.S.; Menezes-Silva, P.E.; Gusman, G.S.; Oliveira, J.A. When Bad Guys Become Good Ones: The Key Role of Reactive Oxygen Species and Nitric Oxide in the Plant Responses to Abiotic Stress. Front. Plant Sci. 2016, 7, 471. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B.; del Rio, L.A. Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci. 2001, 6, 145–150. [Google Scholar] [CrossRef]
- Barroso, J.B.; Corpas, F.J.; Carreras, A.; Sandalio, L.M.; Valderrama, R.; Palma, J.M.; Lupianez, J.A.; del Rio, L.A. Localization of nitric-oxide synthase in plant peroxisomes. J. Biol. Chem. 1999, 274, 36729–36733. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B.; Carreras, A.; Quiros, M.; Leon, A.M.; Romero-Puertas, M.C.; Esteban, F.J.; Valderrama, R.; Palma, J.M.; Sandalio, L.M.; et al. Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiol. 2004, 136, 2722–2733. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B. Nitric oxide synthase-like activity in higher plants. Nitric Oxide 2017, 68, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Chellamuthu, V.R.; Alva, V.; Forchhammer, K. From cyanobacteria to plants: Conservation of PII functions during plastid evolution. Planta 2013, 237, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Ninfa, A.J.; Jiang, P. PII signal transduction proteins: Sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr. Opin. Microbiol. 2005, 8, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Huergo, L.F.; Chandra, G.; Merrick, M. P(II) signal transduction proteins: Nitrogen regulation and beyond. FEMS Microbiol. Rev. 2013, 37, 251–283. [Google Scholar] [CrossRef] [PubMed]
- Zeth, K.; Fokina, O.; Forchhammer, K. Structural basis and target-specific modulation of ADP sensing by the Synechococcus elongatus PII signaling protein. J. Biol. Chem. 2014, 289, 8960–8972. [Google Scholar] [CrossRef] [PubMed]
- Nunes-Nesi, A.; Fernie, A.R.; Stitt, M. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 2010, 3, 973–996. [Google Scholar] [CrossRef] [PubMed]
- Ferrario-Mery, S.; Besin, E.; Pichon, O.; Meyer, C.; Hodges, M. The regulatory PII protein controls arginine biosynthesis in Arabidopsis. FEBS Lett. 2006, 580, 2015–2020. [Google Scholar] [CrossRef] [PubMed]
- Ferrario-Mery, S.; Meyer, C.; Hodges, M. Chloroplast nitrite uptake is enhanced in Arabidopsis PII mutants. FEBS Lett. 2008, 582, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Chellamuthu, V.R.; Ermilova, E.; Lapina, T.; Luddecke, J.; Minaeva, E.; Herrmann, C.; Hartmann, M.D.; Forchhammer, K. A widespread glutamine-sensing mechanism in the plant kingdom. Cell 2014, 159, 1188–1199. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Ballester, D.; Sanz-Luque, E.; Galvan, A.; Fernandez, E.; de Montaigu, A. Arginine is a component of the ammonium-CYG56 signalling cascade that represses genes of the nitrogen assimilation pathway in Chlamydomonas reinhardtii. PLoS ONE 2018, 13, e0196167. [Google Scholar] [CrossRef] [PubMed]
- Zalutskaya, Z.; Kochemasova, L.; Ermilova, E. Dual positive and negative control of Chlamydomonas PII signal transduction protein expression by nitrate/nitrite and NO via the components of nitric oxide cycle. BMC Plant Biol. 2018, 18, 305. [Google Scholar] [CrossRef] [PubMed]
- De Montaigu, A.; Sanz-Luque, E.; Macias, M.I.; Galvan, A.; Fernandez, E. Transcriptional regulation of CDP1 and CYG56 is required for proper NH4+ sensing in Chlamydomonas. J. Exp. Bot. 2011, 62, 1425–1437. [Google Scholar] [CrossRef] [PubMed]
- Rasul, S.; Wendehenne, D.; Jeandroz, S. Study of oligogalacturonides-triggered nitric oxide (NO) production provokes new questioning about the origin of NO biosynthesis in plants. Plant Signal Behav. 2012, 7, 1031–1033. [Google Scholar] [CrossRef] [PubMed]
- Modolo, L.V.; Augusto, O.; Almeida, I.M.; Pinto-Maglio, C.A.; Oliveira, H.C.; Seligman, K.; Salgado, I. Decreased arginine and nitrite levels in nitrate reductase-deficient Arabidopsis thaliana plants impair nitric oxide synthesis and the hypersensitive response to Pseudomonas syringae. Plant Sci. 2006, 171, 34–40. [Google Scholar] [CrossRef]
- Zalutskaya, Z.; Ostroukhova, M.; Filina, V.; Ermilova, E. Nitric oxide upregulates expression of alternative oxidase 1 in Chlamydomonas reinhardtii. J. Plant Physiol. 2017, 219, 123–127. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tejada-Jimenez, M.; Llamas, A.; Galván, A.; Fernández, E. Role of Nitrate Reductase in NO Production in Photosynthetic Eukaryotes. Plants 2019, 8, 56. https://doi.org/10.3390/plants8030056
Tejada-Jimenez M, Llamas A, Galván A, Fernández E. Role of Nitrate Reductase in NO Production in Photosynthetic Eukaryotes. Plants. 2019; 8(3):56. https://doi.org/10.3390/plants8030056
Chicago/Turabian StyleTejada-Jimenez, Manuel, Angel Llamas, Aurora Galván, and Emilio Fernández. 2019. "Role of Nitrate Reductase in NO Production in Photosynthetic Eukaryotes" Plants 8, no. 3: 56. https://doi.org/10.3390/plants8030056




